Chromatin modulation and DNA damage response A De












- Slides: 20

Chromatin modulation and DNA damage response A- De visu: chromatin decondensation in mammalian cells B- Chromatin modulation 1 - Covalent histones modifications 2 - Histone exchange 3 - ATP-dependent chromatin remodeling C- Models of dynamic chromatin remodeling at sites of DNA damage

Nucleosome and chromatin structure Costelloe et al. Exp. Cell. Res. 2006 312: 2677 -2686.

chromatin decondensation in mammalian cells Kruhlak et al. JCB 2006 172: 823 -834

GR-PAGFP H 2 B-PAGFP H 3 -PAGFP GR-PAGFP Kruhlak et al. JCB 2006 172: 823 -834

Covalent histones modifications in DNA repair - Histones are rich in polar aminoacids: Lysine, Arginine, Serine, Threonine -> located on the exposed exterior surface of histone octamers. - Differences in electrostatic properties between the modified and unmodified forms of these residues can significantly affect the interaction between histones and DNA or other proteins. - Four modifications: Phosphorylation (kinase/phosphatase), acetylation (HAT/HDAC), methylation (methyl transferase/demethylase), ubiquitylation (ubiquitin ligase/deubiquitinase)

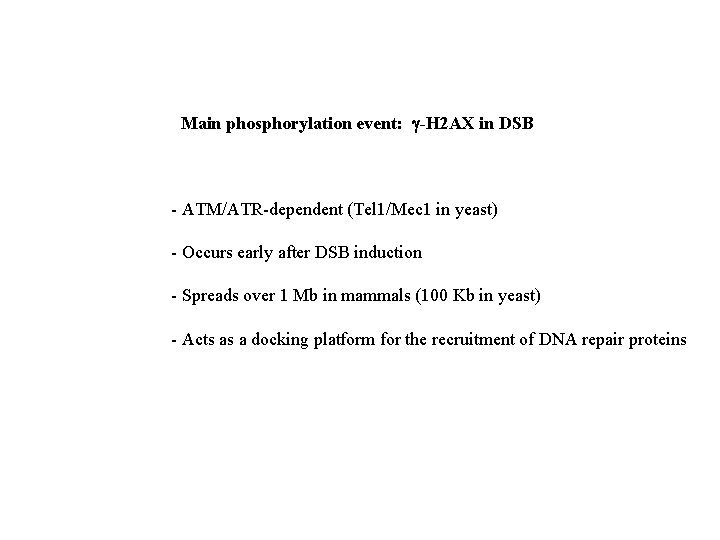
Main phosphorylation event: g-H 2 AX in DSB - ATM/ATR-dependent (Tel 1/Mec 1 in yeast) - Occurs early after DSB induction - Spreads over 1 Mb in mammals (100 Kb in yeast) - Acts as a docking platform for the recruitment of DNA repair proteins

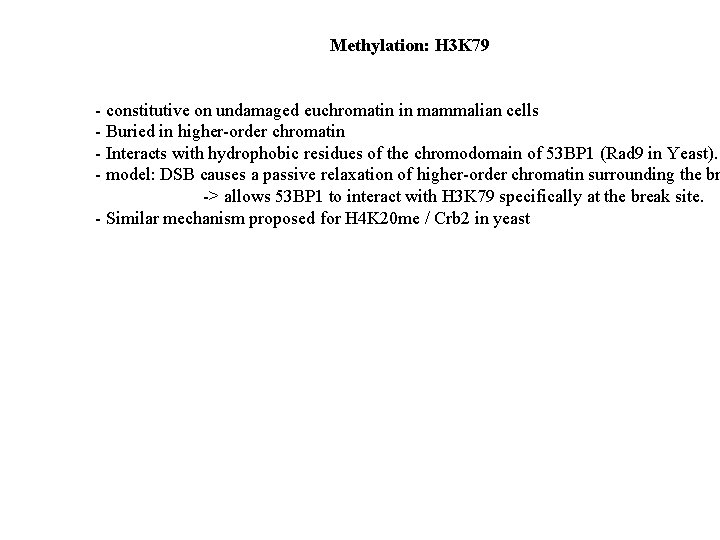
Methylation: H 3 K 79 - constitutive on undamaged euchromatin in mammalian cells - Buried in higher-order chromatin - Interacts with hydrophobic residues of the chromodomain of 53 BP 1 (Rad 9 in Yeast). - model: DSB causes a passive relaxation of higher-order chromatin surrounding the br -> allows 53 BP 1 to interact with H 3 K 79 specifically at the break site. - Similar mechanism proposed for H 4 K 20 me / Crb 2 in yeast

Acetylation - Acetylation of lysines in the N-terminal tails of H 3 and H 4 removes the positive charges on the side chain -> destabilizes higher-order chromatin structure. - Acetylated lysines can be recognized and recruit bromodomain-containing proteins such as Tip 60 - Regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs)

Composition of the yeast SWR 1, INO 80, Nu. A 4 and human TIP 60 chromatin remodeling/histone acetyltransferase complexes involved in DNA repair (Chromatin remodeling) (HAT) (Chromatin remodeling/HAT) : Tip 60 Yeast SWR 1, INO 80 and Nu. A 4 complexes share many subunits with human TIP 60. It is postulated that TIP 60 is a hybrid of at least two and possibly all three yeast complexes.

Proposed role for Tip 60 and the Nu. A 4 complex in chromatin remodeling during DNA repair Recruitment of MRX to the DSB Tip 60 -dependent acetylation/ phosphorylation of ATM Phosphorylation of NBS 1, H 2 AX g H 2 AX acts as a docking platform, recruits DDR proteins (eg MDC 1) and spreads the signal further downstream from the DSB Nu. A 4 might recognize g H 2 AX, facilitating Tip 60 acetylation of Histones H 4 and g H 2 AX P 400 might catalyze histone exchange between g H 2 AX and unmodified H 2 AZ. Squatrito et al. , Trends Cell Biol. 2006, 16: 433 -442.

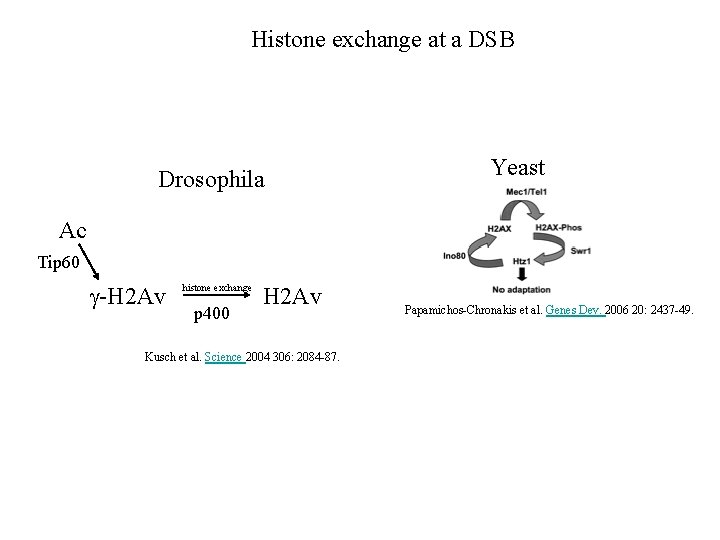
Histone exchange at a DSB Drosophila Yeast Ac Tip 60 g-H 2 Av histone exchange p 400 H 2 Av Kusch et al. Science 2004 306: 2084 -87. Papamichos-Chronakis et al. Genes Dev. 2006 20: 2437 -49.

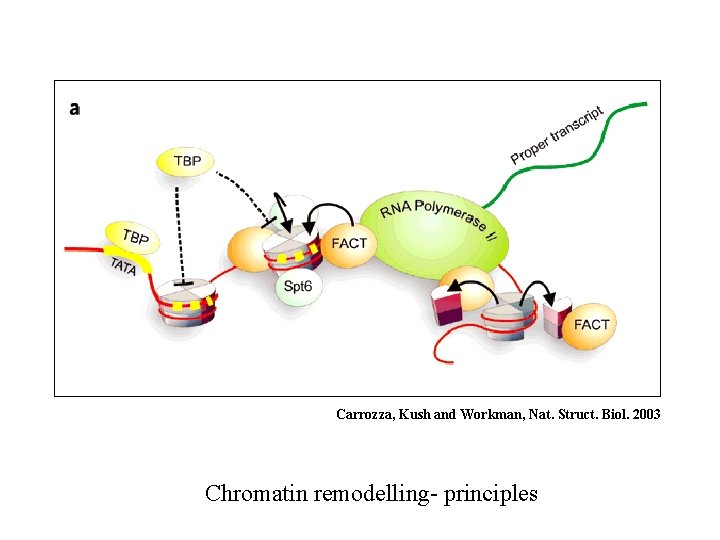
Carrozza, Kush and Workman, Nat. Struct. Biol. 2003 Chromatin remodelling- principles

Composition of the yeast SWR 1, INO 80, Nu. A 4 and human TIP 60 chromatin remodeling/histone acetyltransferase complexes involved in DNA repair (Chromatin remodeling) (HAT) (Chromatin remodeling/HAT) : Tip 60 Yeast SWR 1, INO 80 and Nu. A 4 complexes share many subunits with human TIP 60. It is postulated that TIP 60 is a hybrid of at least two and possibly all three yeast complexes.

Studies in yeast: the MAT/HO system Homologous recombination Southern blot Papamichos-Chronakis et al. Genes Dev. 2006 20: 2437 -49.

NHEJ Papamichos-Chronakis et al. Genes Dev. 2006 20: 2437 -49.

Kb a, A DSB was induced at MAT in strains expressing Flag–H 2 B or Flag–H 3, and Ch. IP was done with antibodies against -H 2 A or Flag. DNA was analysed by real-time PCR using primers corresponding to sequences on the left (- ) or the right (+ ) side of the DSB (0), and results were normalized to the ratio of immunoprecipitation (IP) to input DNA at time 0. Data are the mean s. e. m. b, Nuclei were prepared after DSB induction, and chromatin was digested with MNase and subjected to Southern blot analysis using a probe for MAT DNA. The triangles denote increasing times of MNase digestion. M indicates a 1 -kb DNA ladder. Wt: loss of histones and nucleosome integrity near the DSB Figure 1: Chromatin changes at the MAT DSB. Tsukuda et al. Nature 2005 438: 379 -383.

a, A DSB was induced at MAT, and Ch. IP was done with antibodies against Flag in an mre 11: : Kan-MX strain expressing Flag–H 2 B and an hta 1/hta 2 -S 129* strain expressing Flag –H 3. DNA was analysed by real-time PCR on both the left (- ) and the right (+ ) side of the DSB (0). Data are the mean s. e. m. b, MNase analysis was done on nuclei isolated from the mre 11: : Kan-MX strain by Southern blot analysis as described in Fig. 1 b. Figure 2: MRX is involved in histone loss at the MAT DSB. Tsukuda et al. Nature 2005 438: 379 -383.

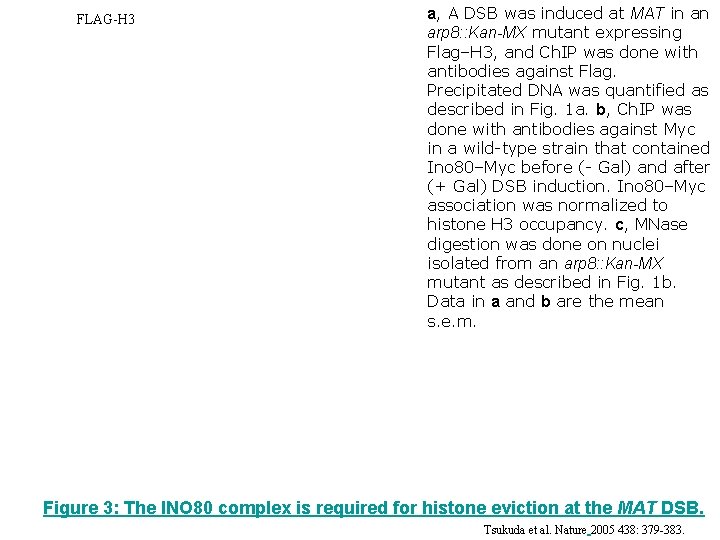
FLAG-H 3 a, A DSB was induced at MAT in an arp 8: : Kan-MX mutant expressing Flag–H 3, and Ch. IP was done with antibodies against Flag. Precipitated DNA was quantified as described in Fig. 1 a. b, Ch. IP was done with antibodies against Myc in a wild-type strain that contained Ino 80–Myc before (- Gal) and after (+ Gal) DSB induction. Ino 80–Myc association was normalized to histone H 3 occupancy. c, MNase digestion was done on nuclei isolated from an arp 8: : Kan-MX mutant as described in Fig. 1 b. Data in a and b are the mean s. e. m. Figure 3: The INO 80 complex is required for histone eviction at the MAT DSB. Tsukuda et al. Nature 2005 438: 379 -383.

Chromatin remodeling factors in non-DSB repair Blue: ATP-dependent chromatin remodelers Histone modifying enzymes: yellow Chromatin assembly factors: magenta Ataian and Krebs, Biochem. Cell. Biol. 2006, 84: 490 -504.

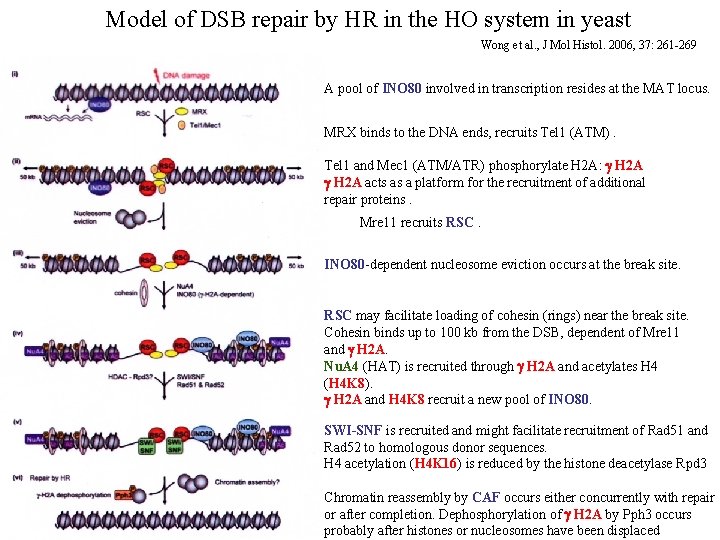
Model of DSB repair by HR in the HO system in yeast Wong et al. , J Mol Histol. 2006, 37: 261 -269 A pool of INO 80 involved in transcription resides at the MAT locus. MRX binds to the DNA ends, recruits Tel 1 (ATM). Tel 1 and Mec 1 (ATM/ATR) phosphorylate H 2 A: g H 2 A acts as a platform for the recruitment of additional repair proteins. Mre 11 recruits RSC. INO 80 -dependent nucleosome eviction occurs at the break site. RSC may facilitate loading of cohesin (rings) near the break site. Cohesin binds up to 100 kb from the DSB, dependent of Mre 11 and g H 2 A. Nu. A 4 (HAT) is recruited through g H 2 A and acetylates H 4 (H 4 K 8). g H 2 A and H 4 K 8 recruit a new pool of INO 80. SWI-SNF is recruited and might facilitate recruitment of Rad 51 and Rad 52 to homologous donor sequences. H 4 acetylation (H 4 K 16) is reduced by the histone deacetylase Rpd 3 Chromatin reassembly by CAF occurs either concurrently with repair or after completion. Dephosphorylation of g H 2 A by Pph 3 occurs probably after histones or nucleosomes have been displaced