Epigenetics RNA interference Discovery of RNA interference 1998







- Slides: 41

Epigenetics - RNA interference

Discovery of RNA interference (1998) - silencing of gene expression with ds. RNA Cenorhabditis elegans

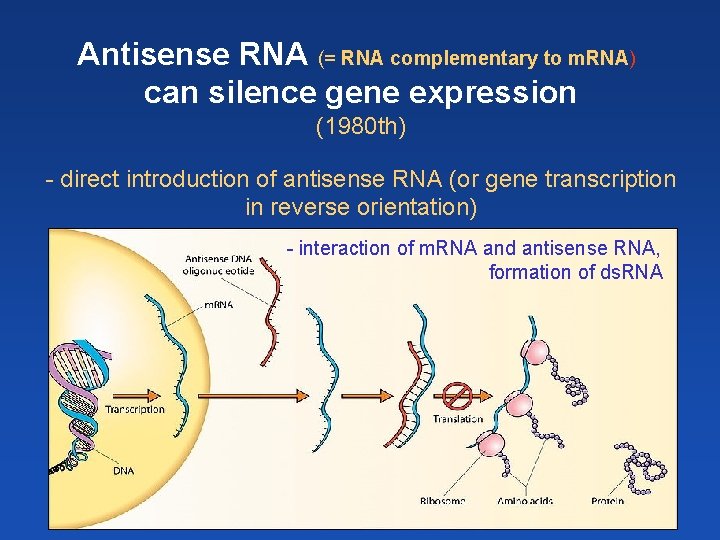
Antisense RNA (= RNA complementary to m. RNA) can silence gene expression (1980 th) - direct introduction of antisense RNA (or gene transcription in reverse orientation) - interaction of m. RNA and antisense RNA, formation of ds. RNA

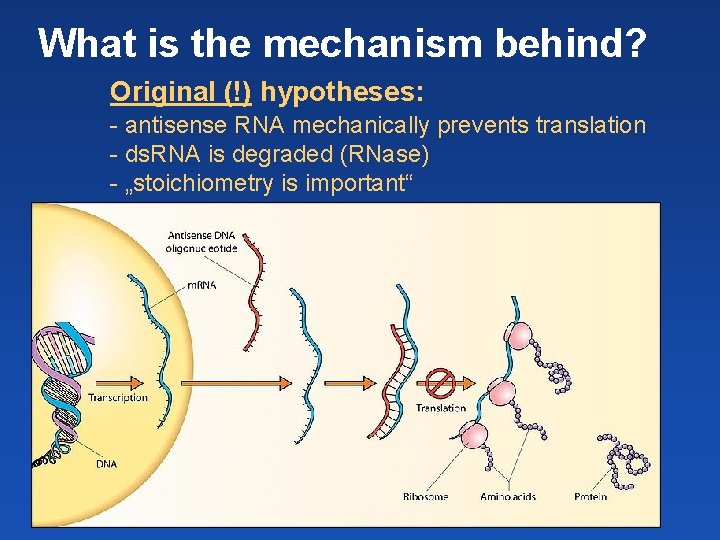
What is the mechanism behind? Original (!) hypotheses: - antisense RNA mechanically prevents translation - ds. RNA is degraded (RNase) - „stoichiometry is important“

What they get the Nobel prize for? - making proper controls pays off! - Introduction of even very small amount of ds. RNA induce specific silencing (antisence RNA is less efficient!) ds. RNA has to be a trigger/signal! - for sequence specific silencing Andrew J. Hamilton, David C. Baulcombe*(1999): A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants, Science 286 (5441): 950 -952

RNA interference (RNAi) = silencing of gene expression mediated by small RNAs (small RNA, s. RNA) in plants predominantly - 21 -24 nt The precise role of 25 -nt RNA in PTGS remains to be determined. However, because they are long enough to convey sequence specificity yet small enough to move through plasmodesmata, it is possible that they are components of the systemic signal and specificity determinants of PTGS (Hamilton and Baulcombe, 1999).

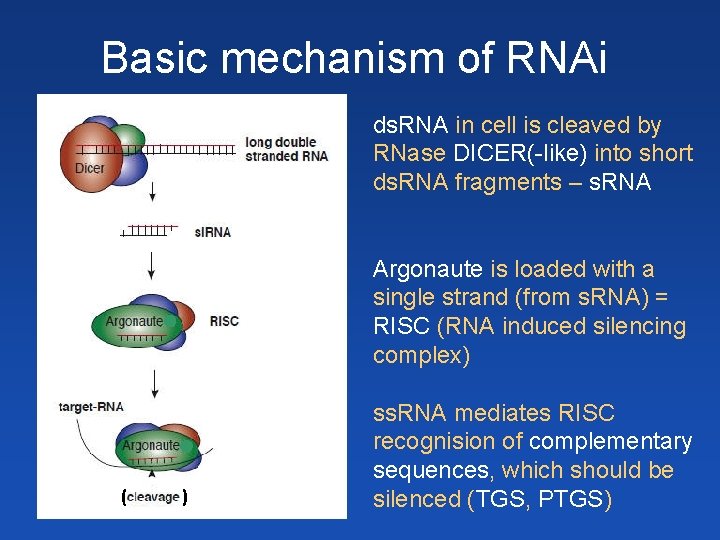
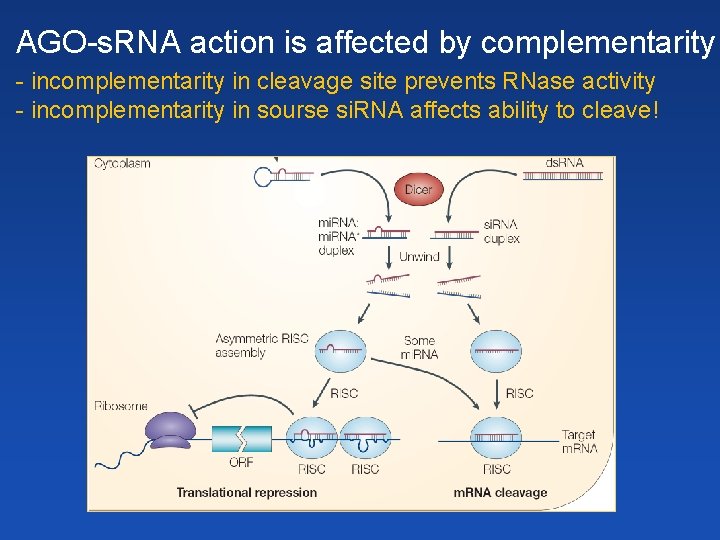
Basic mechanism of RNAi ds. RNA v buňce je štěpena enzymem Dicer(-like) na krátké ds. RNA fragmenty – malé RNA (s. RNA 21 -24 nt) protein Argonaut váže jedno z vláken malé RNA (RISC: RNA induced silencing complex) ss. RNA zprostředkovává rozpoznání komplementárních ( ) (cílových) sekvencí ds. RNA in cell is cleaved by RNase DICER(-like) into short ds. RNA fragments – s. RNA Argonaute is loaded with a single strand (from s. RNA) = RISC (RNA induced silencing complex) ss. RNA mediates RISC recognision of complementary sequences, which should be silenced (TGS, PTGS)

RNA interference (RNAi) gene silencing at • transcriptional level (TGS) (transcriptional gene silencing) - induction of DNA methylation (m. RNA not formed) • posttranscriptional level (PTGS) (posttranscriptional gene silencing) - transcript cleavage - block of translation

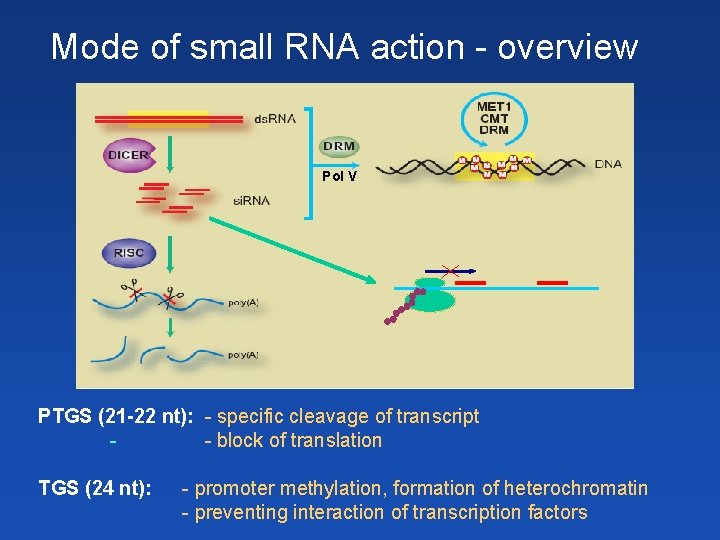
Mode of small RNA action - overview Pol V PTGS (21 -22 nt): - specific cleavage of transcript - block of translation TGS (24 nt): - promoter methylation, formation of heterochromatin - preventing interaction of transcription factors

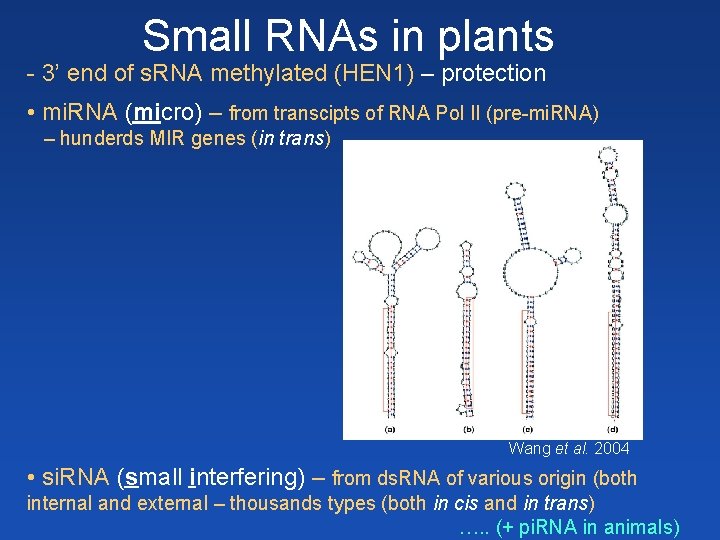
Small RNAs in plants - 3’ end of s. RNA methylated (HEN 1) – protection • mi. RNA (micro) – from transcipts of RNA Pol II (pre-mi. RNA) – hunderds MIR genes (in trans) pre-mi. RNA Wang et al. 2004 • si. RNA (small interfering) – from ds. RNA of various origin (both internal and external – thousands types (both in cis and in trans) …. . (+ pi. RNA in animals)

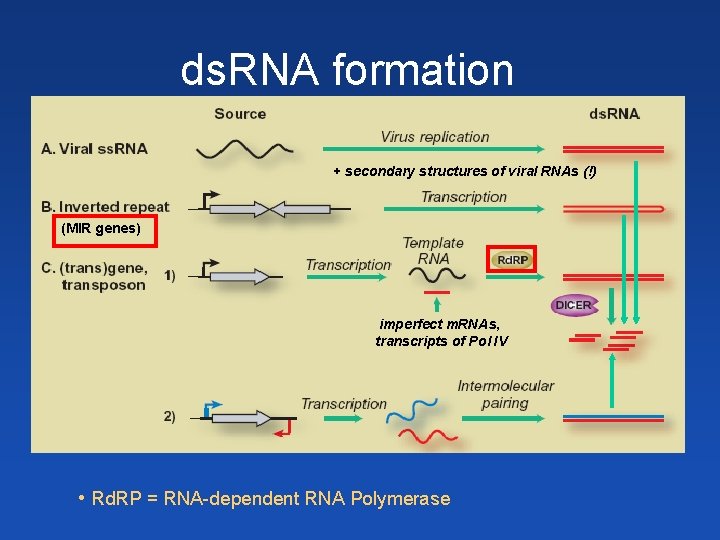
ds. RNA formation + secondary structures of viral RNAs (!) (MIR genes) ? imperfect m. RNAs, transcripts of Pol IV • Rd. RP = RNA-dependent RNA Polymerase

AGO-s. RNA action is affected by complementarity - incomplementarity in cleavage site prevents RNase activity - incomplementarity in sourse si. RNA affects ability to cleave!

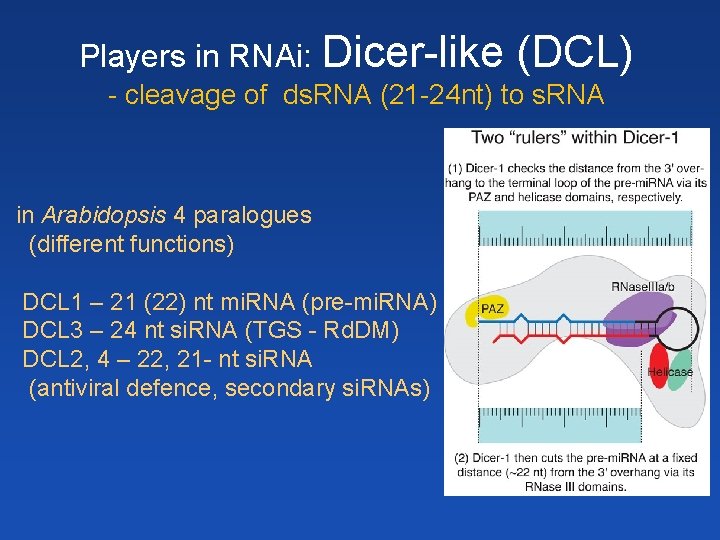
Players in RNAi: Dicer-like (DCL) - cleavage of ds. RNA (21 -24 nt) to s. RNA in Arabidopsis 4 paralogues (different functions) DCL 1 – 21 (22) nt mi. RNA (pre-mi. RNA) DCL 3 – 24 nt si. RNA (TGS - Rd. DM) DCL 2, 4 – 22, 21 - nt si. RNA (antiviral defence, secondary si. RNAs)

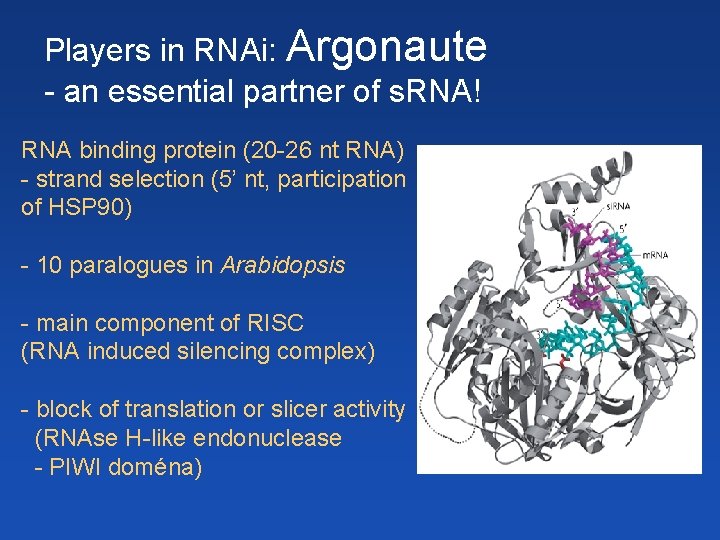
Players in RNAi: Argonaute - an essential partner of s. RNA! RNA binding protein (20 -26 nt RNA) - strand selection (5’ nt, participation of HSP 90) - 10 paralogues in Arabidopsis - main component of RISC (RNA induced silencing complex) - block of translation or slicer activity (RNAse H-like endonuclease - PIWI doména)

RNA polymerases in plant RNAi DNA-dependent RNA polymerases - DNA template - derived from RNA polymerase II (share common subunits) - plant specific Pol IV, Pol V, - Pol II RNA-dependent RNA polymerases (Rd. RP, RDR) - RNA template

RNA polymerases in plant RNAi DNA dependent RNA polymerases RNA polymerase IV - transcribes chromatin with H 3 K 9 me 2 and non-met H 3 K 4 (SHH 1) - short transcripts (hundreds nt), with cap, without poly. A - all transcripts used for generation of si. RNAs RNA polymerase V - necessary for RNA-directed DNA methylation (Rd. DM) - transcribes methylated DNA (SUVH 2/9) - direct interaction with AGO 4 – identification of target seq. RNA polymeráza II – unclear, important role in generation of si. RNAs and likely also in navigation of AGO to target DNA

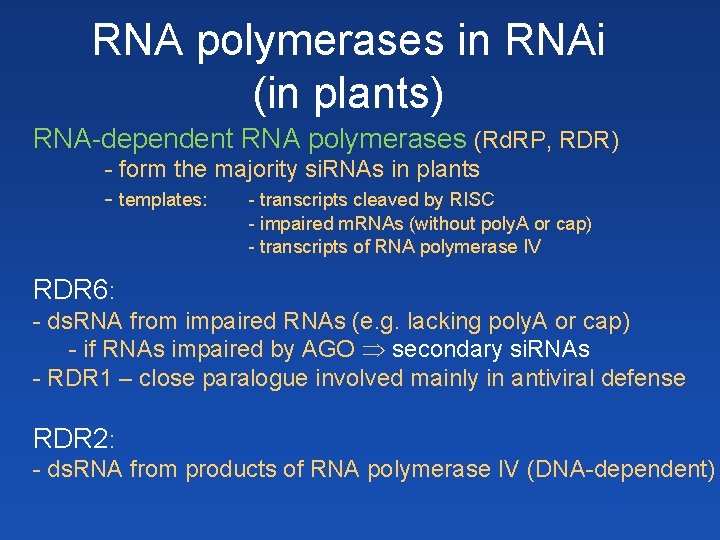
RNA polymerases in RNAi (in plants) RNA-dependent RNA polymerases (Rd. RP, RDR) - form the majority si. RNAs in plants - templates: - transcripts cleaved by RISC - impaired m. RNAs (without poly. A or cap) - transcripts of RNA polymerase IV RDR 6: - ds. RNA from impaired RNAs (e. g. lacking poly. A or cap) - if RNAs impaired by AGO secondary si. RNAs - RDR 1 – close paralogue involved mainly in antiviral defense RDR 2: - ds. RNA from products of RNA polymerase IV (DNA-dependent)

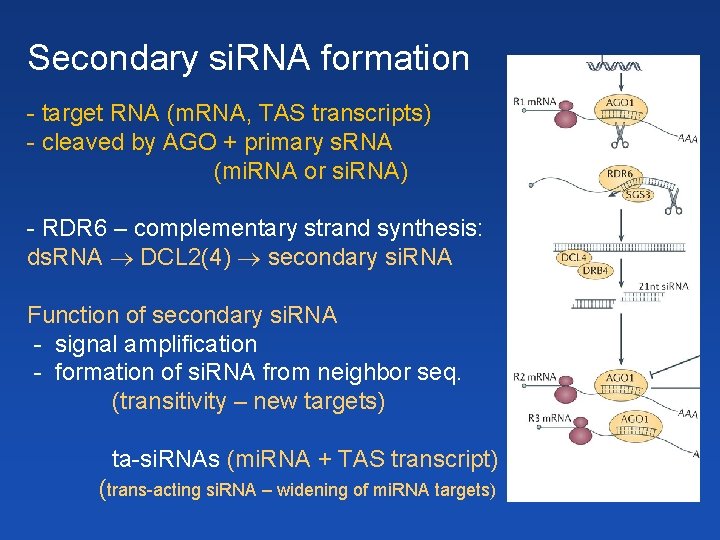
Secondary si. RNA formation - target RNA (m. RNA, TAS transcripts) - cleaved by AGO + primary s. RNA (mi. RNA or si. RNA) - RDR 6 – complementary strand synthesis: ds. RNA DCL 2(4) secondary si. RNA Function of secondary si. RNA - signal amplification - formation of si. RNA from neighbor seq. (transitivity – new targets) ta-si. RNAs (mi. RNA + TAS transcript) (trans-acting si. RNA – widening of mi. RNA targets)

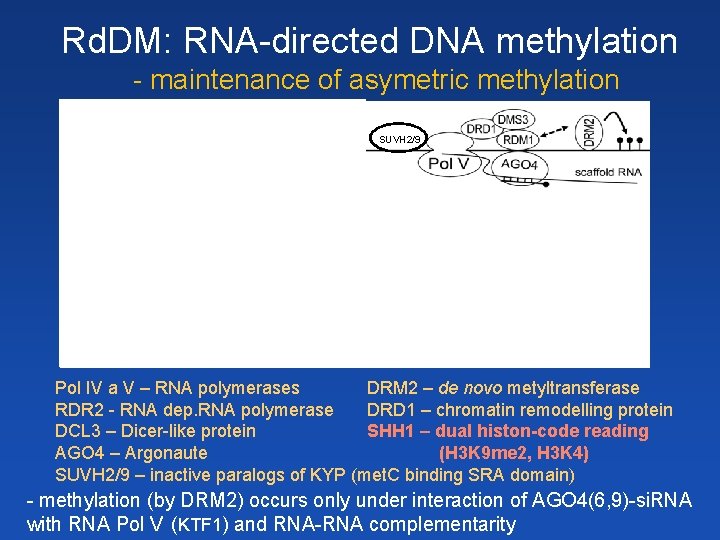
Rd. DM: RNA-directed DNA methylation - maintenance of asymetric methylation SHH 1 SUVH 2/9 Zhang et al. 2013 Pol IV a V – RNA polymerases DRM 2 – de novo metyltransferase RDR 2 - RNA dep. RNA polymerase DRD 1 – chromatin remodelling protein DCL 3 – Dicer-like protein SHH 1 – dual histon-code reading AGO 4 – Argonaute (H 3 K 9 me 2, H 3 K 4) SUVH 2/9 – inactive paralogs of KYP (met. C binding SRA domain) - methylation (by DRM 2) occurs only under interaction of AGO 4(6, 9)-si. RNA with RNA Pol V (KTF 1) and RNA-RNA complementarity

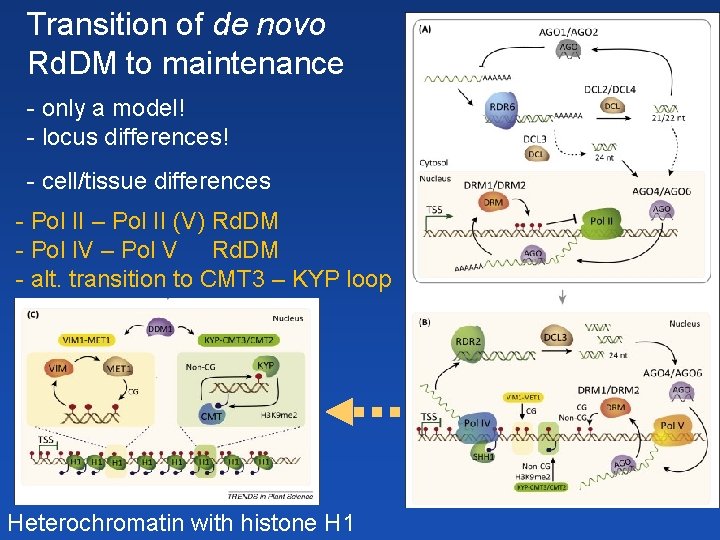
Transition of de novo Rd. DM to maintenance - only a model! - locus differences! - cell/tissue differences - Pol II – Pol II (V) Rd. DM - Pol IV – Pol V Rd. DM - alt. transition to CMT 3 – KYP loop Heterochromatin with histone H 1

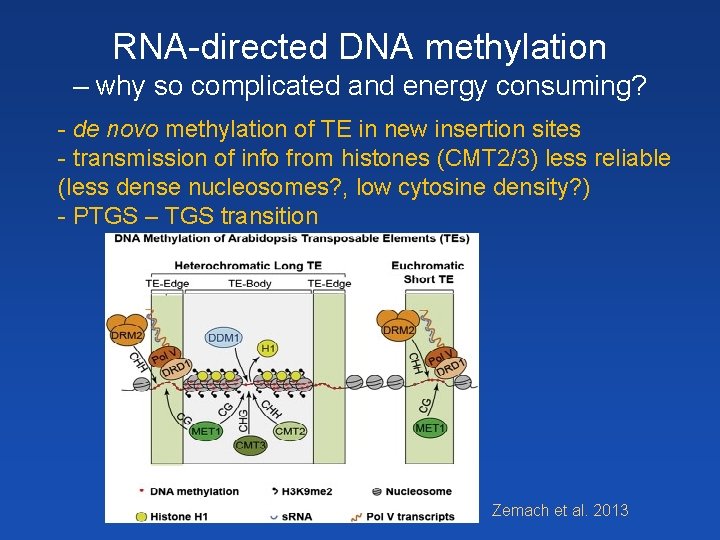
RNA-directed DNA methylation – why so complicated and energy consuming? - de novo methylation of TE in new insertion sites - transmission of info from histones (CMT 2/3) less reliable (less dense nucleosomes? , low cytosine density? ) - PTGS – TGS transition Zemach et al. 2013

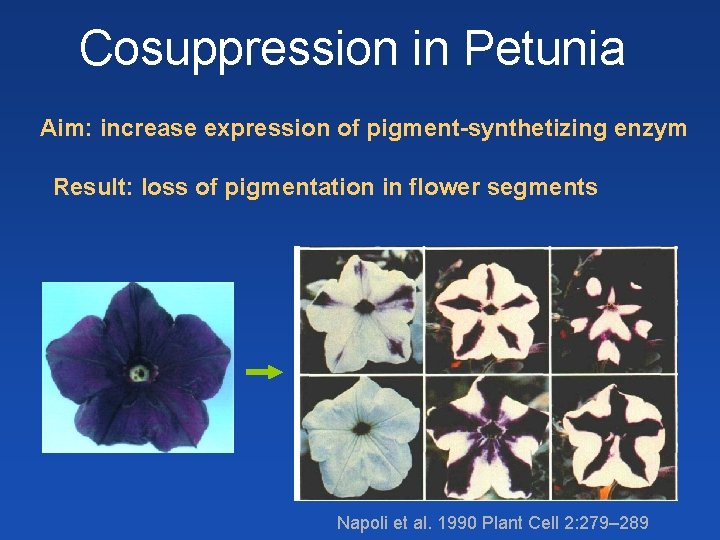
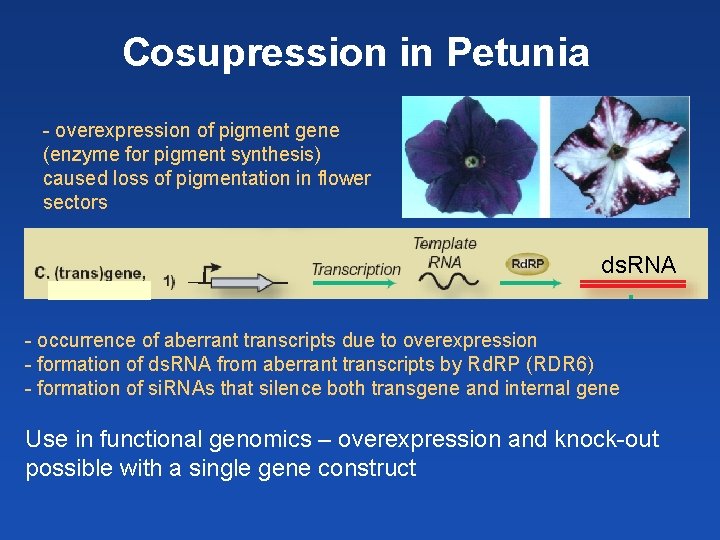
Cosuppression in Petunia Aim: increase expression of pigment-synthetizing enzym Result: loss of pigmentation in flower segments Napoli et al. 1990 Plant Cell 2: 279– 289

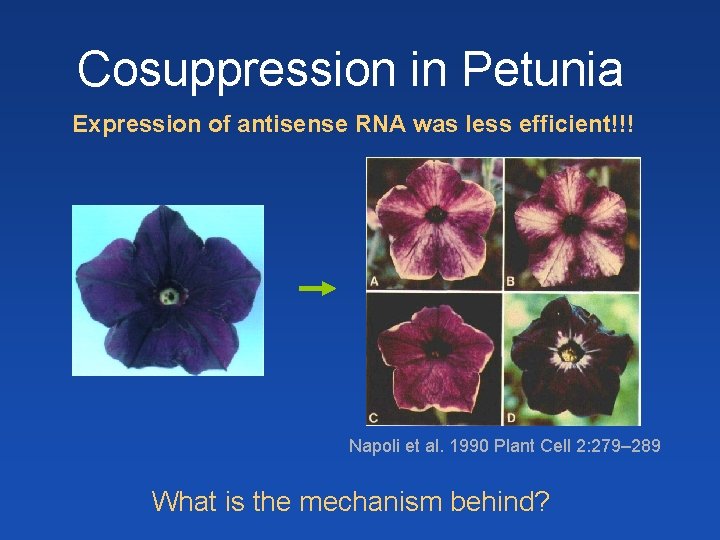
Cosuppression in Petunia Expression of antisense RNA was less efficient!!! Napoli et al. 1990 Plant Cell 2: 279– 289 What is the mechanism behind?

Cosupression in Petunia - overexpression of pigment gene (enzyme for pigment synthesis) caused loss of pigmentation in flower sectors ds. RNA - occurrence of aberrant transcripts due to overexpression - formation of ds. RNA from aberrant transcripts by Rd. RP (RDR 6) - formation of si. RNAs that silence both transgene and internal gene Use in functional genomics – overexpression and knock-out possible with a single gene construct

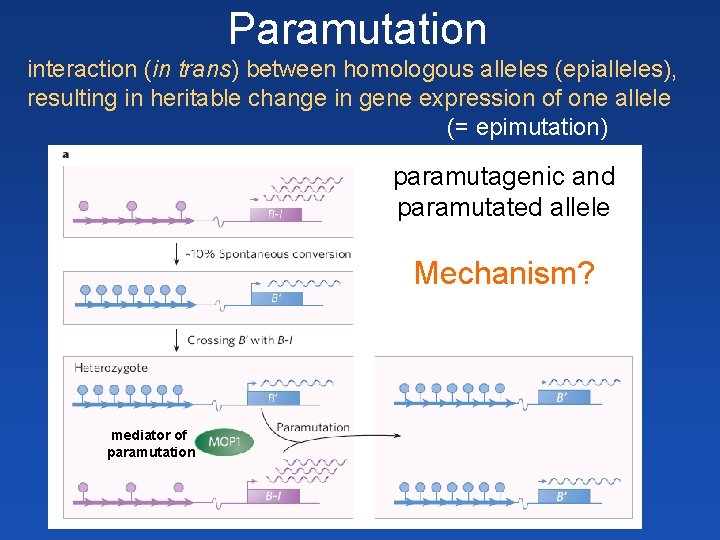
Paramutation interaction (in trans) between homologous alleles (epialleles), resulting in heritable change in gene expression of one allele (= epimutation) paramutagenic and paramutated allele Mechanism? mediator of paramutation

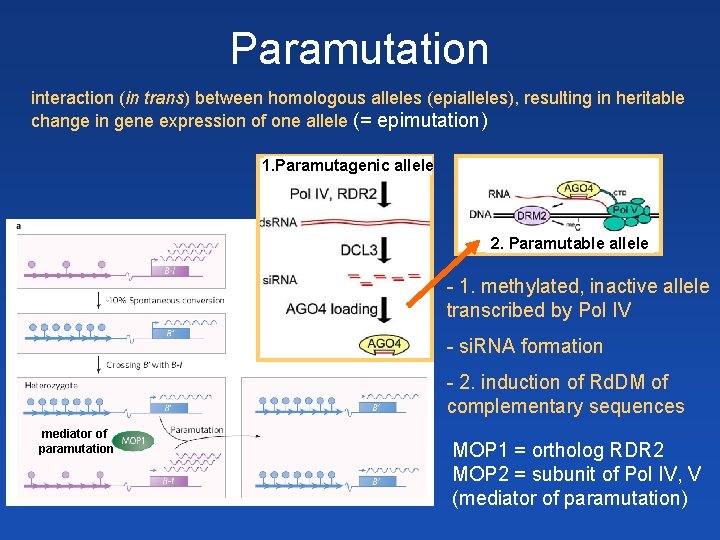
Paramutation interaction (in trans) between homologous alleles (epialleles), resulting in heritable change in gene expression of one allele (= epimutation) 1. Paramutagenic allele 2. Paramutable allele - 1. methylated, inactive allele transcribed by Pol IV - si. RNA formation - 2. induction of Rd. DM of complementary sequences mediator of paramutation MOP 1 = ortholog RDR 2 MOP 2 = subunit of Pol IV, V (mediator of paramutation)

Antiviral systemic resistance - newly developed leaves resistant against infection (1928) Mechanism?

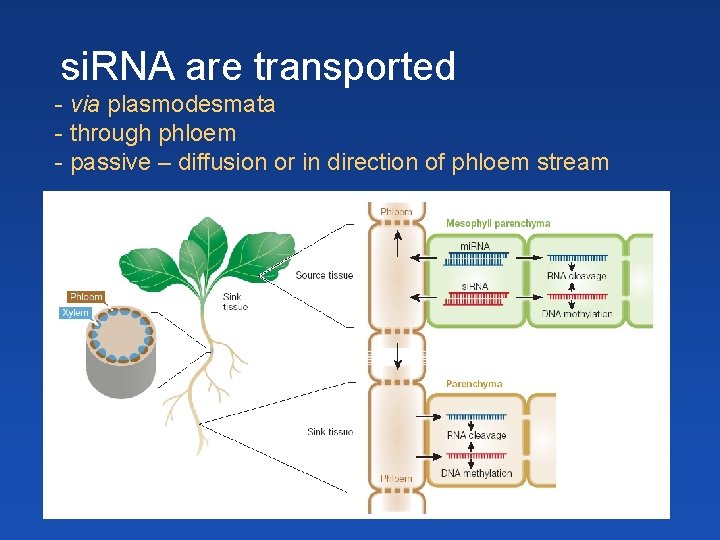
si. RNA are transported - via plasmodesmata - through phloem - passive – diffusion or in direction of phloem stream

Spreading of silencing - GFP silencing by si. RNAs (coming through vasculature) green = GFP fluorescence, red = chlorophyll autofluorescence

Function of RNAi and systemic spreading of s. RNAs - antiviral defense – preventing of spreading of infection and new infection - TE defense – keeping genome integrity, heterochromatin structure - regulation of development – practically all phases - regulation of nutrient uptake – e. g. phosphorus - stress response (even heritable changes – see bellow) - epigenetic modulation of genetic information in meristems (heritable changes) - environmental adaptations

Epigenetic regulation of gene expression in plant development - regulation mainly by mi. RNA (21 nt) (hundreds of MIR genes) - sometimes mediated with ta-si. RNA (non-coding TAS transcript cleaved by RISC with mi. RNA, RDR 6, …) - frequently transported between neighbouring cells (non-cell autonomous) - targets – genes for regulatory proteins e. g. transcription factors, components of ubiquitination pathway, … What is the output from Promoter : : Reporter gene fusion experiments?

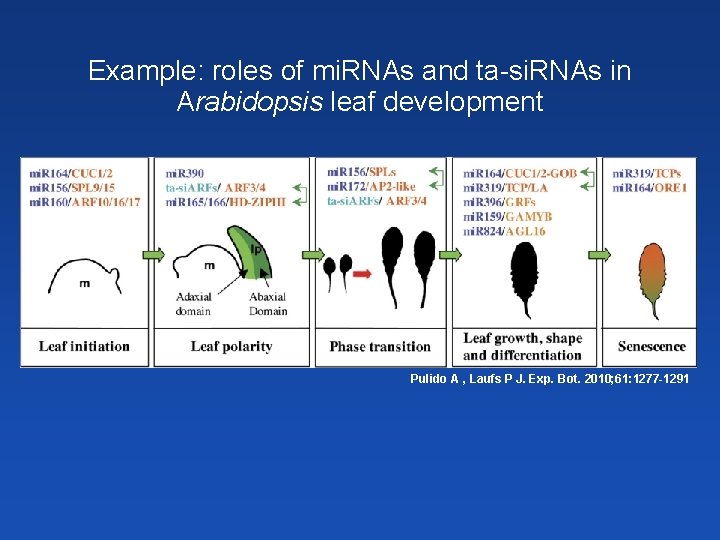
Example: roles of mi. RNAs and ta-si. RNAs in Arabidopsis leaf development Pulido A , Laufs P J. Exp. Bot. 2010; 61: 1277 -1291

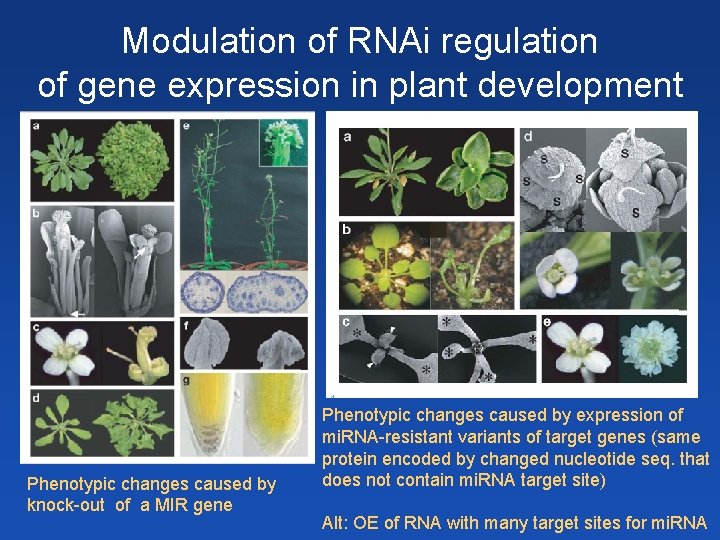
Modulation of RNAi regulation of gene expression in plant development Phenotypic changes caused by knock-out of a MIR gene Phenotypic changes caused by expression of mi. RNA-resistant variants of target genes (same protein encoded by changed nucleotide seq. that does not contain mi. RNA target site) Alt: OE of RNA with many target sites for mi. RNA

Parental imprinting - varying epigenetic modification of alleles inherited from father and mather (parental imprint) – it results in differencial expression of these alleles in zygote = alleles of some genes are methylated in one or the other gamet - evolved in mammals and angiosperm plants – WHY? - hemizygote“ state can serve to ensure reasonable feeding of embryo (in mather body) – prevents evolution of paternal alleles causing excessive embryo feeding

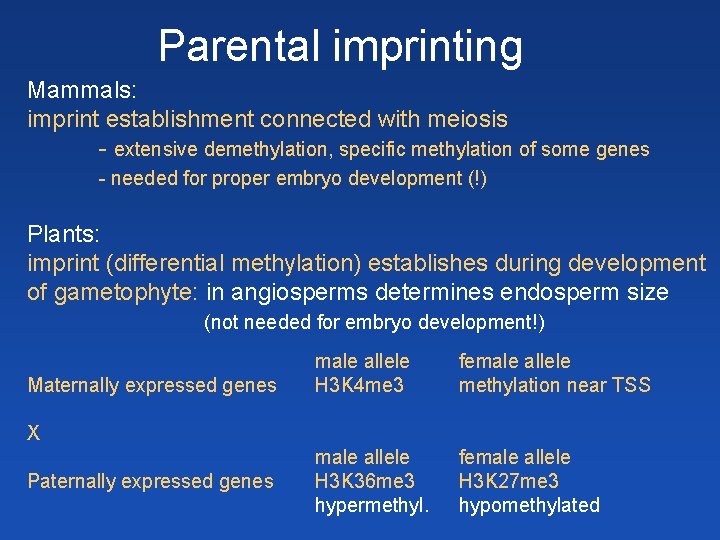
Parental imprinting Mammals: imprint establishment connected with meiosis - extensive demethylation, specific methylation of some genes - needed for proper embryo development (!) Plants: imprint (differential methylation) establishes during development of gametophyte: in angiosperms determines endosperm size (not needed for embryo development!) Maternally expressed genes male allele H 3 K 4 me 3 female allele methylation near TSS male allele H 3 K 36 me 3 hypermethyl. female allele H 3 K 27 me 3 hypomethylated X Paternally expressed genes

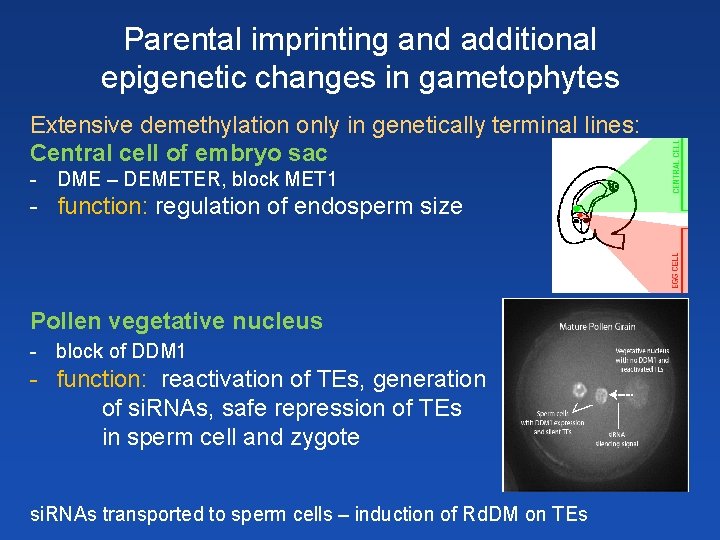
Parental imprinting and additional epigenetic changes in gametophytes Extensive demethylation only in genetically terminal lines: Central cell of embryo sac - DME – DEMETER, block MET 1 - function: regulation of endosperm size Pollen vegetative nucleus - block of DDM 1 - function: reactivation of TEs, generation of si. RNAs, safe repression of TEs in sperm cell and zygote si. RNAs transported to sperm cells – induction of Rd. DM on TEs


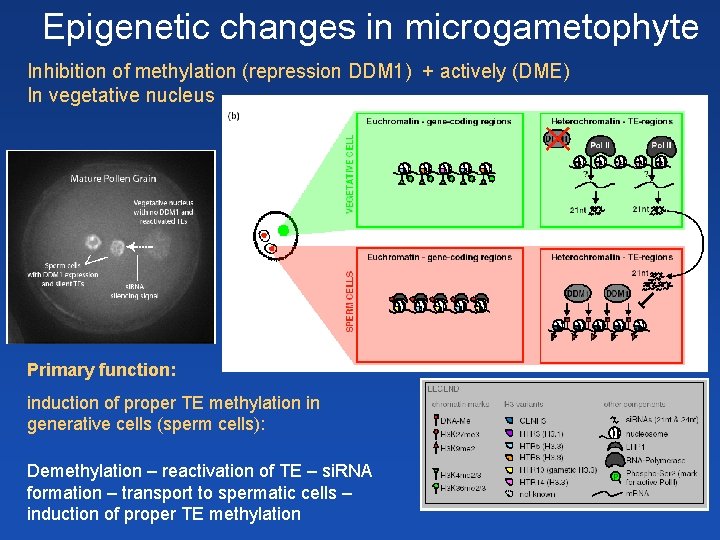
Epigenetic changes in microgametophyte Inhibition of methylation (repression DDM 1) + actively (DME) In vegetative nucleus Primary function: induction of proper TE methylation in generative cells (sperm cells): Demethylation – reactivation of TE – si. RNA formation – transport to spermatic cells – induction of proper TE methylation

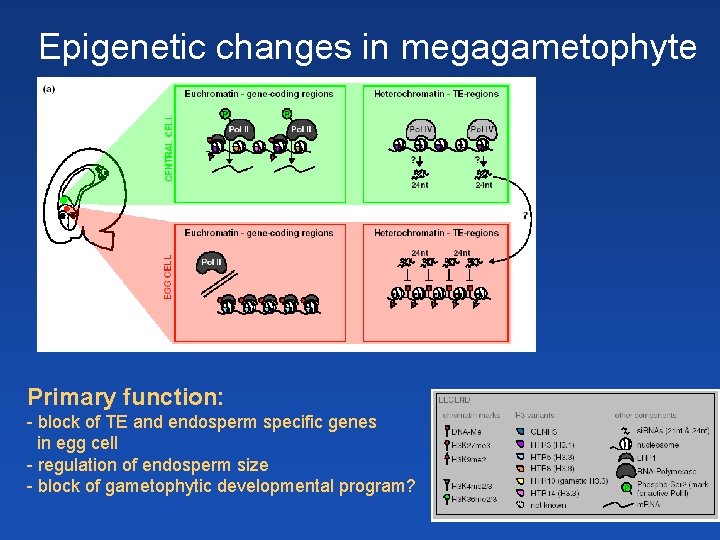
Epigenetic changes in megagametophyte Primary function: - block of TE and endosperm specific genes in egg cell - regulation of endosperm size - block of gametophytic developmental program?

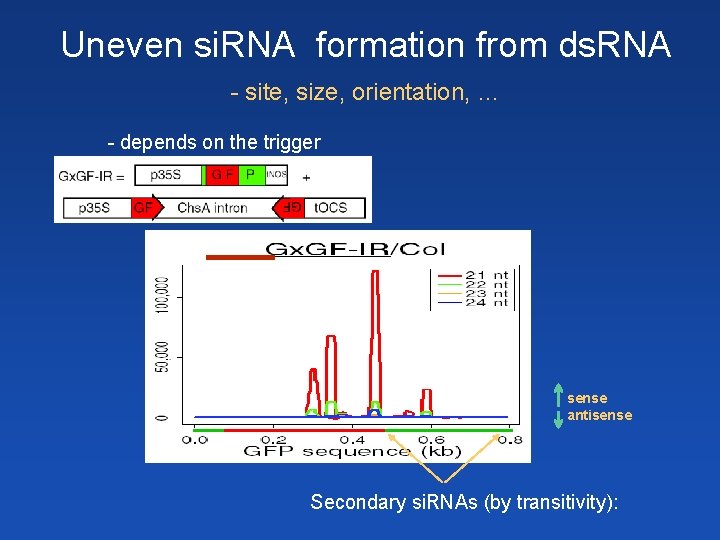
Uneven si. RNA formation from ds. RNA - site, size, orientation, … - depends on the trigger sense antisense Secondary si. RNAs (by transitivity):

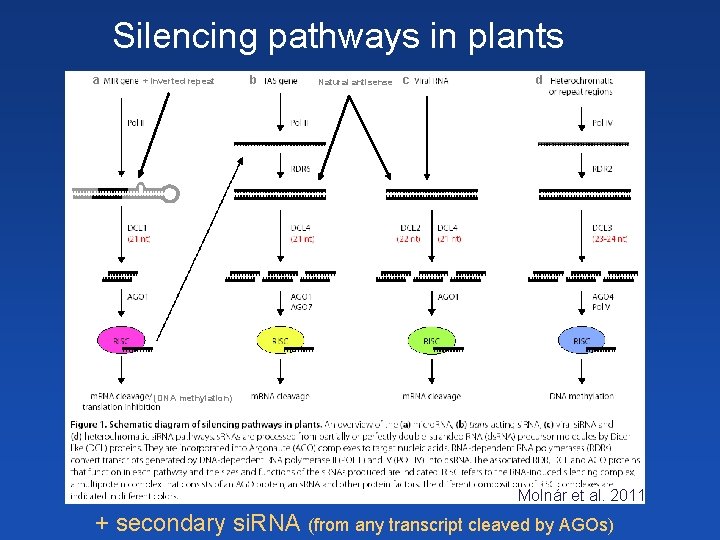
Silencing pathways in plants a + Inverted repeat b Natural antisense c d (DNA methylation) Molnár et al. 2011 + secondary si. RNA (from any transcript cleaved by AGOs)