RNA Interference Iain Fraser Model for RNAi mechanism



- Slides: 23

RNA Interference Iain Fraser

Model for RNAi mechanism Initiation step (RNA-induced silencing complex) Dicer Effector step Hammond et al. , 2001

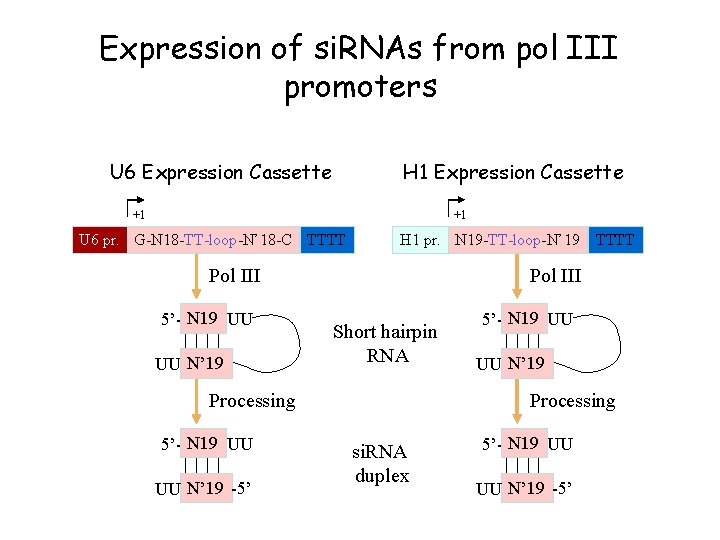
Expression of si. RNAs from pol III promoters U 6 Expression Cassette H 1 Expression Cassette +1 +1 U 6 pr. G-N 18 -TT-loop-N’ 18 -C TTTT H 1 pr. N 19 -TT-loop-N’ 19 Pol III 5’- N 19 UU UU N’ 19 Pol III Short hairpin RNA Processing 5’- N 19 UU UU N’ 19 -5’ TTTT 5’- N 19 UU UU N’ 19 Processing si. RNA duplex 5’- N 19 UU UU N’ 19 -5’

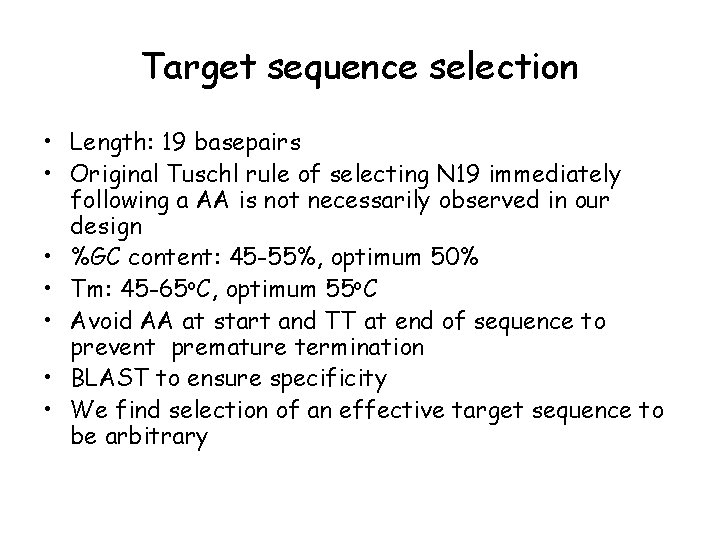
Target sequence selection • Length: 19 basepairs • Original Tuschl rule of selecting N 19 immediately following a AA is not necessarily observed in our design • %GC content: 45 -55%, optimum 50% • Tm: 45 -65 o. C, optimum 55 o. C • Avoid AA at start and TT at end of sequence to prevent premature termination • BLAST to ensure specificity • We find selection of an effective target sequence to be arbitrary

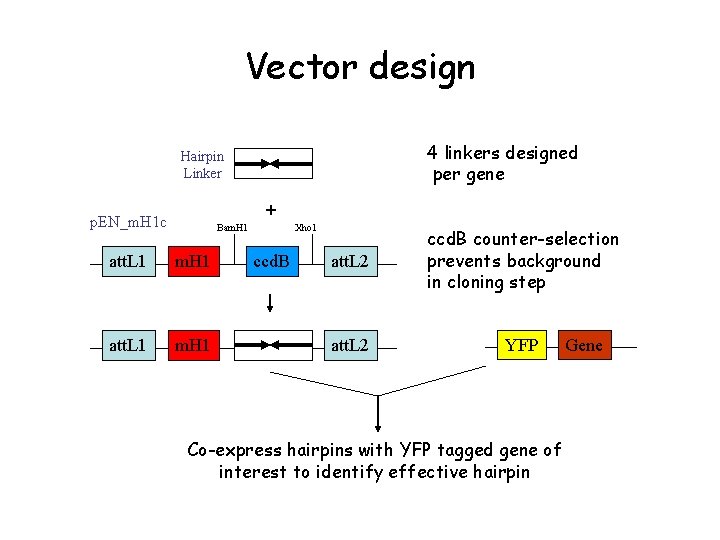
Vector design 4 linkers designed per gene Hairpin Linker + p. EN_m. H 1 c Bam. H 1 att. L 1 m. H 1 Xho 1 ccd. B att. L 2 ccd. B counter-selection prevents background in cloning step att. L 2 YFP Co-express hairpins with YFP tagged gene of interest to identify effective hairpin Gene

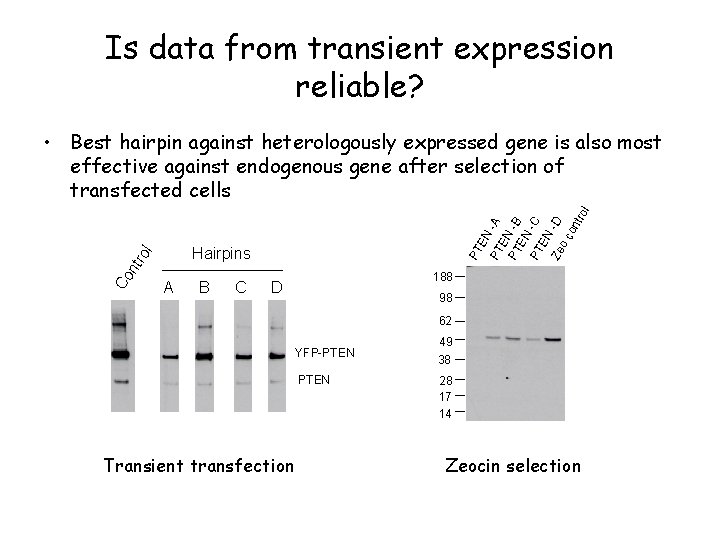
Is data from transient expression reliable? l Hairpins tro Co n PT EN PT -A E PT N -B EN PT -C EN -D Ze oc on tro l • Best hairpin against heterologously expressed gene is also most effective against endogenous gene after selection of transfected cells A B C 188 D 98 62 YFP-PTEN Transient transfection 49 38 28 17 14 Zeocin selection

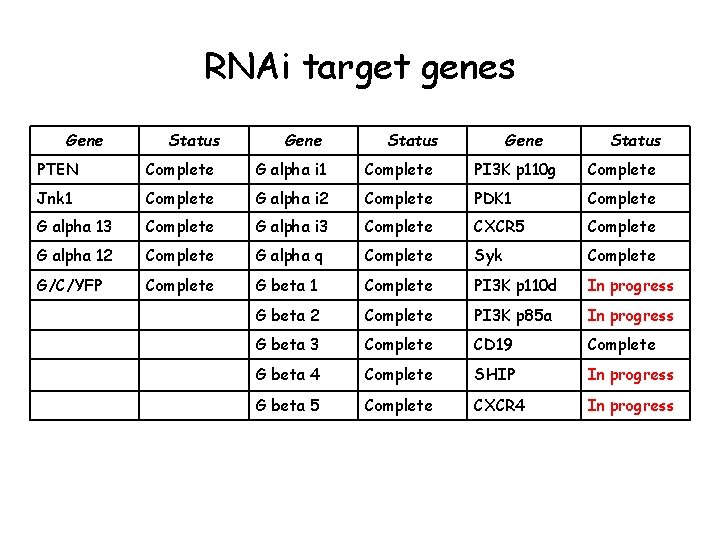
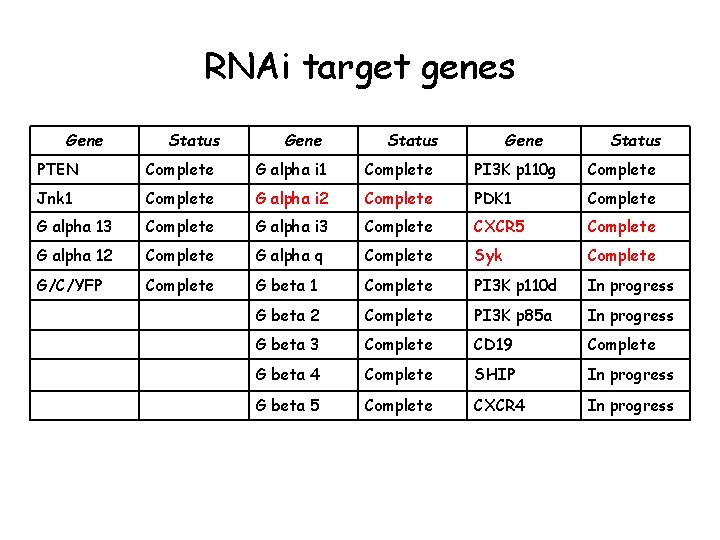
RNAi target genes Gene Status PTEN Complete G alpha i 1 Complete PI 3 K p 110 g Complete Jnk 1 Complete G alpha i 2 Complete PDK 1 Complete G alpha 13 Complete G alpha i 3 Complete CXCR 5 Complete G alpha 12 Complete G alpha q Complete Syk Complete G/C/YFP Complete G beta 1 Complete PI 3 K p 110 d In progress G beta 2 Complete PI 3 K p 85 a In progress G beta 3 Complete CD 19 Complete G beta 4 Complete SHIP In progress G beta 5 Complete CXCR 4 In progress

RNAi target genes Gene Status PTEN Complete G alpha i 1 Complete PI 3 K p 110 g Complete Jnk 1 Complete G alpha i 2 Complete PDK 1 Complete G alpha 13 Complete G alpha i 3 Complete CXCR 5 Complete G alpha 12 Complete G alpha q Complete Syk Complete G/C/YFP Complete G beta 1 Complete PI 3 K p 110 d In progress G beta 2 Complete PI 3 K p 85 a In progress G beta 3 Complete CD 19 Complete G beta 4 Complete SHIP In progress G beta 5 Complete CXCR 4 In progress

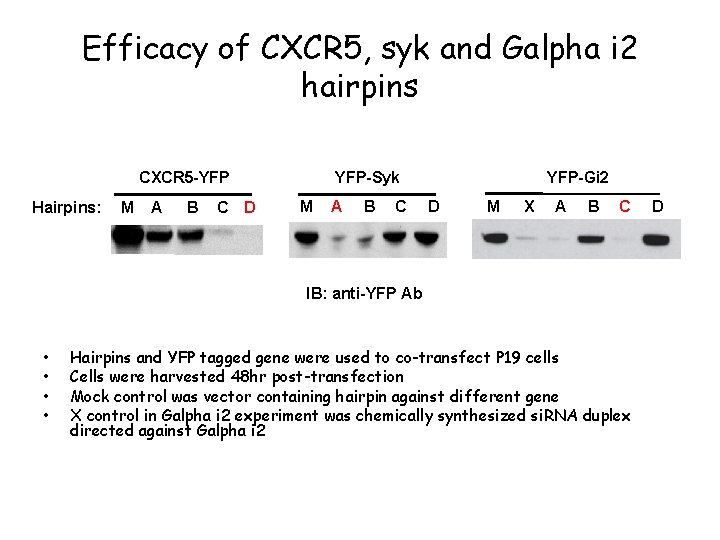
Efficacy of CXCR 5, syk and Galpha i 2 hairpins YFP-Syk CXCR 5 -YFP Hairpins: M A B C D M A B C YFP-Gi 2 D M X A B C IB: anti-YFP Ab • • Hairpins and YFP tagged gene were used to co-transfect P 19 cells Cells were harvested 48 hr post-transfection Mock control was vector containing hairpin against different gene X control in Galpha i 2 experiment was chemically synthesized si. RNA duplex directed against Galpha i 2 D

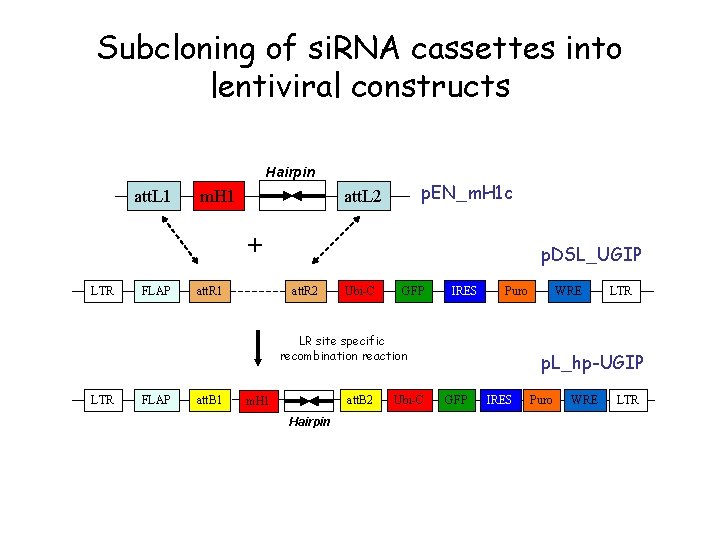
Subcloning of si. RNA cassettes into lentiviral constructs Hairpin att. L 1 m. H 1 p. EN_m. H 1 c att. L 2 + LTR FLAP att. R 1 p. DSL_UGIP att. R 2 Ubi-C GFP IRES Puro LR site specific recombination reaction LTR FLAP att. B 1 att. B 2 m. H 1 Hairpin Ubi-C WRE LTR p. L_hp-UGIP GFP IRES Puro WRE LTR

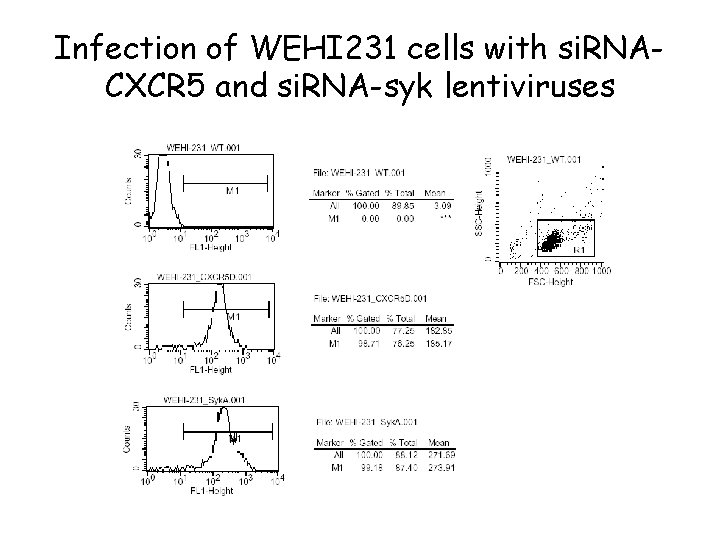
Infection of WEHI 231 cells with si. RNACXCR 5 and si. RNA-syk lentiviruses

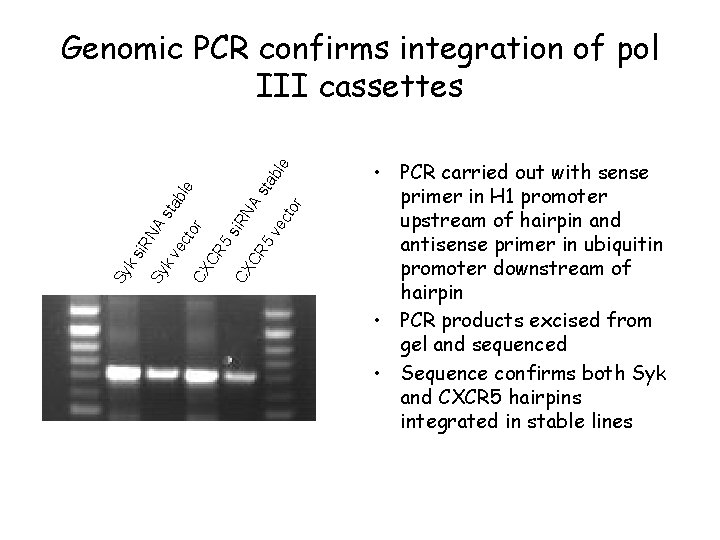
R 5 si. R CX NA CR sta 5 v ble ec tor CX C ec kv Sy Sy ks i. R NA sta ble tor Genomic PCR confirms integration of pol III cassettes • PCR carried out with sense primer in H 1 promoter upstream of hairpin and antisense primer in ubiquitin promoter downstream of hairpin • PCR products excised from gel and sequenced • Sequence confirms both Syk and CXCR 5 hairpins integrated in stable lines

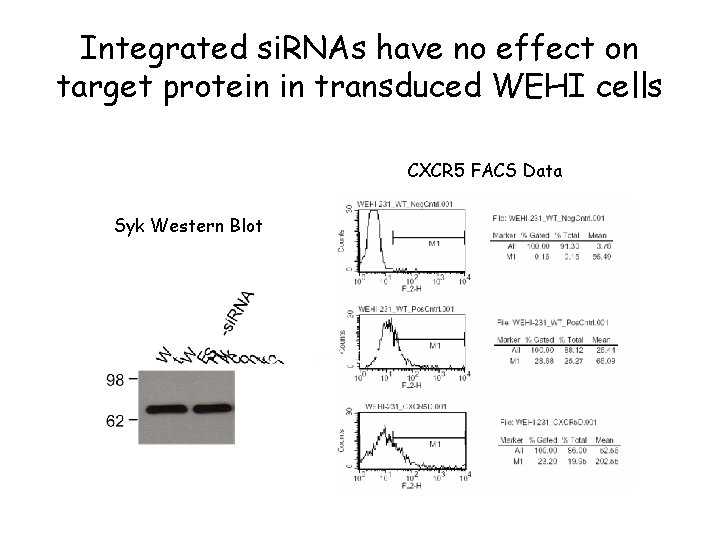
Integrated si. RNAs have no effect on target protein in transduced WEHI cells CXCR 5 FACS Data Syk Western Blot

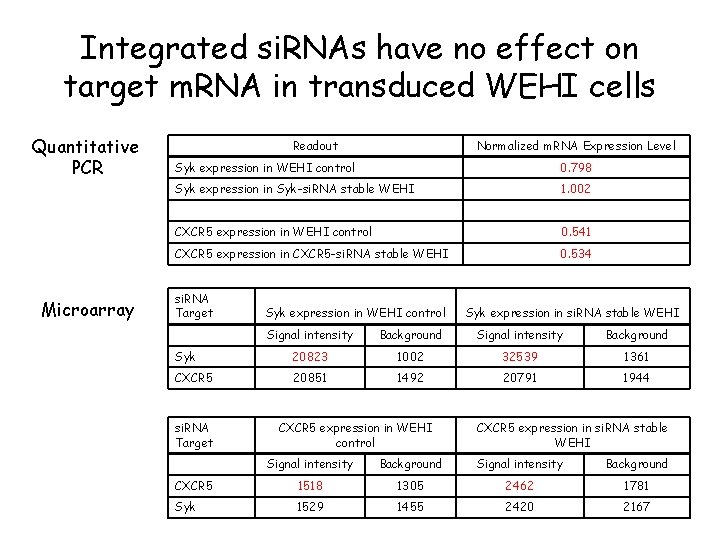
Integrated si. RNAs have no effect on target m. RNA in transduced WEHI cells Quantitative PCR Readout Normalized m. RNA Expression Level Syk expression in WEHI control 0. 798 Syk expression in Syk-si. RNA stable WEHI 1. 002 Microarray CXCR 5 expression in WEHI control 0. 541 CXCR 5 expression in CXCR 5 -si. RNA stable WEHI 0. 534 si. RNA Target Syk expression in WEHI control Signal intensity Background Syk 20823 1002 32539 1361 CXCR 5 20851 1492 20791 1944 si. RNA Target CXCR 5 expression in WEHI control Syk expression in si. RNA stable WEHI CXCR 5 expression in si. RNA stable WEHI Signal intensity Background CXCR 5 1518 1305 2462 1781 Syk 1529 1455 2420 2167

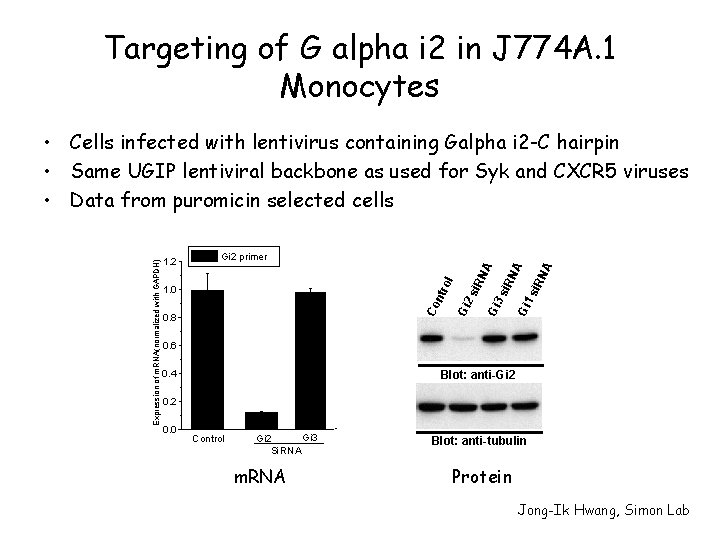
Targeting of G alpha i 2 in J 774 A. 1 Monocytes si. R N A A Gi 1 si. R N Gi 3 0. 8 Gi 2 ntr 1. 0 si. R N A Gi 2 primer ol 1. 2 Co Expression of m. RNA(normalized with GAPDH) • Cells infected with lentivirus containing Galpha i 2 -C hairpin • Same UGIP lentiviral backbone as used for Syk and CXCR 5 viruses • Data from puromicin selected cells 0. 6 0. 4 Blot: anti-Gi 2 0. 0 Control Gi 3 Gi 2 Si. RNA m. RNA Blot: anti-tubulin Protein Jong-Ik Hwang, Simon Lab

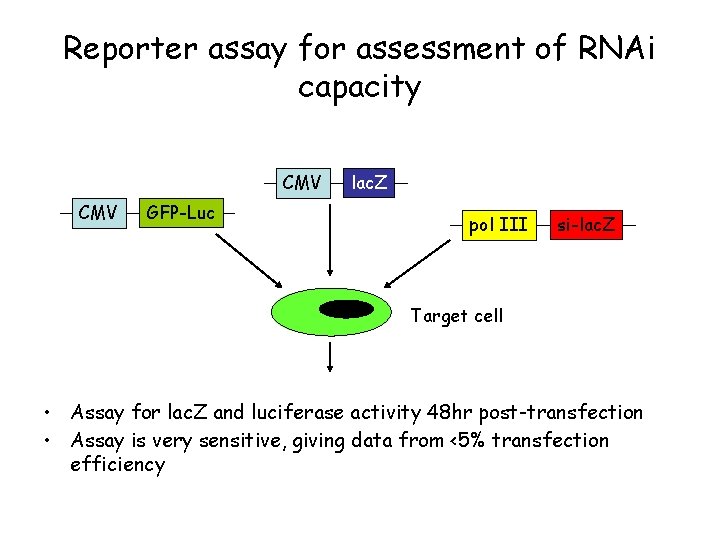
Reporter assay for assessment of RNAi capacity CMV GFP-Luc lac. Z pol III si-lac. Z Target cell • Assay for lac. Z and luciferase activity 48 hr post-transfection • Assay is very sensitive, giving data from <5% transfection efficiency

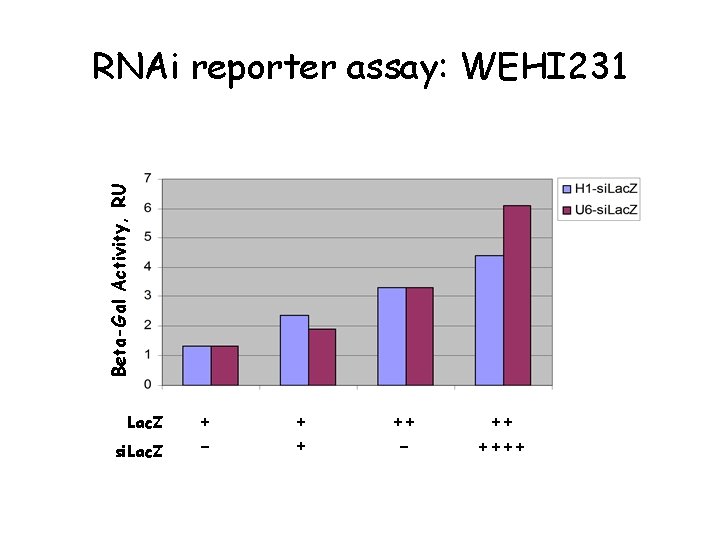
Beta-Gal Activity, RU RNAi reporter assay: WEHI 231 Lac. Z si. Lac. Z + - + + ++ - ++ ++++

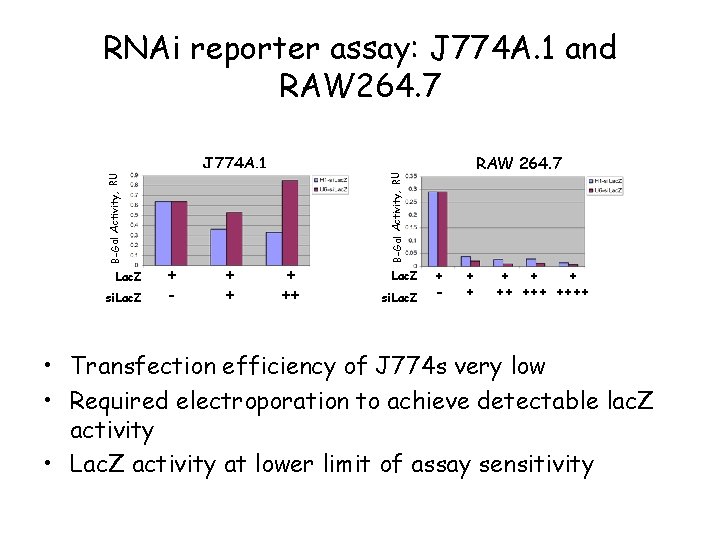
Lac. Z si. Lac. Z J 774 A. 1 + - + ++ RAW 264. 7 B-Gal Activity, RU RNAi reporter assay: J 774 A. 1 and RAW 264. 7 Lac. Z si. Lac. Z + - + + ++ ++++ • Transfection efficiency of J 774 s very low • Required electroporation to achieve detectable lac. Z activity • Lac. Z activity at lower limit of assay sensitivity

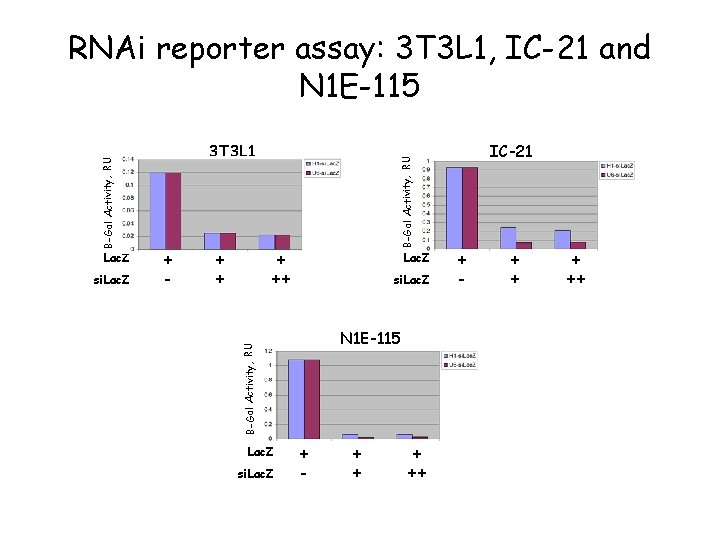
Lac. Z si. Lac. Z + - + + B-Gal Activity, RU 3 T 3 L 1 + ++ Lac. Z si. Lac. Z N 1 E-115 B-Gal Activity, RU RNAi reporter assay: 3 T 3 L 1, IC-21 and N 1 E-115 Lac. Z si. Lac. Z + - + ++ IC-21 + - + ++

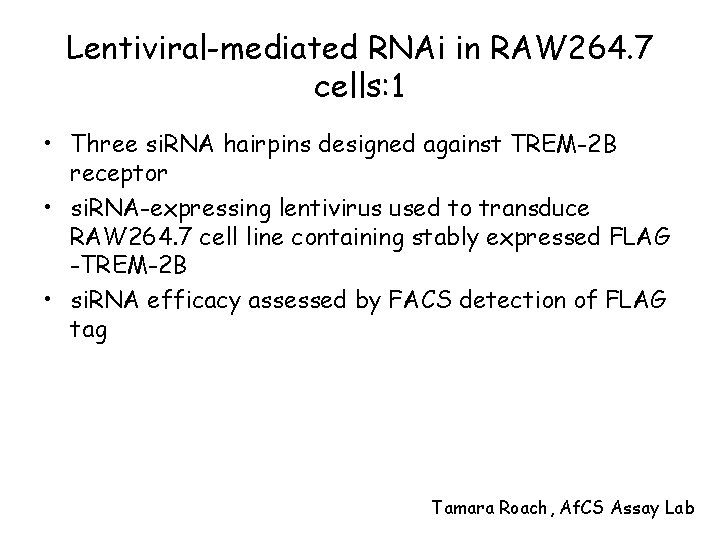
Lentiviral-mediated RNAi in RAW 264. 7 cells: 1 • Three si. RNA hairpins designed against TREM-2 B receptor • si. RNA-expressing lentivirus used to transduce RAW 264. 7 cell line containing stably expressed FLAG -TREM-2 B • si. RNA efficacy assessed by FACS detection of FLAG tag Tamara Roach, Af. CS Assay Lab

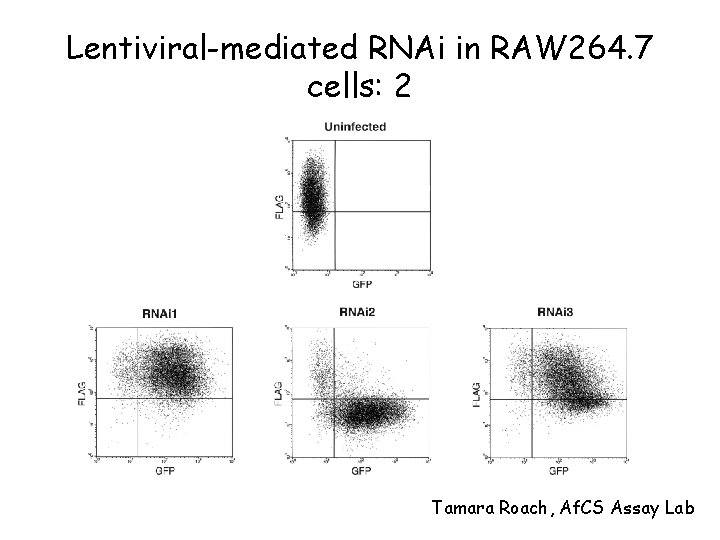
Lentiviral-mediated RNAi in RAW 264. 7 cells: 2 Tamara Roach, Af. CS Assay Lab

Summary/Conclusions • We have established a flexible vector-based system for the development of effective si. RNAs against genes of interest to the Af. CS. • Expression of such si. RNAs from lentiviral vectors permits the transduction of hematopoietic cell lines. • WEHI 231 cells transduced with si. RNAs do not exhibit RNAimediated target knockdown. It remains unclear whether this is due to the si. RNAs not being expressed from pol III promoters, or the absence of the components of the RNAi machinery required to recognize the si. RNAs. • Vector-based RNAi was effective in six other cell lines tested by reporter assay: J 774 A. 1, RAW 264. 7, IC-21, N 1 E-115, 3 T 3 -L 1 and HEK 293. • Lentiviral-mediated RNAi was effective in both J 774 A. 1 and RAW 264. 7 cells

Acknowledgements Molecular Biology Lab Assay Lab Joelle Zavzavadjian Pamela Eversole-Cire Jamie Liu Dan Allen Mei Wang Tamara Roach Sangdun Choi Caltech Jong-Ik Hwang Melvin Simon