RNA interference RNAi 1 Outline Introduction RNA silencing















- Slides: 41

RNA interference (RNAi) 1

Outline • Introduction -RNA silencing -Definition of RNA interference -Discovery of RNAi • • Mechanism of RNA interference Generation of small interfering RNA Small interfering RNA delivery methods Applications of RNA interference -Therapeutic applications -Other applications • Conclusion 2

Introduction 3

RNA silencing • Several terms are used to described RNA silencing; usually there are three phenotypically different but mechanistically similar phenomena: 1. Cosuppression or post-trascriptional gene silencing (PTGS) in plants 2. Quelling in fungi 3. RNA interference in animal kingdom 4

Definition • RNA interference (RNAi) is a mechanism that inhibits gene expression at the stage of translation or by hindering the transcription of specific genes • RNAi targets include RNA from viruses andtransposons 7

Need for interference • Defense Mechanism -Defense against Infection by viruses, etc -As a defense mechanism to protect against transposons and other insertional elements • Genome Wide Regulation -RNAi plays a role in regulating development and genome maintenance -30% of human genome regulated 8

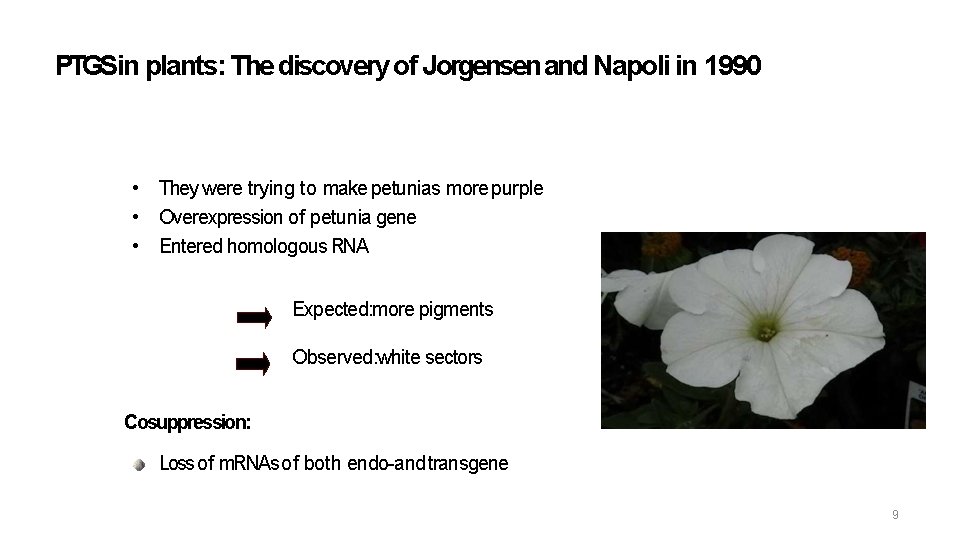
PTGSin plants: The discovery of Jorgensen and Napoli in 1990 • They were trying to make petunias more purple • Overexpression of petunia gene • Entered homologous RNA Expected: more pigments Observed: white sectors Cosuppression: Loss of m. RNAs of both endo-and transgene 9

Andrew Fire Craig C. Mellow 10

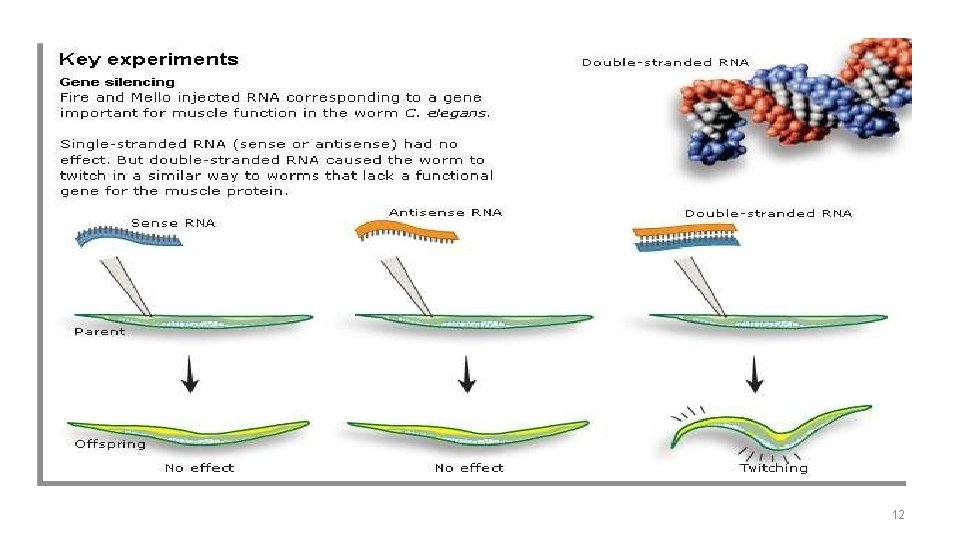
Discovery Inject worms with ds. RNA corresponding to a gene (important for muscle function) involved in wiggling (unc-22) 11

Discovery Inject worms with ds. RNA corresponding to a gene (important for muscle function) involved in wiggling (unc-22) Conclusion: ds. RNA triggers potent and specific gene silencing 12

Mechanism of RNAi 13

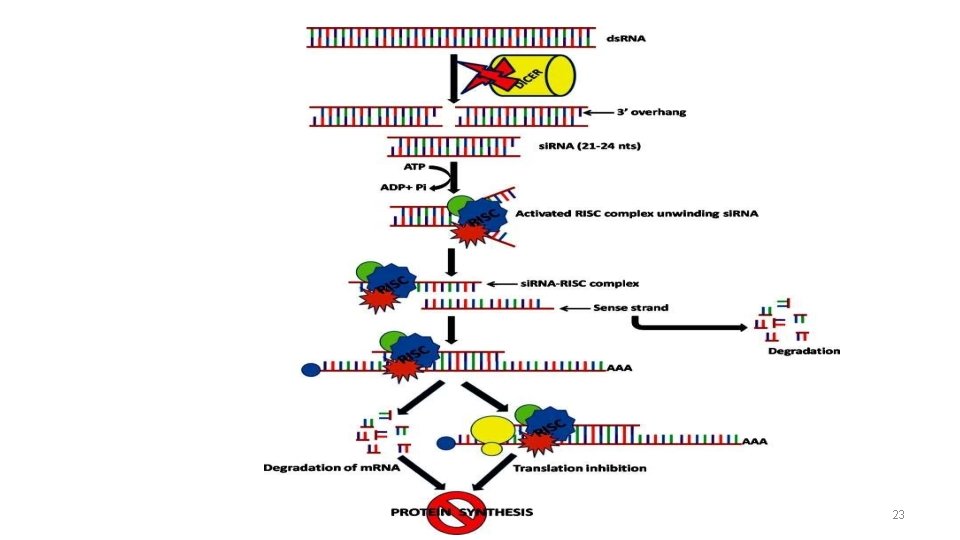
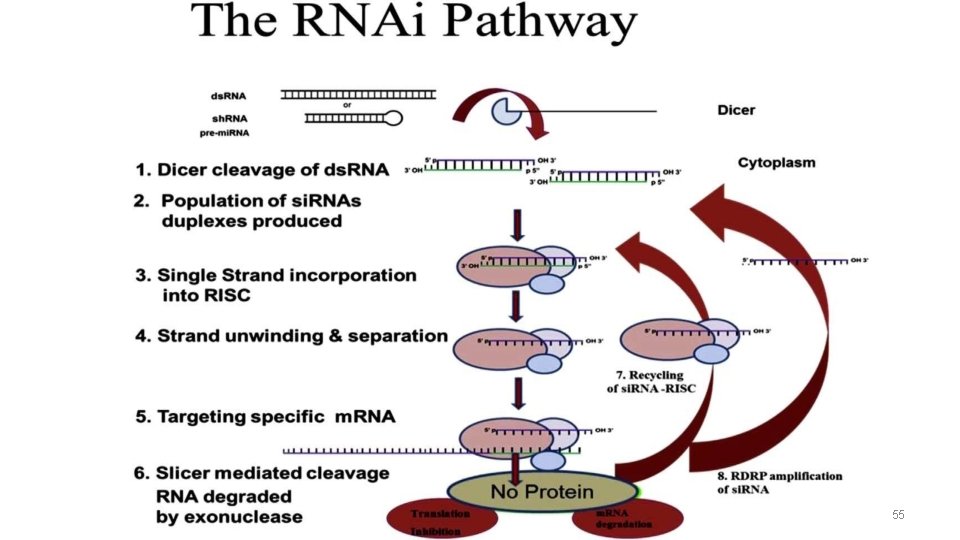
RNAi Overview • During RNAi Double-stranded RNAs cut into short double-stranded RNAs, s(small) i(interfering) RNA's, by an enzyme called Dicer • These then base pair to an m. RNA through a ds. RNA-enzyme complex. This will either lead to degradation of the m. RNA strand • • Highly specific process Very potent activity So far only been seen in eukaryotes Evidence 30% of genome is regulated by RNAi 14

The components In Interference • RNA -si. RNA: ds. RNA 21 -22 nt. -mi. RNA: ss. RNA 19 -25 nt. Encoded by non protein coding genome • RISC: -RNA induced Silencing Complex, that cleavesm. RNA • Enzymes -Dicer : produces 20 -21 nt cleavages that initiate RNAi -Drosha : cleaves base hairpin in to form pre mi. RNA; which is later processed by Dicer 15

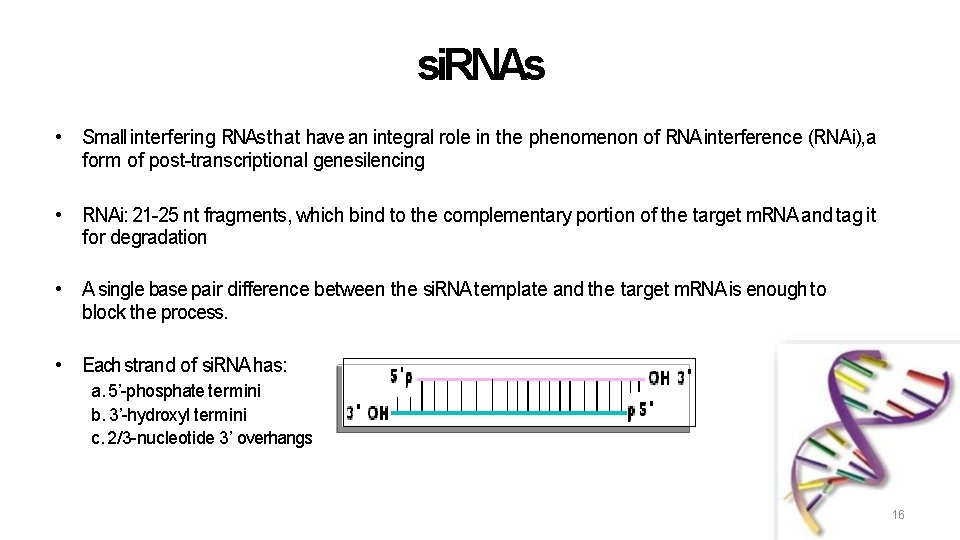
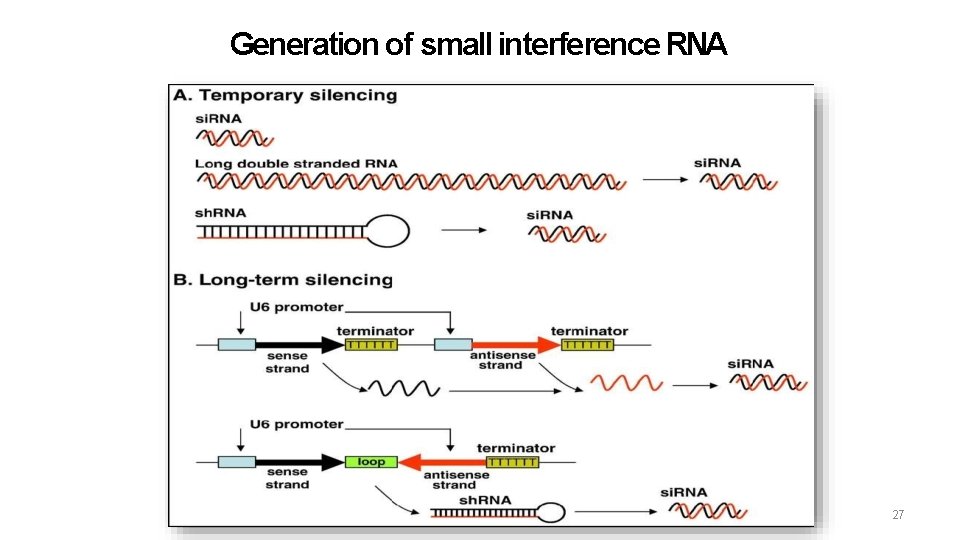
si. RNAs • Small interfering RNAs that have an integral role in the phenomenon of RNA interference (RNAi), a form of post-transcriptional genesilencing • RNAi: 21 -25 nt fragments, which bind to the complementary portion of the target m. RNA and tag it for degradation • A single base pair difference between the si. RNA template and the target m. RNA is enough to block the process. • Each strand of si. RNA has: a. 5’-phosphate termini b. 3’-hydroxyl termini c. 2/3 -nucleotide 3’ overhangs 16

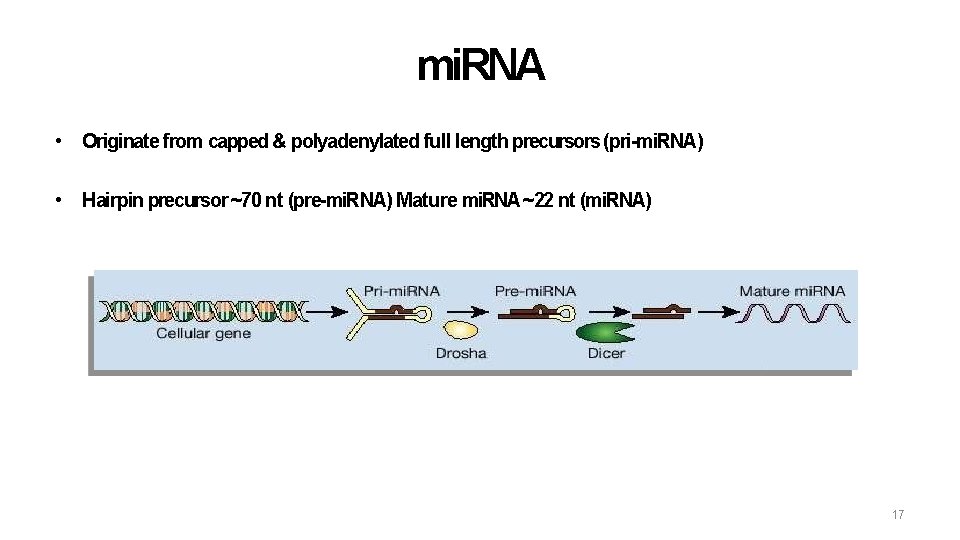
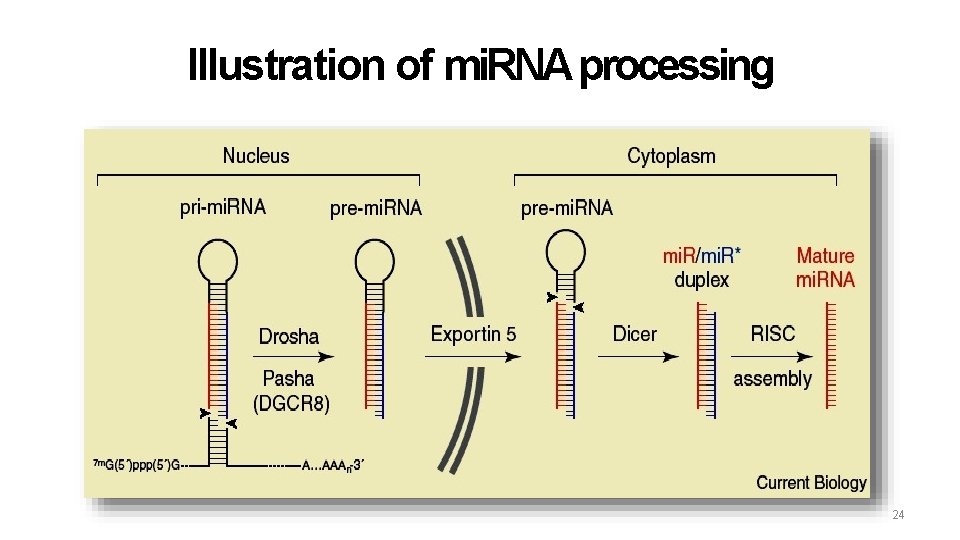
mi. RNA • Originate from capped & polyadenylated full length precursors (pri-mi. RNA) • Hairpin precursor ~70 nt (pre-mi. RNA) Mature mi. RNA ~22 nt (mi. RNA) 17

Difference between mi. RNA and si. RNA Function of both species is regulation of gene expression. Difference is in where they originate. si. RNA originates with ds. RNA. si. RNA is most commonly a response to foreign RNA (usually viral) and is often 100% complementary to the target. mi. RNA originates with ss. RNA that forms a hairpin secondary structure. mi. RNA regulates post-transcriptional gene expression and is often not 100% complementary to the target. Also mi. RNA help to regulate gene expression, particularly during induction of heterochromatin formation serves to downregulate genes pre- transcriptionally (RNA 18 induced transcriptional silencing or RITS)

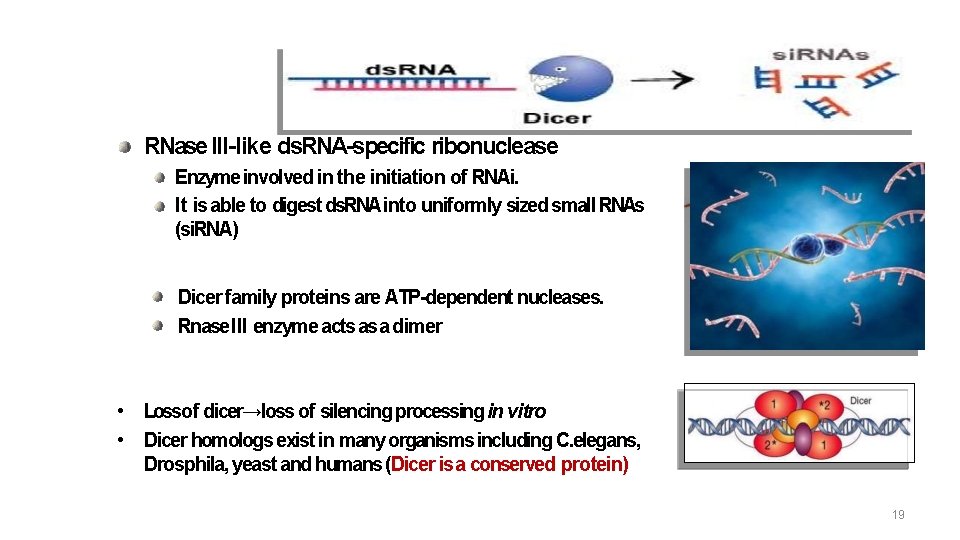
Dicer RNase III-like ds. RNA-specific ribonuclease Enzyme involved in the initiation of RNAi. It is able to digest ds. RNA into uniformly sized small RNAs (si. RNA) Dicer family proteins are ATP-dependent nucleases. Rnase III enzyme acts as a dimer • Lossof dicer→loss of silencing processing in vitro • Dicer homologs exist in many organisms including C. elegans, Drosphila, yeast and humans (Dicer is a conserved protein) 19

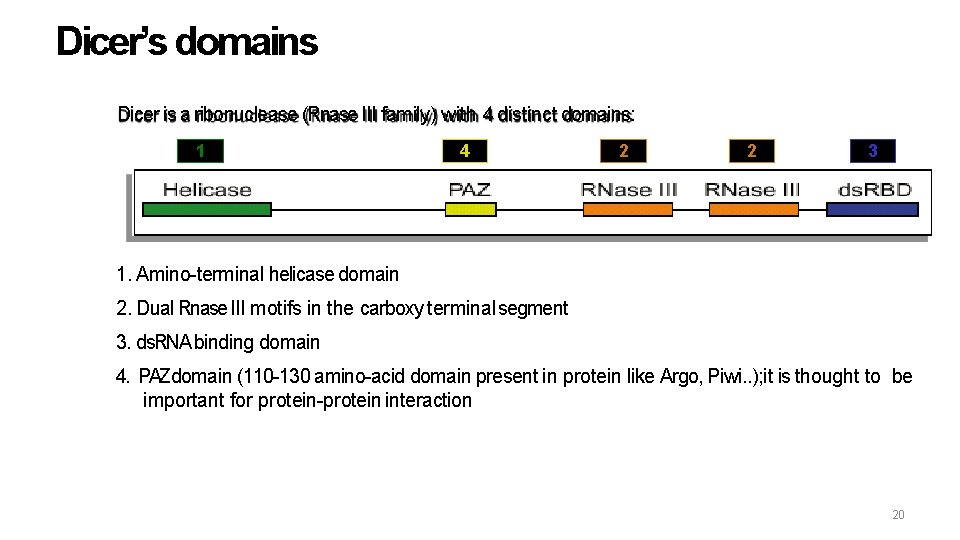
Dicer’s domains Dicer is a ribonuclease (Rnase III family) with 4 distinct domains: 1 4 2 2 3 1. Amino-terminal helicase domain 2. Dual Rnase III motifs in the carboxy terminal segment 3. ds. RNA binding domain 4. PAZdomain (110 -130 amino-acid domain present in protein like Argo, Piwi. . ); it is thought to be important for protein-protein interaction 20

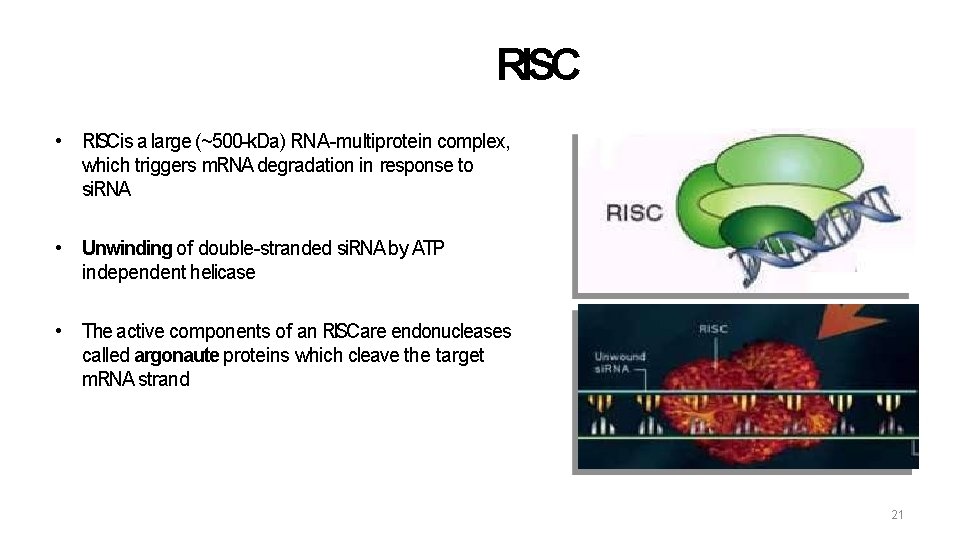
RISC • RISCis a large (~500 -k. Da) RNA-multiprotein complex, which triggers m. RNA degradation in response to si. RNA • Unwinding of double-stranded si. RNA by ATP independent helicase • The active components of an RISCare endonucleases called argonaute proteins which cleave the target m. RNA strand 21

23

Illustration of mi. RNA processing 24

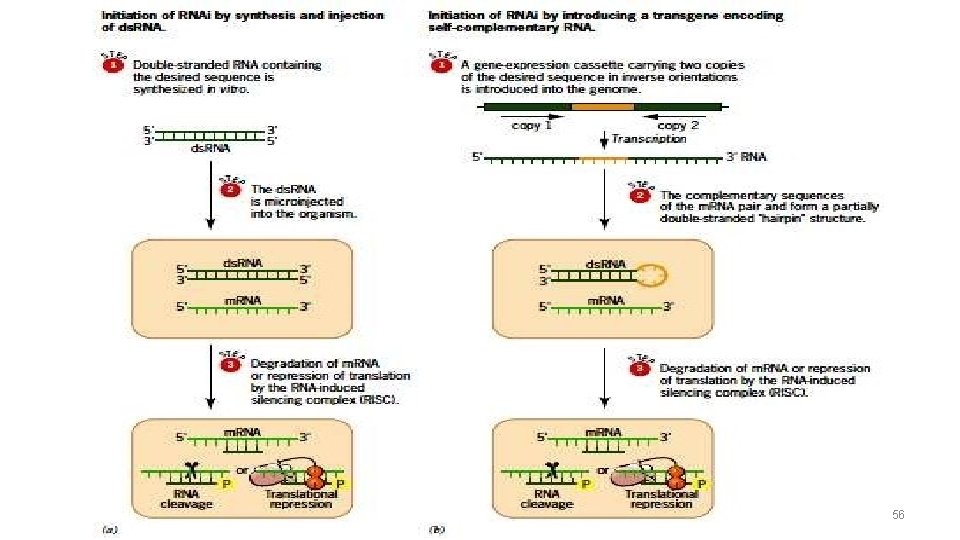
Generation of si. RNA’s 26

Generation of small interference RNA 27

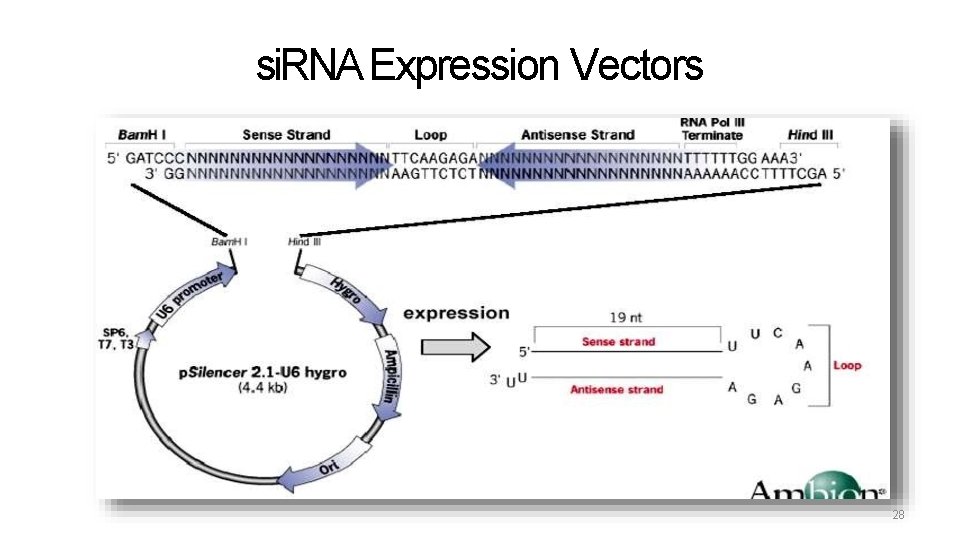
si. RNA Expression Vectors 28

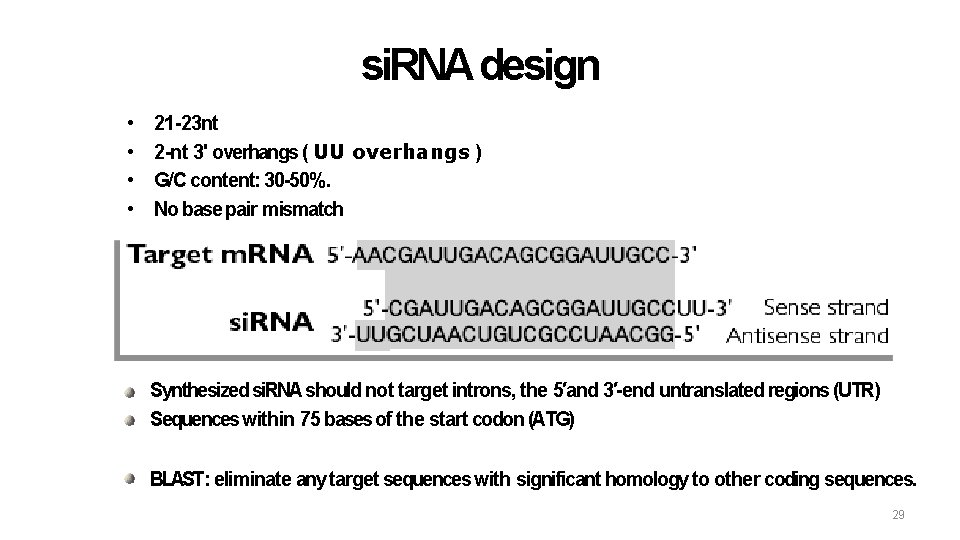
si. RNA design • • 21 -23 nt 2 -nt 3' overhangs ( UU overhangs ) G/C content: 30 -50%. No base pair mismatch Synthesized si. RNA should not target introns, the 5′and 3′-end untranslated regions (UTR) Sequences within 75 bases of the start codon (ATG) BLAST: eliminate any target sequences with significant homology to other coding sequences. 29

si. RNA delivery methods 30

• Effective methods for the delivery of small RNA to allow a sufficient silencing effect in the target organ(s) and/or cells are yet to be developed • In particular, toxicity and side effects of RNAi must be well characterized and limited • Therefore, careful design and selection of target sequence and quantification of the effect on the expression of target protein and m. RNA are essential for success of gene interfering approaches 31

High-pressure injection “High-pressure injection” was the first strategy to demonstrate successful delivery of si. RNA in vivo A large volume (1– 2 m. L) of saline containing unmodified si. RNA is injected intravenously into the tail vein of mice within very short time (in less than 7 sec), which presumably results in the si. RNA molecules being forced into several organs mainly the liver, kidney and to a lesser degree the lung Certainly, such an approach seems to be impossible in human subjects (1000 m. L saline solution containing si. RNA per 10 kg of weight) 32

Applications of RNAi 42

Hematology (blood) • Hematologic disorders result from – Loss of gene function – Mutant gene function – Absent gene function • RNAi – May be used to create models of disease processes – Could help to develop pharmacologic and genetic therapeutic targets 43

Oncology (cancer) • Targeting of oncogenes – Dominant mutant oncogenes, amplified oncogenes, viral oncogenes • Define role of signaling molecules in tumor-creation • Improvement efficacy of chemotherapy and radiotherapy • Tumor regression through creation of potentially new mode of chemotherapy 44

Stem cell biology • Mouse research -Knock out tumor-suppression gene in mouse embryonic stem cell -Observe tumor phenotype -Positive correlation between extent of Trp 53 (suppression gene) inhibition and severity of disease 45

Infectious Diseases • Virus targeting -RNAi – inhibit cellular and viral factors of disease -RNA transcriptase is RNAi target -Inhibition of replication Main goal -Render cells resistant to infectious organisms 46

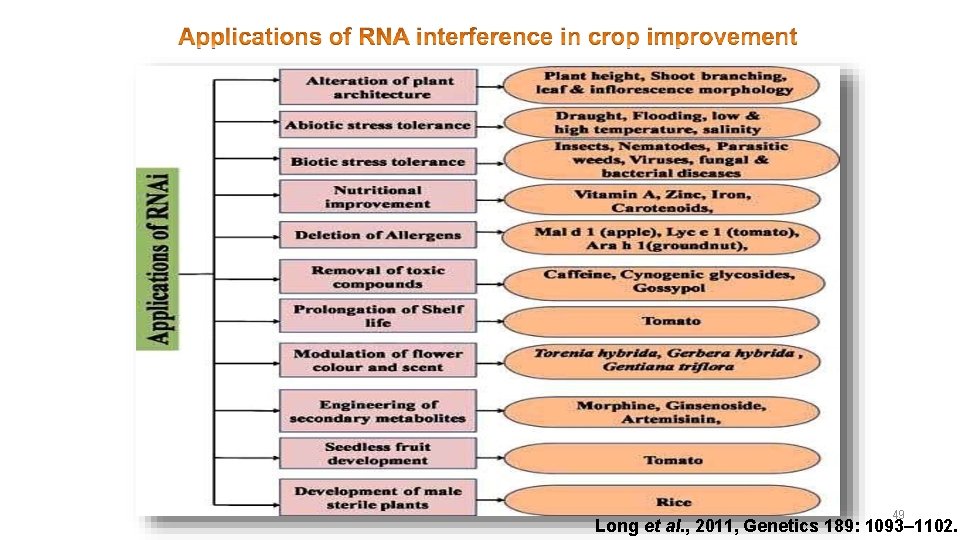
49 Long et al. , 2011, Genetics 189: 1093– 1102.

Conclusion of RNA i 50

RNA interference characteristics • ds. RNA needs to be directed against an exon, not an intron in order to be effective • Homology of the ds. RNA and the target gene/m. RNA is required • Targeted m. RNA is lost (degraded) after RNAi • ss. RNA does not work as well asds. RNA 51

Advantage of RNAi • Down regulation of gene expression simplifies "knockout" analysis. • Easier than use of antisense oligonucleotides. si. RNA more effective and sensitive at lower concentration. • Cost effective • High Specificity • Middle region 9 -14 are most sensitive • With si. RNA, the researcher can simultaneously perform experiments in any cell type of interest • Can be labelled • Ease of transfection by use of vector 52

Importance of RNAi • Powerful for analyzing unknown genes in sequenced genomes. • More efforts are being undertaken to target every human gene via si. RNAs • Faster identification of gene function • Gene therapy: down-regulation of certain genes/ mutated alleles • Cancer treatments – knock-out of genes required for cell proliferation – knock-out of genes encoding key structural proteins • Agriculture 53

Thank You 54

55

56