Reactive Molecular Dynamics Progress Report Mar 7 th


- Slides: 41

Reactive Molecular Dynamics Progress Report Mar. 7 th 2008 Andrei. Smirnov, Jaggu Nanduri, and R. A. Carreno-Chavez Jaggu Nanduri

Objectives • To develop the capabilities of simulating 107 molecules and more on a workstation.

Progress • Segmentation algorithm implemented. • Chemical reactions between species implemented • Surface reactions between species and active surface sites implemented • Simulations of several million molecules on a single workstation have been conducted.

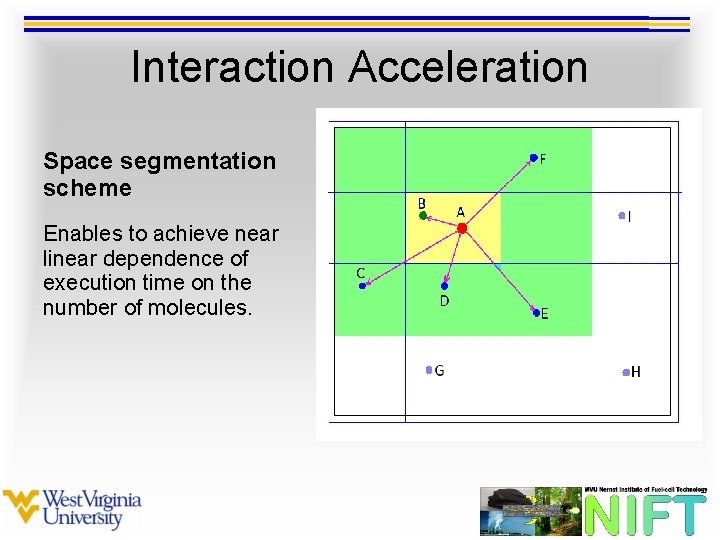
Interaction Acceleration Space segmentation scheme Enables to achieve near linear dependence of execution time on the number of molecules.

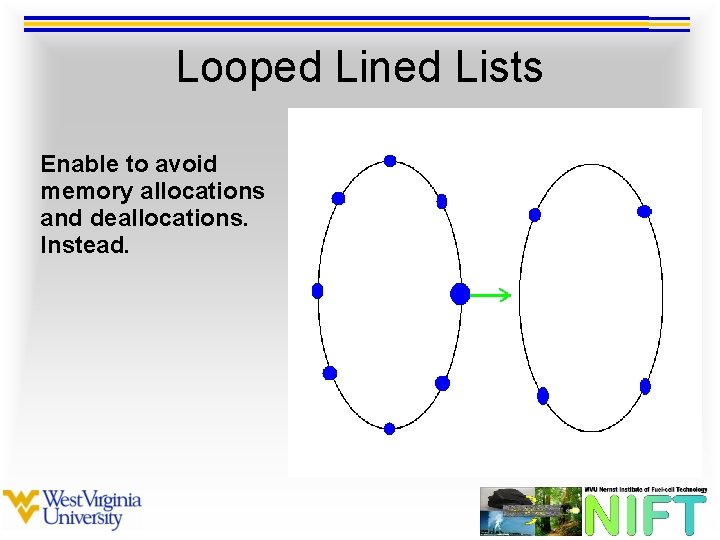
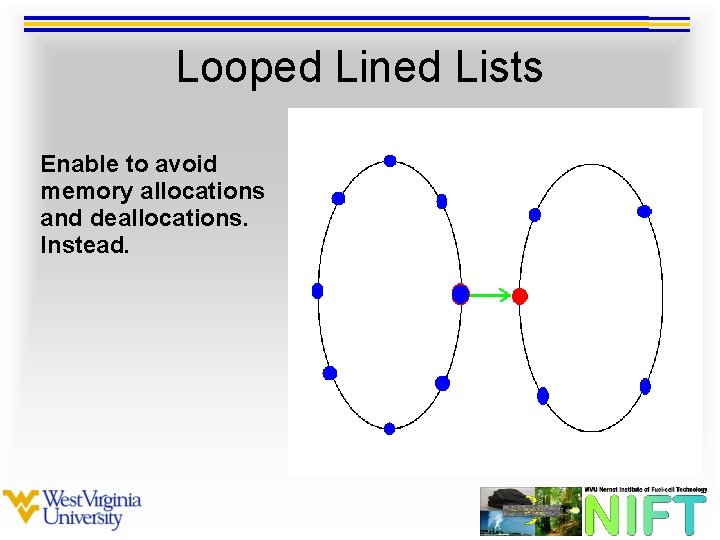
Looped Lined Lists Enable to avoid memory allocations and deallocations. Instead.

Looped Lined Lists Enable to avoid memory allocations and deallocations. Instead.

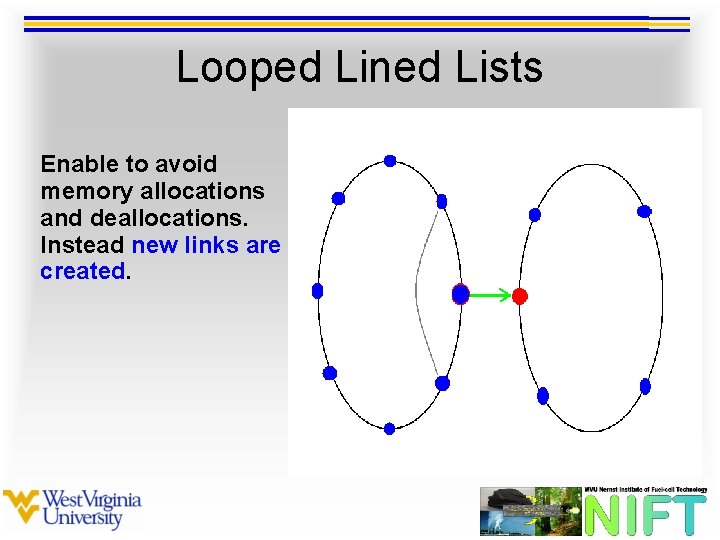
Looped Lined Lists Enable to avoid memory allocations and deallocations. Instead new links are created.

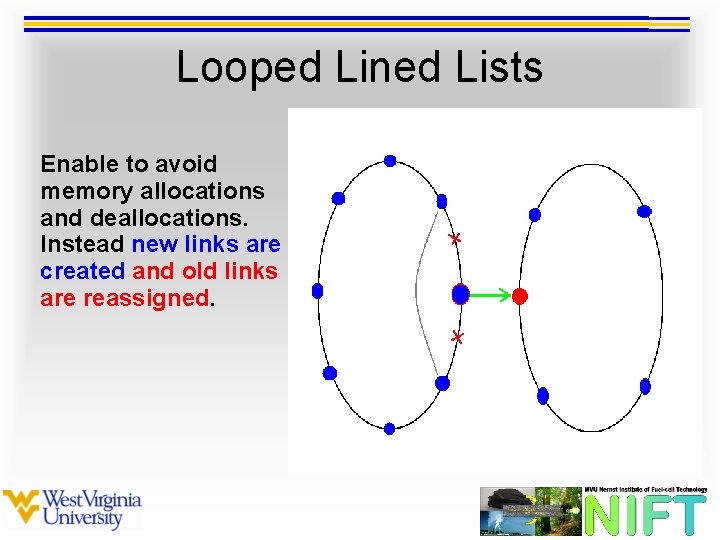
Looped Lined Lists Enable to avoid memory allocations and deallocations. Instead new links are created and old links are reassigned.

Looped Lined Lists Enable to avoid memory allocations and deallocations. Instead new links are created and old links are reassigned.

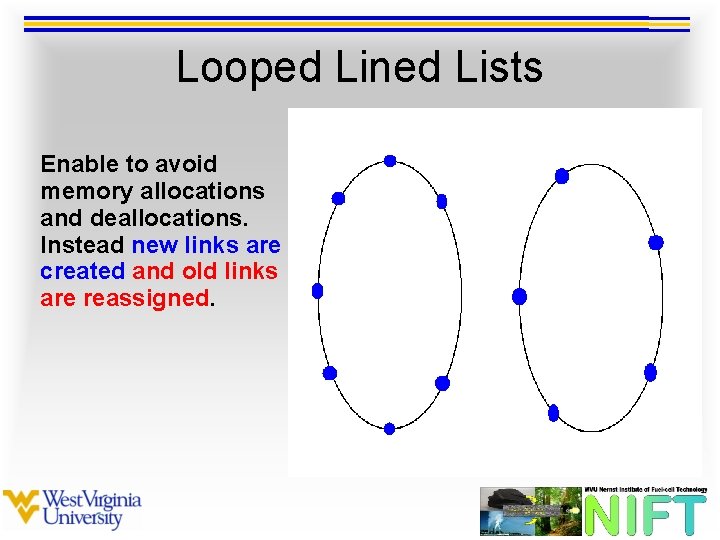
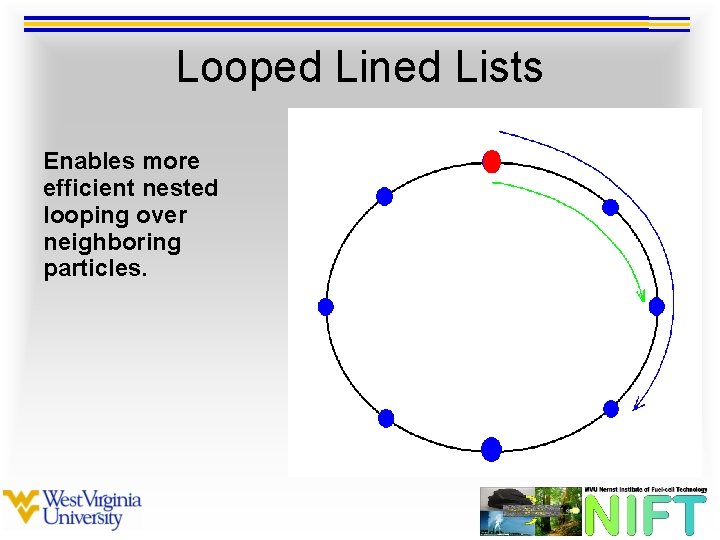
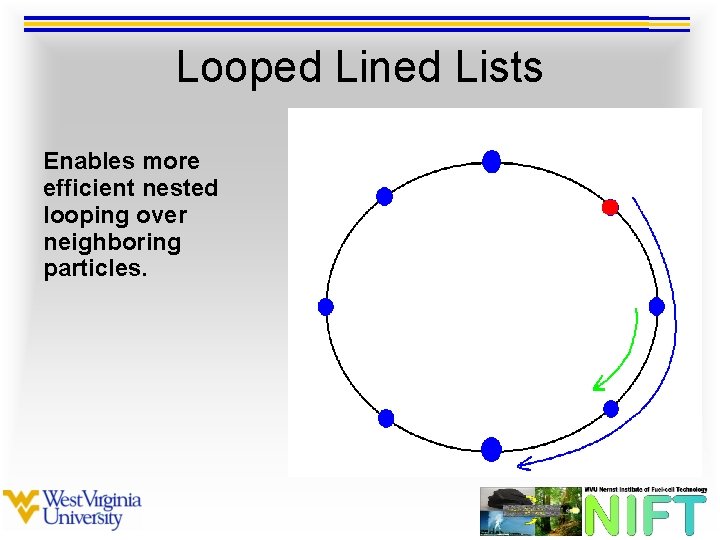
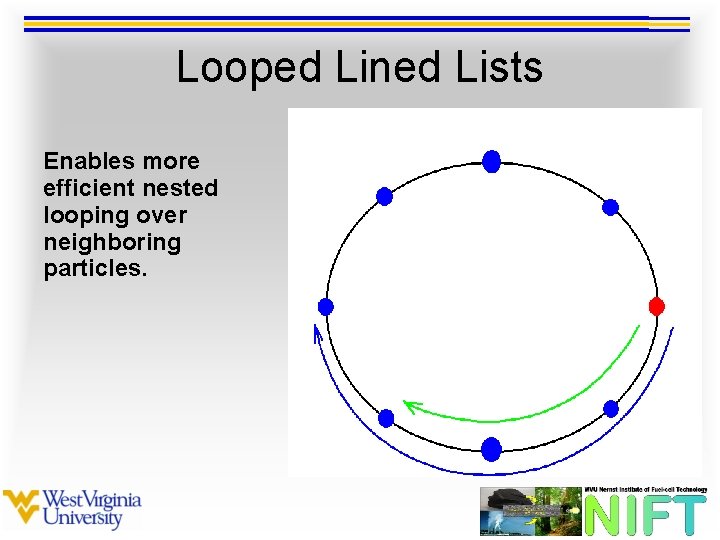
Looped Lined Lists Enables more efficient nested looping over neighboring particles.

Looped Lined Lists Enables more efficient nested looping over neighboring particles.

Looped Lined Lists Enables more efficient nested looping over neighboring particles.

O 2 + H(s) Time: 2. 00 e-01 ns, Molecules: 117946, Temperature: 300 K Species: O 2: 100. 00%

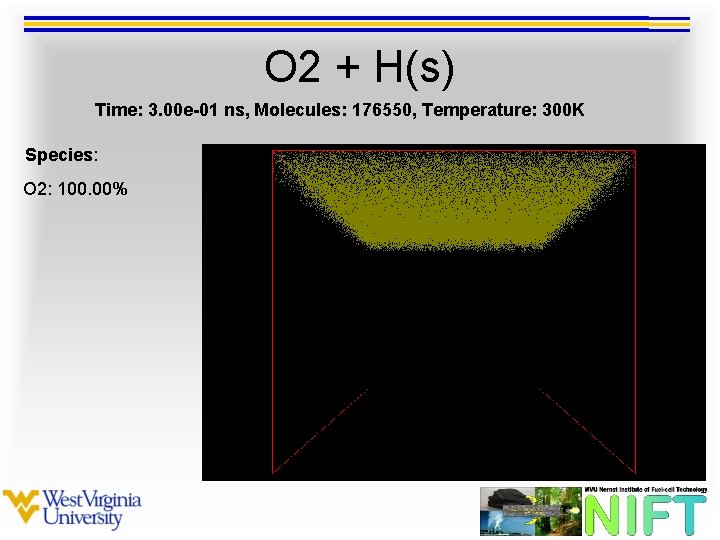
O 2 + H(s) Time: 3. 00 e-01 ns, Molecules: 176550, Temperature: 300 K Species: O 2: 100. 00%

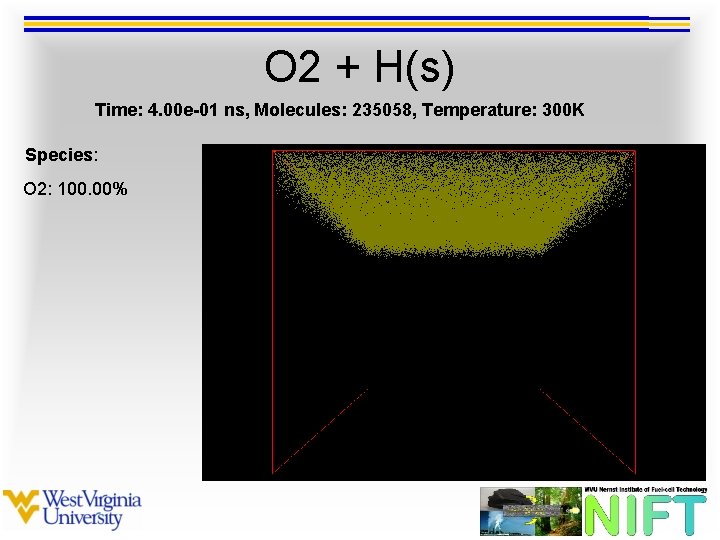
O 2 + H(s) Time: 4. 00 e-01 ns, Molecules: 235058, Temperature: 300 K Species: O 2: 100. 00%

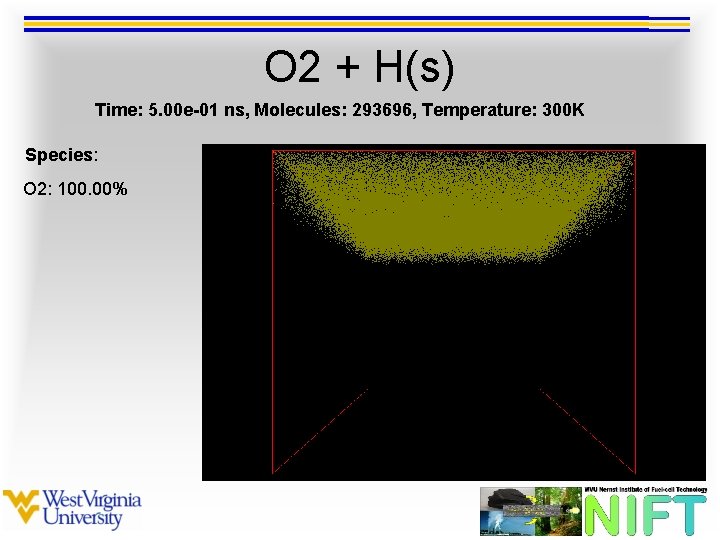
O 2 + H(s) Time: 5. 00 e-01 ns, Molecules: 293696, Temperature: 300 K Species: O 2: 100. 00%

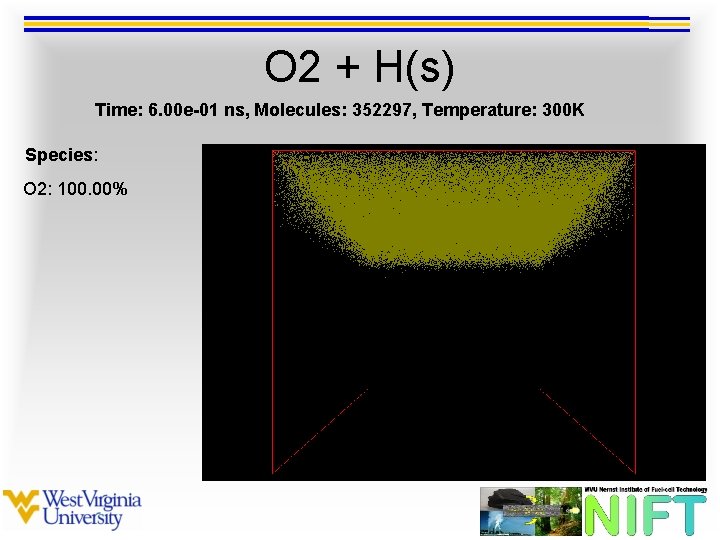
O 2 + H(s) Time: 6. 00 e-01 ns, Molecules: 352297, Temperature: 300 K Species: O 2: 100. 00%

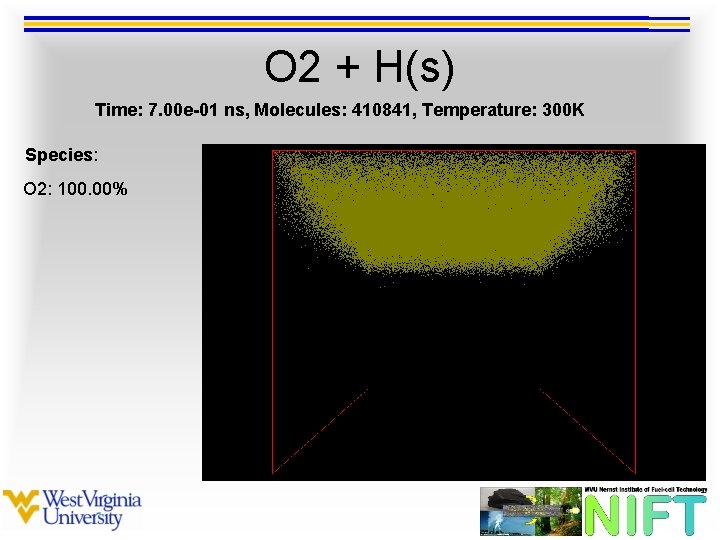
O 2 + H(s) Time: 7. 00 e-01 ns, Molecules: 410841, Temperature: 300 K Species: O 2: 100. 00%

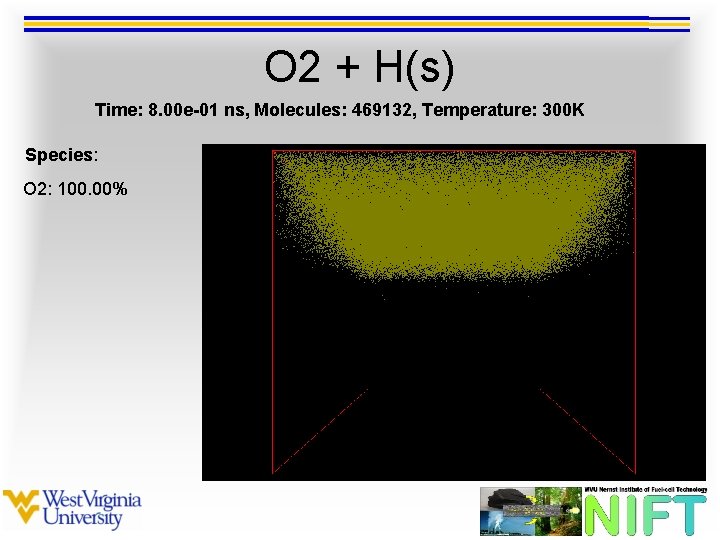
O 2 + H(s) Time: 8. 00 e-01 ns, Molecules: 469132, Temperature: 300 K Species: O 2: 100. 00%

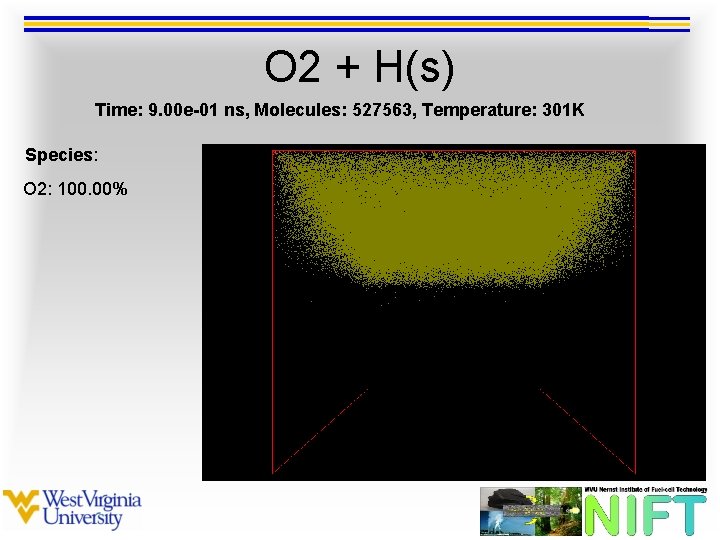
O 2 + H(s) Time: 9. 00 e-01 ns, Molecules: 527563, Temperature: 301 K Species: O 2: 100. 00%

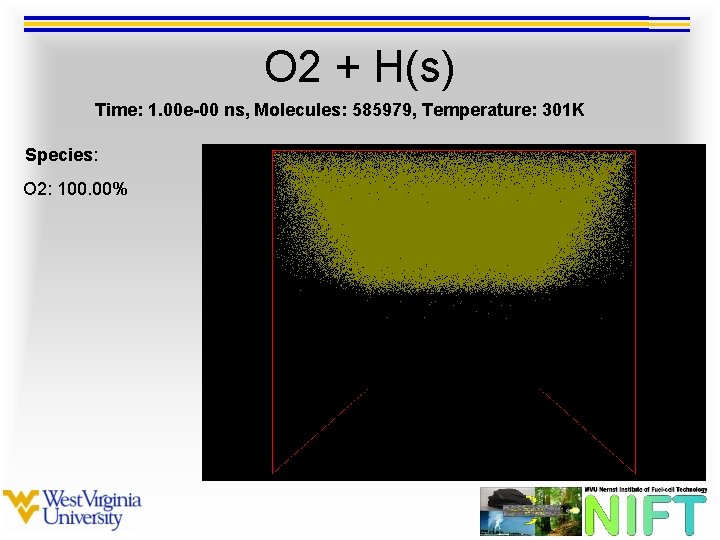
O 2 + H(s) Time: 1. 00 e-00 ns, Molecules: 585979, Temperature: 301 K Species: O 2: 100. 00%

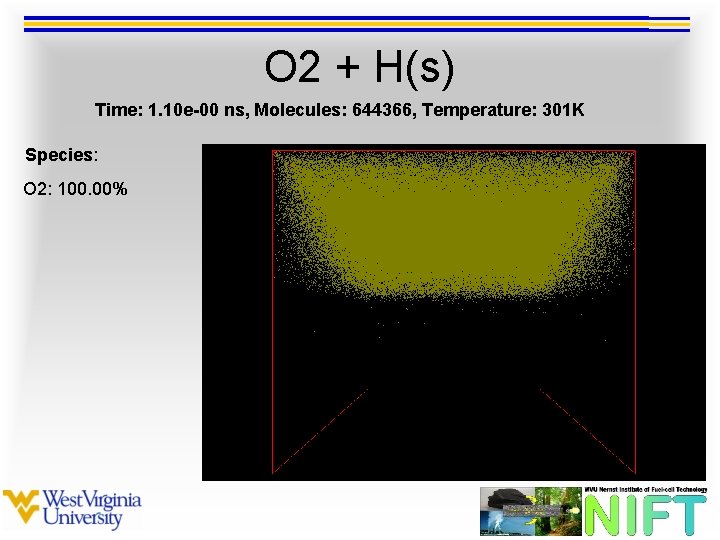
O 2 + H(s) Time: 1. 10 e-00 ns, Molecules: 644366, Temperature: 301 K Species: O 2: 100. 00%

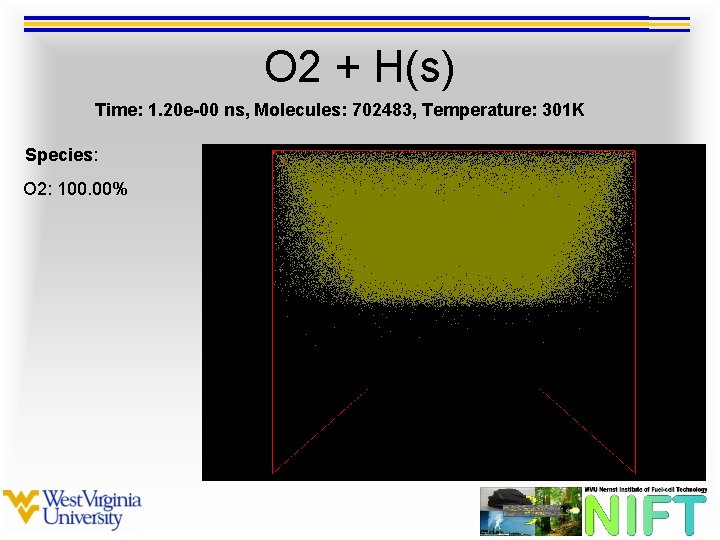
O 2 + H(s) Time: 1. 20 e-00 ns, Molecules: 702483, Temperature: 301 K Species: O 2: 100. 00%

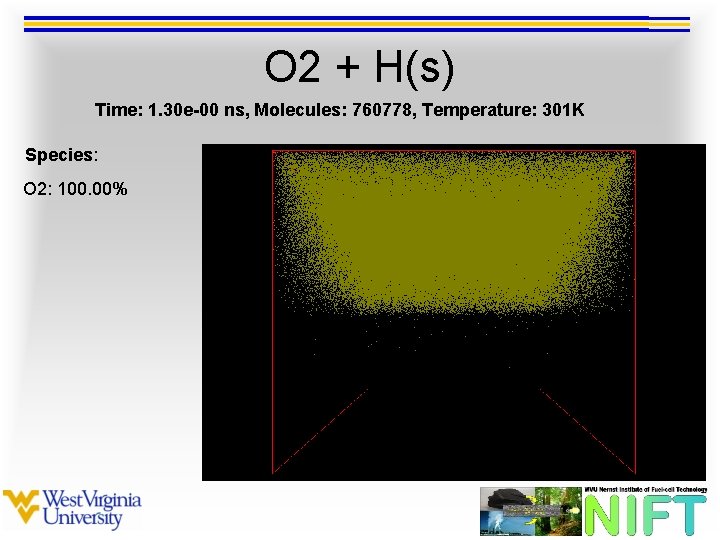
O 2 + H(s) Time: 1. 30 e-00 ns, Molecules: 760778, Temperature: 301 K Species: O 2: 100. 00%

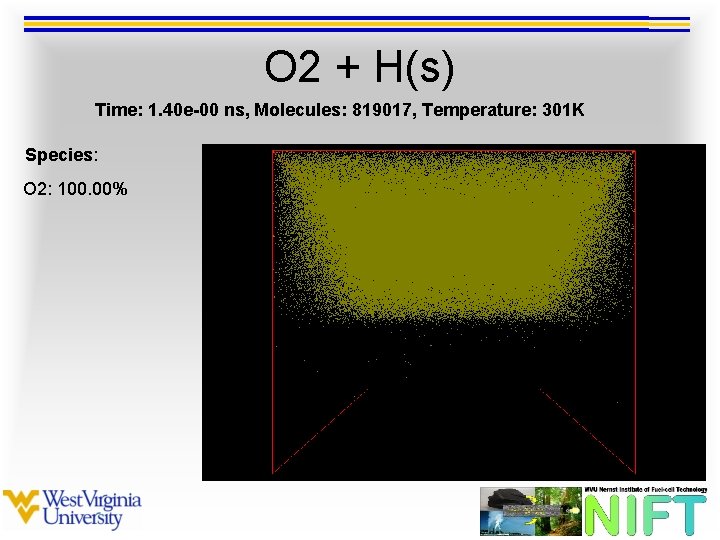
O 2 + H(s) Time: 1. 40 e-00 ns, Molecules: 819017, Temperature: 301 K Species: O 2: 100. 00%

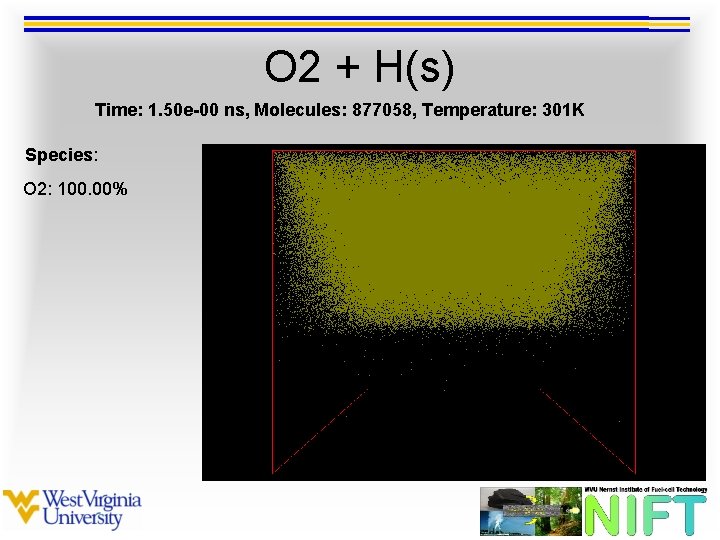
O 2 + H(s) Time: 1. 50 e-00 ns, Molecules: 877058, Temperature: 301 K Species: O 2: 100. 00%

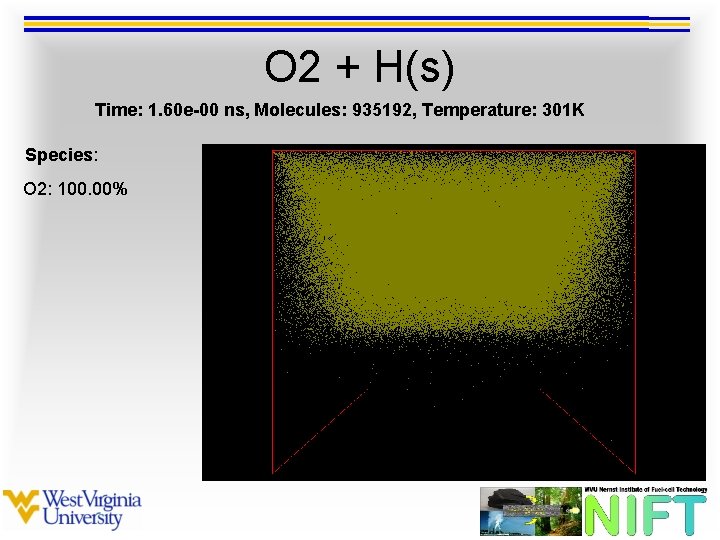
O 2 + H(s) Time: 1. 60 e-00 ns, Molecules: 935192, Temperature: 301 K Species: O 2: 100. 00%

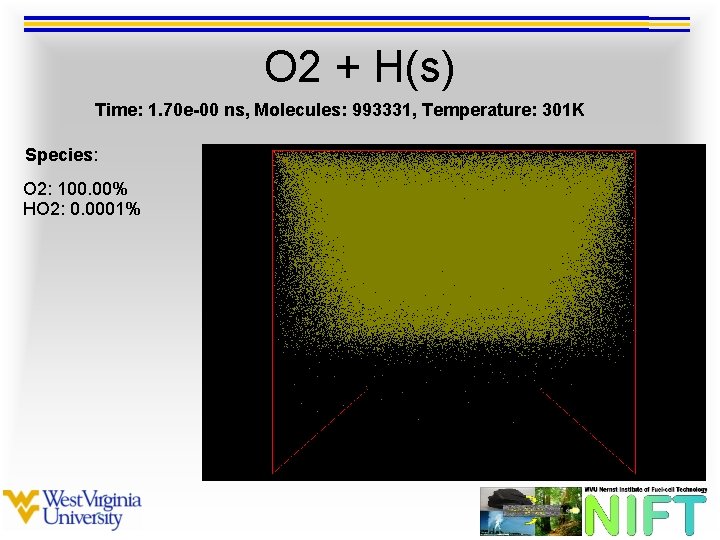
O 2 + H(s) Time: 1. 70 e-00 ns, Molecules: 993331, Temperature: 301 K Species: O 2: 100. 00% HO 2: 0. 0001%

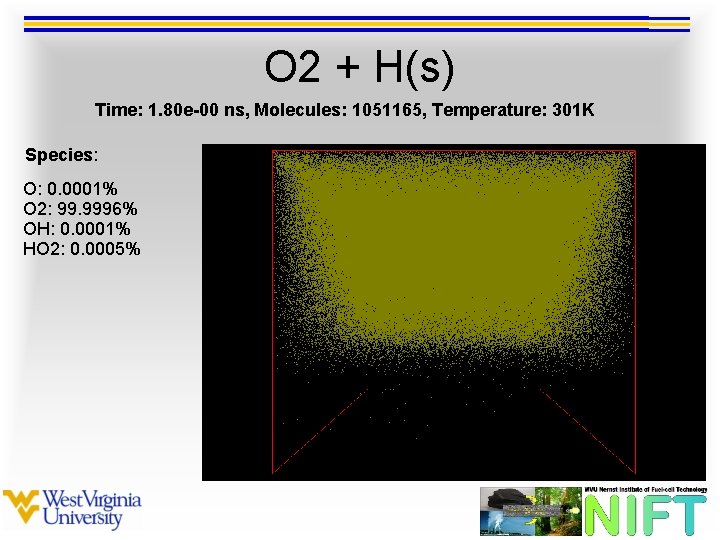
O 2 + H(s) Time: 1. 80 e-00 ns, Molecules: 1051165, Temperature: 301 K Species: O: 0. 0001% O 2: 99. 9996% OH: 0. 0001% HO 2: 0. 0005%

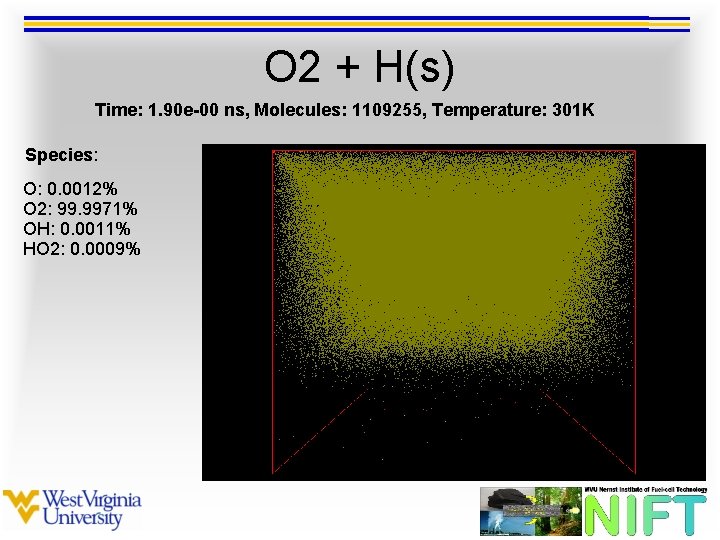
O 2 + H(s) Time: 1. 90 e-00 ns, Molecules: 1109255, Temperature: 301 K Species: O: 0. 0012% O 2: 99. 9971% OH: 0. 0011% HO 2: 0. 0009%

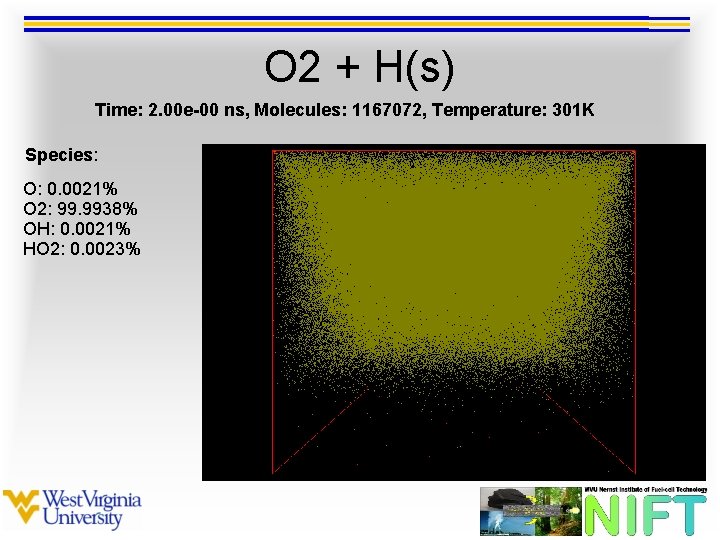
O 2 + H(s) Time: 2. 00 e-00 ns, Molecules: 1167072, Temperature: 301 K Species: O: 0. 0021% O 2: 99. 9938% OH: 0. 0021% HO 2: 0. 0023%

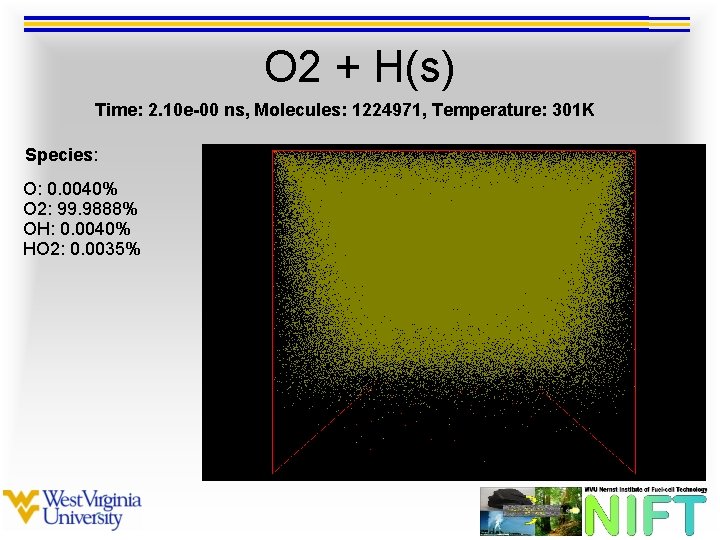
O 2 + H(s) Time: 2. 10 e-00 ns, Molecules: 1224971, Temperature: 301 K Species: O: 0. 0040% O 2: 99. 9888% OH: 0. 0040% HO 2: 0. 0035%

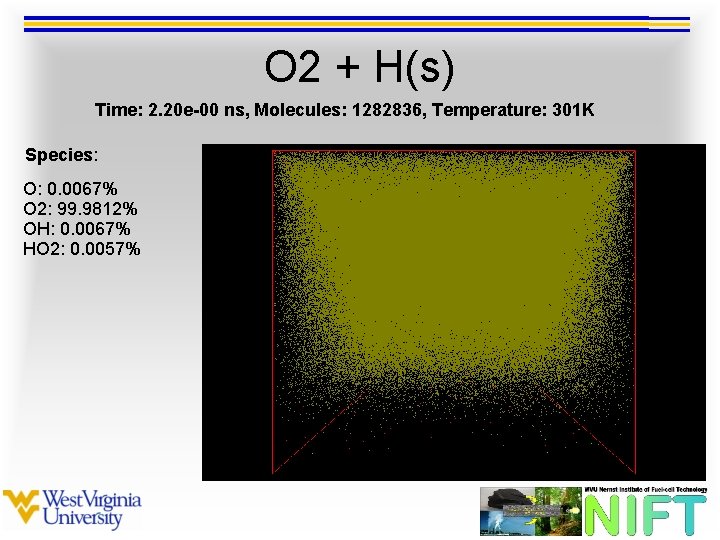
O 2 + H(s) Time: 2. 20 e-00 ns, Molecules: 1282836, Temperature: 301 K Species: O: 0. 0067% O 2: 99. 9812% OH: 0. 0067% HO 2: 0. 0057%

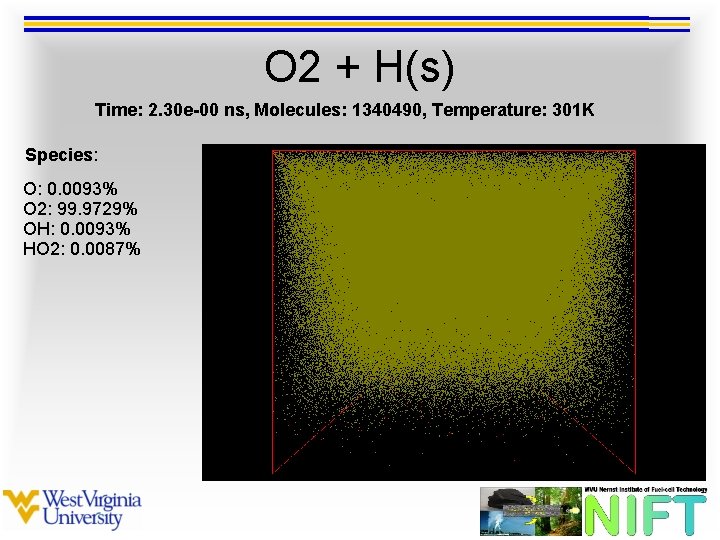
O 2 + H(s) Time: 2. 30 e-00 ns, Molecules: 1340490, Temperature: 301 K Species: O: 0. 0093% O 2: 99. 9729% OH: 0. 0093% HO 2: 0. 0087%

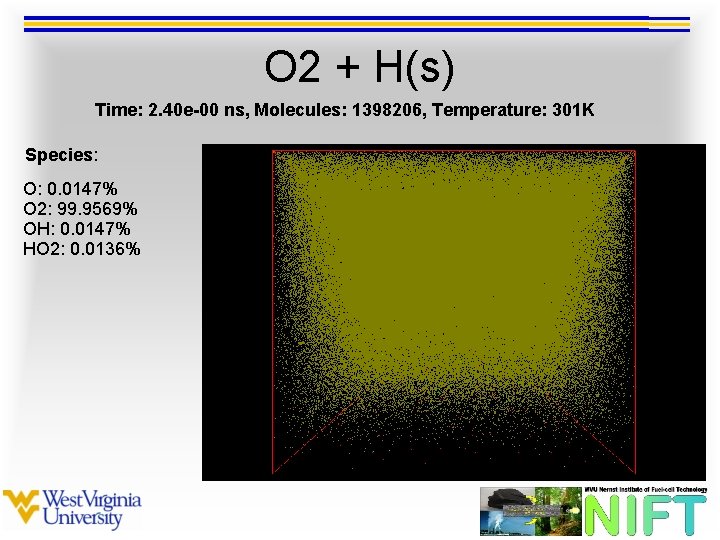
O 2 + H(s) Time: 2. 40 e-00 ns, Molecules: 1398206, Temperature: 301 K Species: O: 0. 0147% O 2: 99. 9569% OH: 0. 0147% HO 2: 0. 0136%

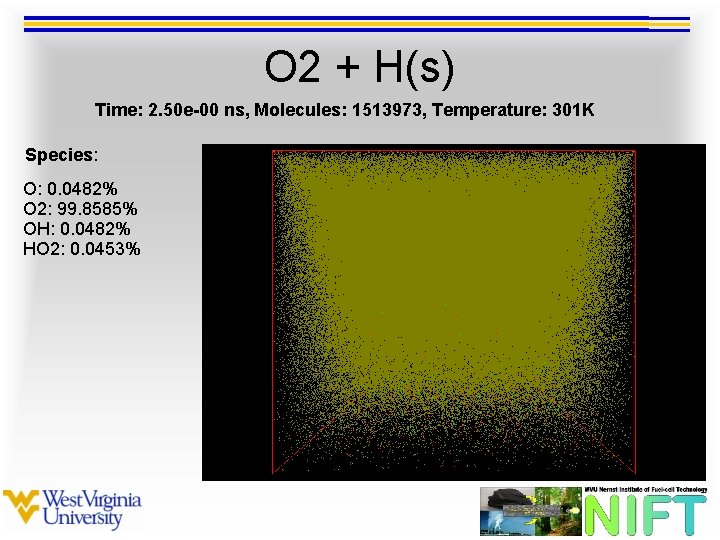
O 2 + H(s) Time: 2. 50 e-00 ns, Molecules: 1513973, Temperature: 301 K Species: O: 0. 0482% O 2: 99. 8585% OH: 0. 0482% HO 2: 0. 0453%

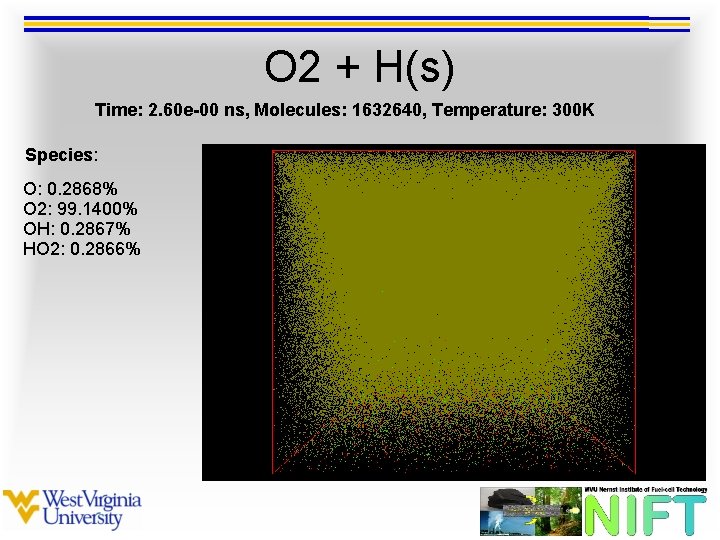
O 2 + H(s) Time: 2. 60 e-00 ns, Molecules: 1632640, Temperature: 300 K Species: O: 0. 2868% O 2: 99. 1400% OH: 0. 2867% HO 2: 0. 2866%

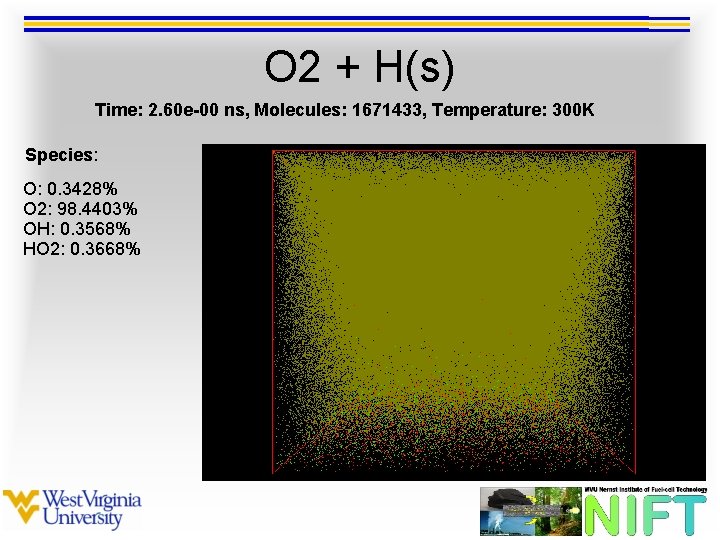
O 2 + H(s) Time: 2. 60 e-00 ns, Molecules: 1671433, Temperature: 300 K Species: O: 0. 3428% O 2: 98. 4403% OH: 0. 3568% HO 2: 0. 3668%

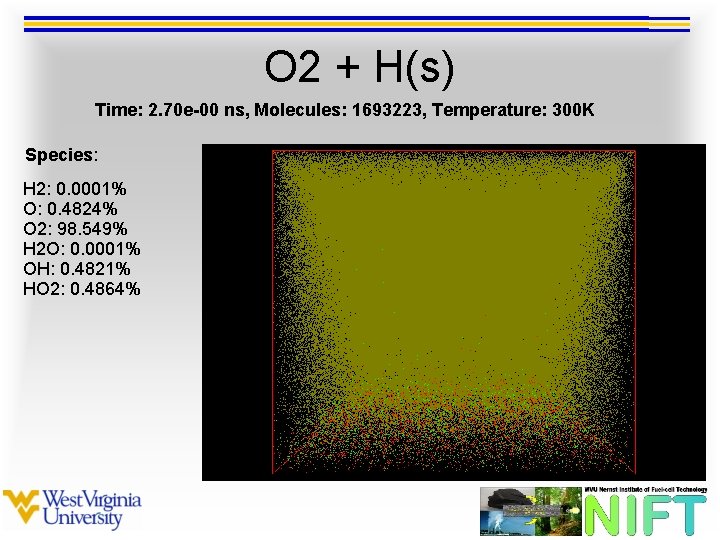
O 2 + H(s) Time: 2. 70 e-00 ns, Molecules: 1693223, Temperature: 300 K Species: H 2: 0. 0001% O: 0. 4824% O 2: 98. 549% H 2 O: 0. 0001% OH: 0. 4821% HO 2: 0. 4864%

Chemical Reactions Bulk reactions Surface reactions

Conclusions • The efficiency bursting measures are working. • Tens of millions of particles can be accommodated on a single workstation. • Time of execution becomes one day for 10 M particles and 1000 time-steps. • Effect of interaction cutoff should be investigated.