Picornaviridae Molecular Virology Plus Strand RNA Virus Families


- Slides: 38

Picornaviridae Molecular Virology

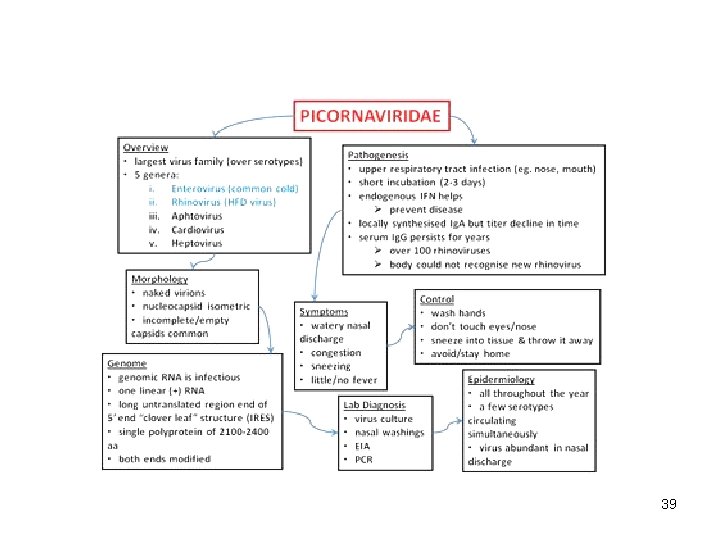
Plus Strand RNA Virus Families PICORNAVIRIDAE • More than 200 viruses prevalent world-wide. • cause many serious diseases of animals and man. • Foot and mouth virus first animal virus described (1898). • Poliovirus is an important model: - first virus purified and crystallized. - first inactivated vaccine used (Salk 1950’s). - first picornavirus to be sequenced. - first infectious c. DNA clone of an animal virus. - first picornavirus structure to be solved. 2

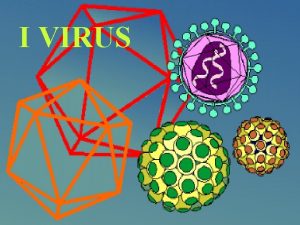
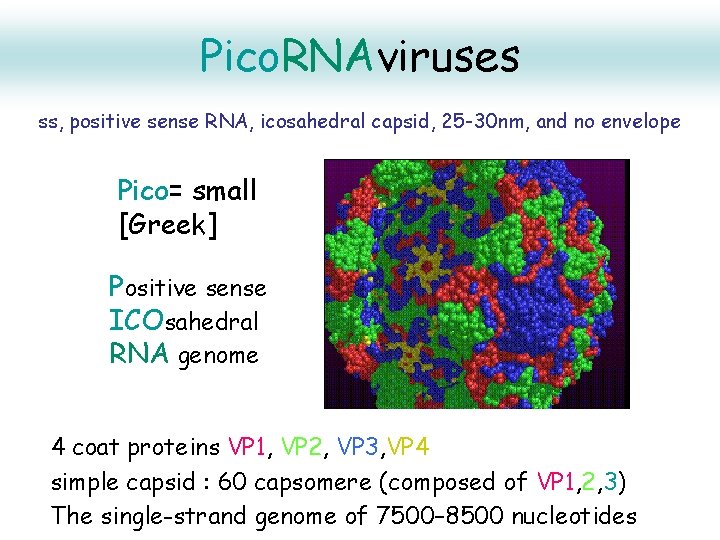
Pico. RNAviruses ss, positive sense RNA, icosahedral capsid, 25 -30 nm, and no envelope Pico= small [Greek] Positive sense ICOsahedral RNA genome 4 coat proteins VP 1, VP 2, VP 3, VP 4 simple capsid : 60 capsomere (composed of VP 1, 2, 3) The single-strand genome of 7500– 8500 nucleotides

Properties • Picornaviruses are among the smallest pathogens of vertebrates and are responsible for many important diseases in humans and animals. • Picornaviruses are responsible for a wide range of clinical diseases resulting from multiple factors such as receptor specificity, tissue-specific susceptibility, virulence and the mechanisms of transmission. • Picornaviruses bind to cell specific surface receptors and this interaction is an important factor in determining host and tissue specificity of each virus. 4

Properties § replicate in cytoplasm § Positive sense genome (genome RNA is infectious) directly as a messenger RNA to template § encode viral polymerase § mature viral proteins generated through proteolytic cleavage § virions released on cell lysis

Properties • Picornaviruses cannot gain entry to cells by membrane fusion, as is the case for enveloped viruses, and require special mechanisms to breach cellular membranes and safely deliver the genome into the host cell. • Genome replication occurs in association with virus modified cellular membranes. • Viral RNA templates complementary negative strand molecules which in turn template multiple positive strand copies. • The synthesis of all RNA molecules is initiated by an uridylated peptide primer. • The mechanisms of transmission of infection play key roles 6 in the epidemiology of picornavirus infections.

Evolutionary relationships 7

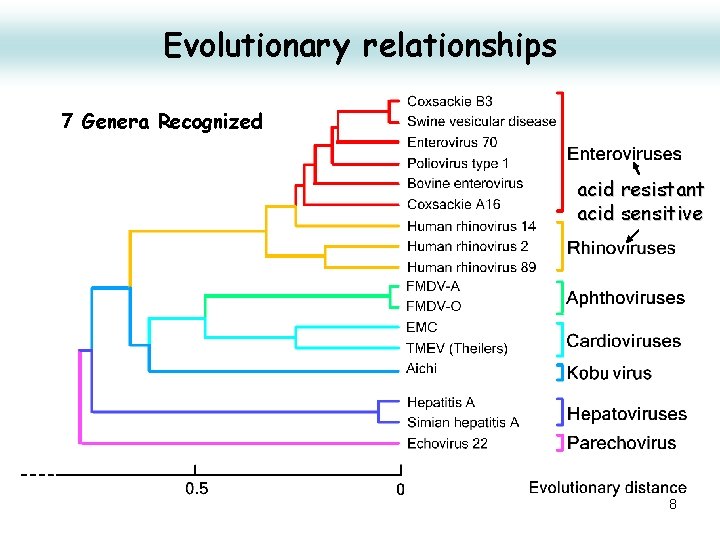
Evolutionary relationships 7 Genera Recognized acid resistant acid sensitive 8

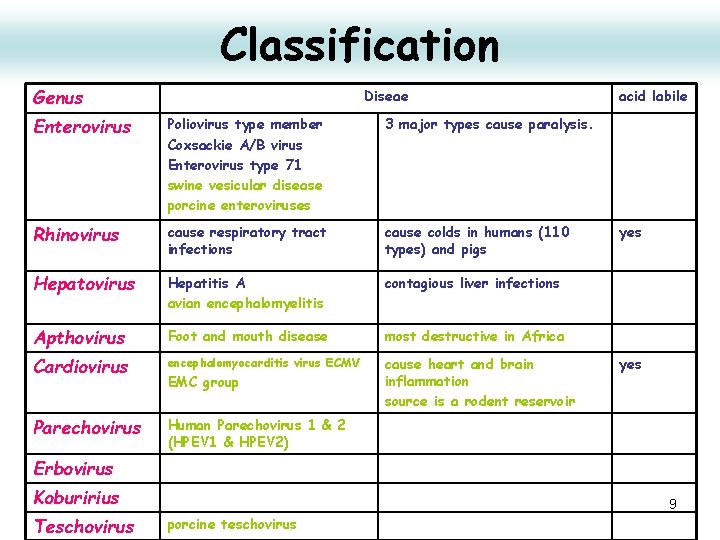
Classification Genus Diseae Enterovirus Poliovirus type member Coxsackie A/B virus Enterovirus type 71 swine vesicular disease porcine enteroviruses 3 major types cause paralysis. Rhinovirus cause respiratory tract infections cause colds in humans (110 types) and pigs Hepatovirus Hepatitis A avian encephalomyelitis contagious liver infections Apthovirus Foot and mouth disease most destructive in Africa Cardiovirus encephalomyocarditis virus ECMV cause heart and brain inflammation source is a rodent reservoir Parechovirus Human Parechovirus 1 & 2 (HPEV 1 & HPEV 2) EMC group acid labile yes Erbovirus Koburirius Teschovirus 9 porcine teschovirus

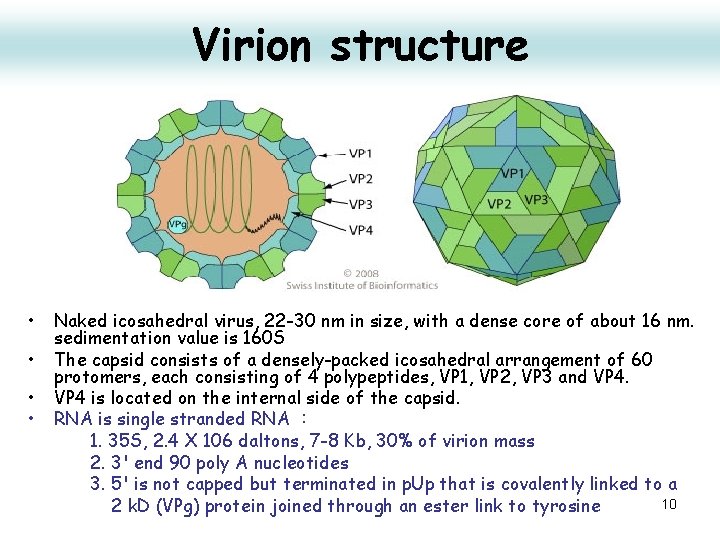
Virion structure • • Naked icosahedral virus, 22 -30 nm in size, with a dense core of about 16 nm. sedimentation value is 160 S The capsid consists of a densely-packed icosahedral arrangement of 60 protomers, each consisting of 4 polypeptides, VP 1, VP 2, VP 3 and VP 4 is located on the internal side of the capsid. RNA is single stranded RNA : 1. 35 S, 2. 4 X 106 daltons, 7 -8 Kb, 30% of virion mass 2. 3' end 90 poly A nucleotides 3. 5' is not capped but terminated in p. Up that is covalently linked to a 10 2 k. D (VPg) protein joined through an ester link to tyrosine

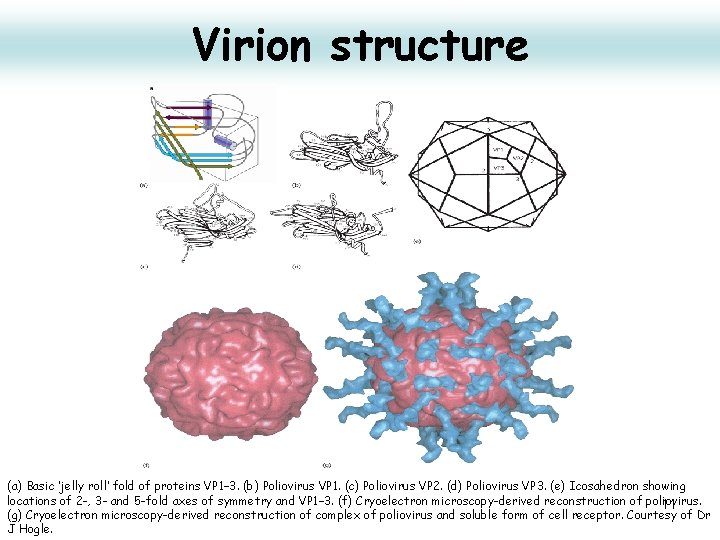
Virion structure (a) Basic ‘jelly roll’ fold of proteins VP 1– 3. (b) Poliovirus VP 1. (c) Poliovirus VP 2. (d) Poliovirus VP 3. (e) Icosahedron showing locations of 2 -, 3 - and 5 -fold axes of symmetry and VP 1– 3. (f) Cryoelectron microscopy-derived reconstruction of poliovirus. 11 (g) Cryoelectron microscopy-derived reconstruction of complex of poliovirus and soluble form of cell receptor. Courtesy of Dr J Hogle.

Genome 14

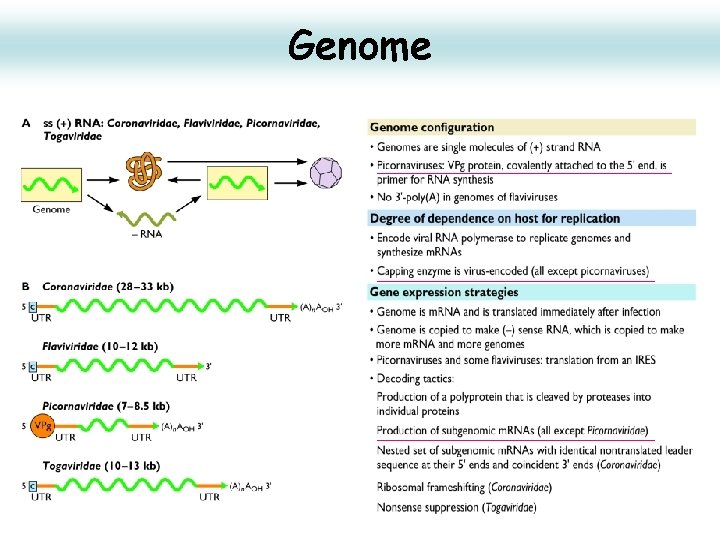
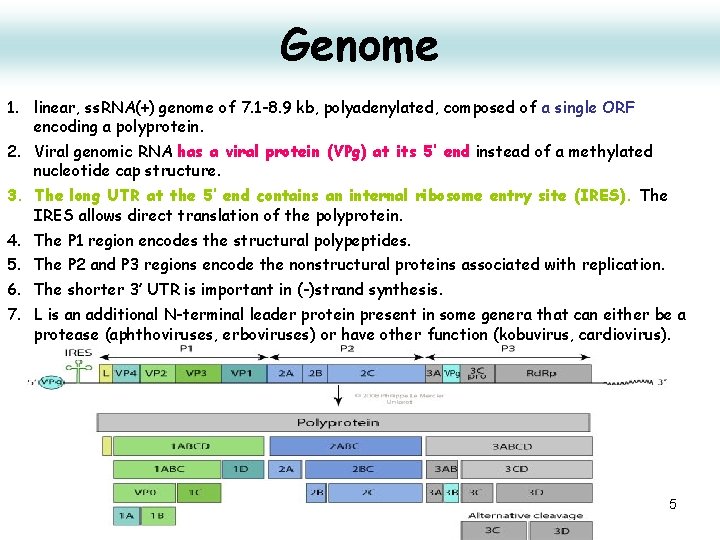
Genome 1. linear, ss. RNA(+) genome of 7. 1 -8. 9 kb, polyadenylated, composed of a single ORF encoding a polyprotein. 2. Viral genomic RNA has a viral protein (VPg) at its 5’ end instead of a methylated nucleotide cap structure. 3. The long UTR at the 5’ end contains an internal ribosome entry site (IRES). The IRES allows direct translation of the polyprotein. 4. The P 1 region encodes the structural polypeptides. 5. The P 2 and P 3 regions encode the nonstructural proteins associated with replication. 6. The shorter 3’ UTR is important in (-)strand synthesis. 7. L is an additional N-terminal leader protein present in some genera that can either be a protease (aphthoviruses, erboviruses) or have other function (kobuvirus, cardiovirus). 15

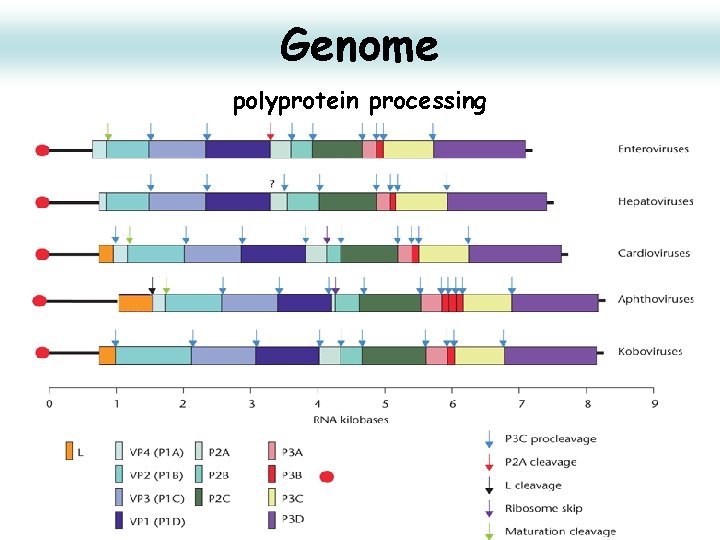
Genome polyprotein processing 16

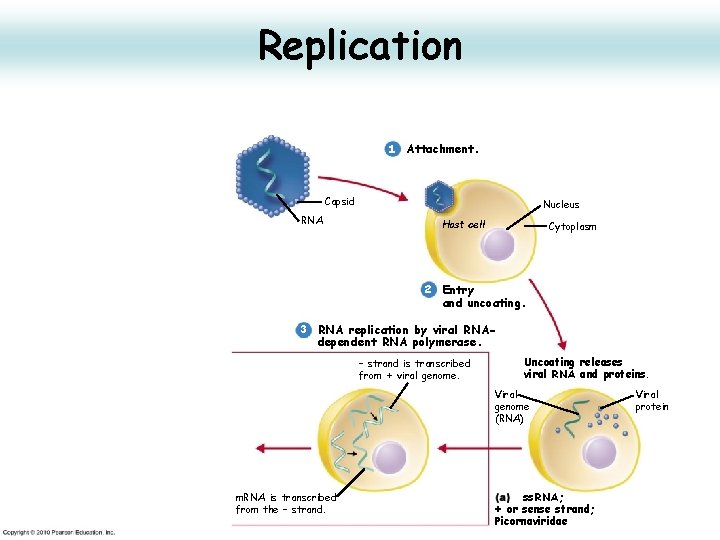
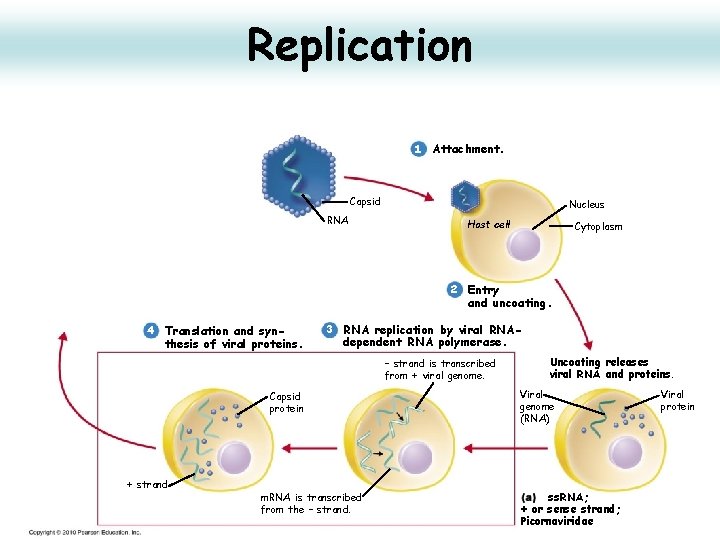
Attachment 1 Attachment. Capsid RNA

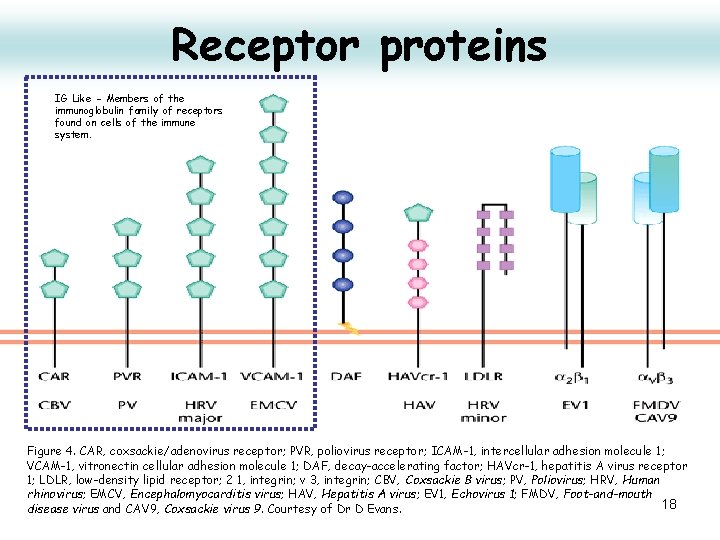
Receptor proteins IG Like - Members of the immunoglobulin family of receptors found on cells of the immune system. Figure 4. CAR, coxsackie/adenovirus receptor; PVR, poliovirus receptor; ICAM-1, intercellular adhesion molecule 1; VCAM-1, vitronectin cellular adhesion molecule 1; DAF, decay-accelerating factor; HAVcr-1, hepatitis A virus receptor 1; LDLR, low-density lipid receptor; 2 1, integrin; v 3, integrin; CBV, Coxsackie B virus; PV, Poliovirus; HRV, Human rhinovirus; EMCV, Encephalomyocarditis virus; HAV, Hepatitis A virus; EV 1, Echovirus 1; FMDV, Foot-and-mouth 18 disease virus and CAV 9, Coxsackie virus 9. Courtesy of Dr D Evans.

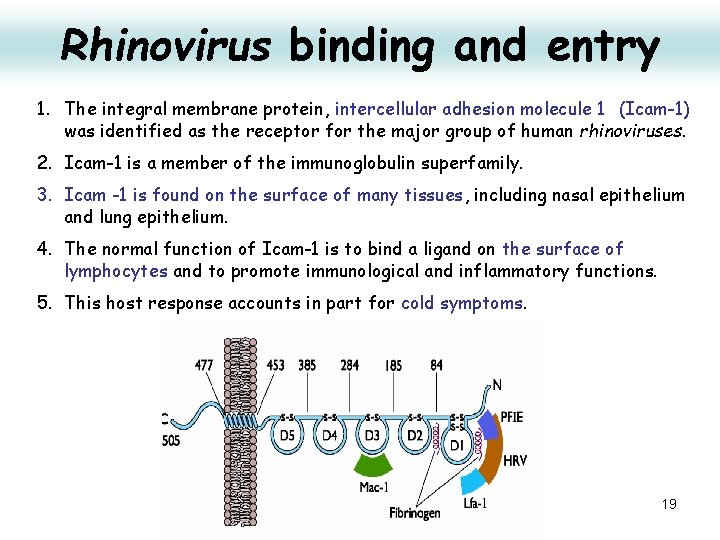
Rhinovirus binding and entry 1. The integral membrane protein, intercellular adhesion molecule 1 (Icam-1) was identified as the receptor for the major group of human rhinoviruses. 2. Icam-1 is a member of the immunoglobulin superfamily. 3. Icam -1 is found on the surface of many tissues, including nasal epithelium and lung epithelium. 4. The normal function of Icam-1 is to bind a ligand on the surface of lymphocytes and to promote immunological and inflammatory functions. 5. This host response accounts in part for cold symptoms. 19

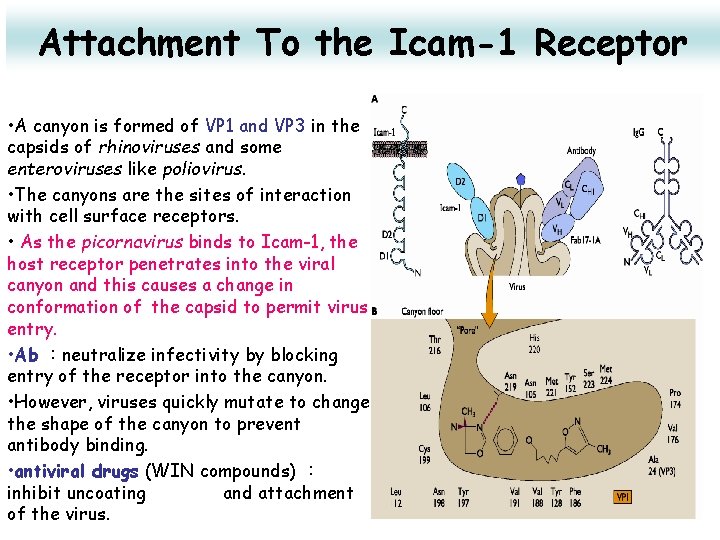
Attachment To the Icam-1 Receptor • A canyon is formed of VP 1 and VP 3 in the capsids of rhinoviruses and some enteroviruses like poliovirus. • The canyons are the sites of interaction with cell surface receptors. • As the picornavirus binds to Icam-1, the host receptor penetrates into the viral canyon and this causes a change in conformation of the capsid to permit virus entry. • Ab :neutralize infectivity by blocking entry of the receptor into the canyon. • However, viruses quickly mutate to change the shape of the canyon to prevent antibody binding. • antiviral drugs (WIN compounds) : inhibit uncoating and attachment of the virus. 20

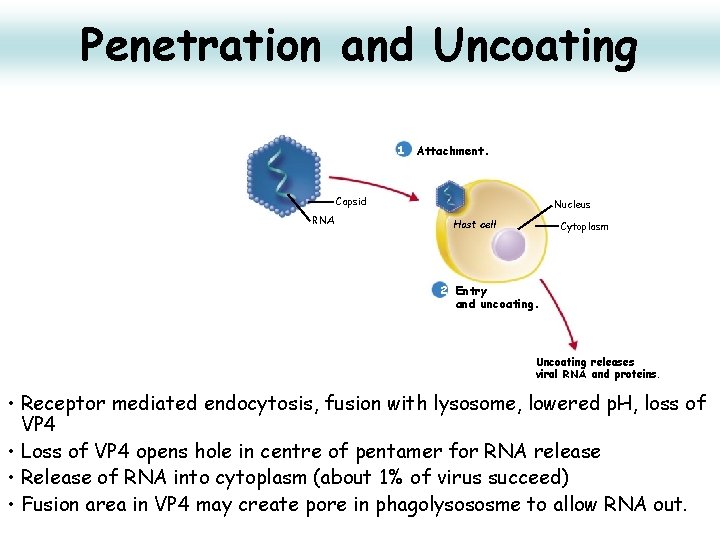
Penetration and Uncoating 1 Attachment. Capsid RNA Nucleus Host cell Cytoplasm 2 Entry and uncoating. Uncoating releases viral RNA and proteins. • Receptor mediated endocytosis, fusion with lysosome, lowered p. H, loss of VP 4 • Loss of VP 4 opens hole in centre of pentamer for RNA release • Release of RNA into cytoplasm (about 1% of virus succeed) • Fusion area in VP 4 may create pore in phagolysososme to allow RNA out.

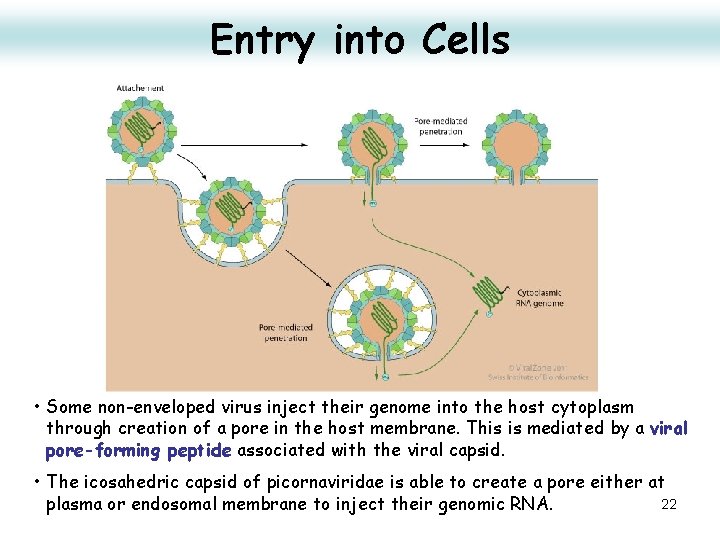
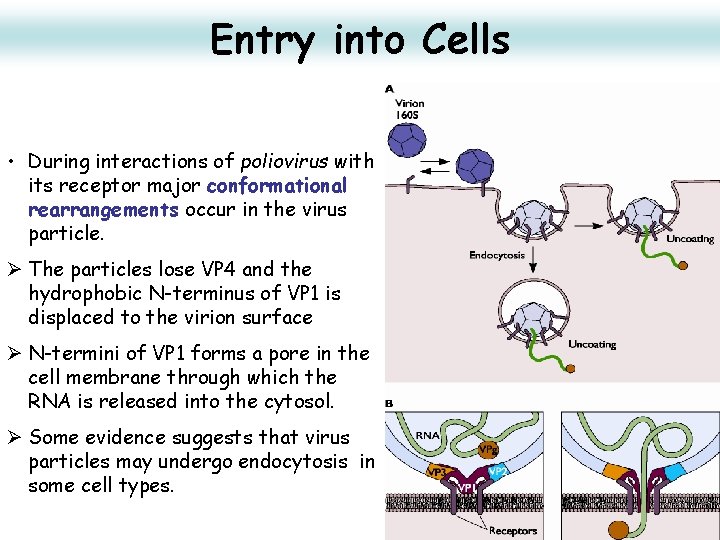
Entry into Cells • Some non-enveloped virus inject their genome into the host cytoplasm through creation of a pore in the host membrane. This is mediated by a viral pore-forming peptide associated with the viral capsid. • The icosahedric capsid of picornaviridae is able to create a pore either at 22 plasma or endosomal membrane to inject their genomic RNA.

Entry into Cells • During interactions of poliovirus with its receptor major conformational rearrangements occur in the virus particle. Ø The particles lose VP 4 and the hydrophobic N-terminus of VP 1 is displaced to the virion surface Ø N-termini of VP 1 forms a pore in the cell membrane through which the RNA is released into the cytosol. Ø Some evidence suggests that virus particles may undergo endocytosis in some cell types. 23

Replication 1 Attachment. Capsid RNA Nucleus Host cell Cytoplasm 2 Entry and uncoating. 3 RNA replication by viral RNA- dependent RNA polymerase. – strand is transcribed from + viral genome. Uncoating releases viral RNA and proteins. Viral genome (RNA) m. RNA is transcribed from the – strand. ss. RNA; + or sense strand; Picornaviridae Viral protein

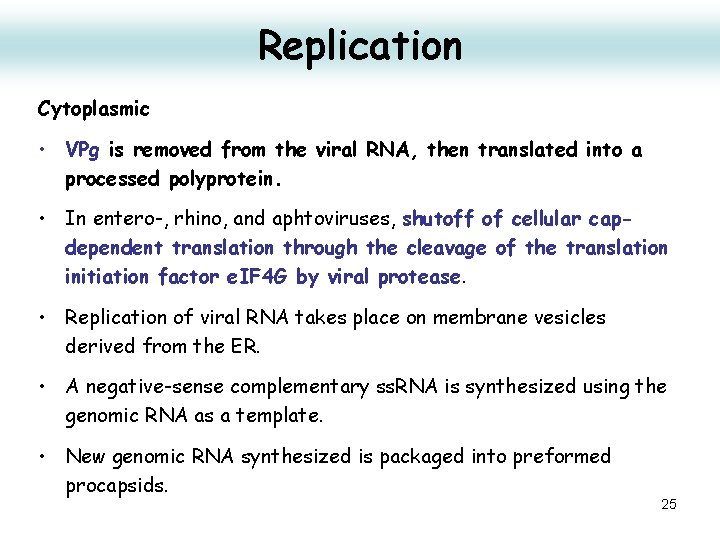
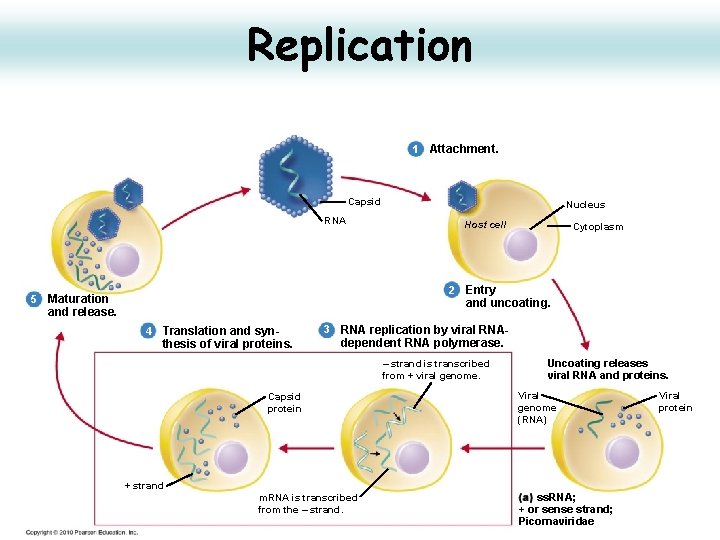
Replication Cytoplasmic • VPg is removed from the viral RNA, then translated into a processed polyprotein. • In entero-, rhino, and aphtoviruses, shutoff of cellular capdependent translation through the cleavage of the translation initiation factor e. IF 4 G by viral protease. • Replication of viral RNA takes place on membrane vesicles derived from the ER. • A negative-sense complementary ss. RNA is synthesized using the genomic RNA as a template. • New genomic RNA synthesized is packaged into preformed procapsids. 25

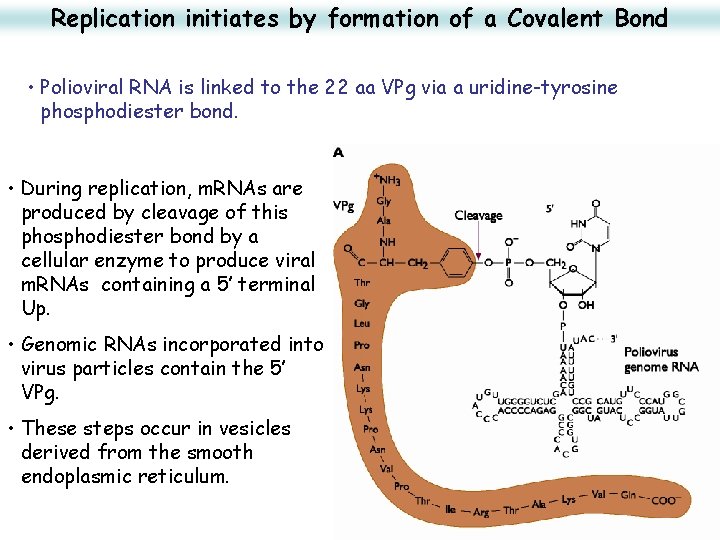
Replication initiates by formation of a Covalent Bond • Polioviral RNA is linked to the 22 aa VPg via a uridine-tyrosine phosphodiester bond. • During replication, m. RNAs are produced by cleavage of this phosphodiester bond by a cellular enzyme to produce viral m. RNAs containing a 5’ terminal Up. • Genomic RNAs incorporated into virus particles contain the 5’ VPg. • These steps occur in vesicles derived from the smooth endoplasmic reticulum. 26

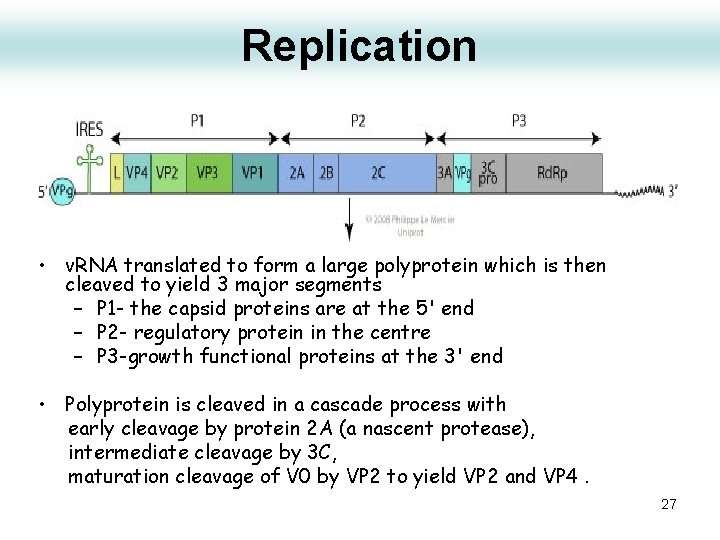
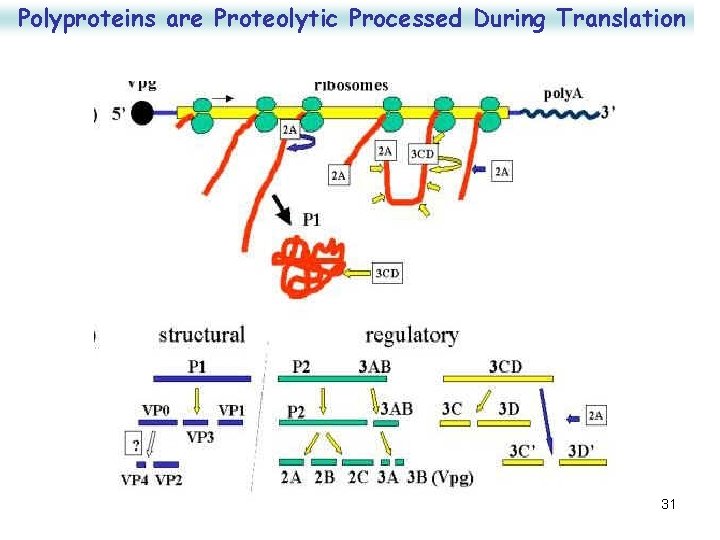
Replication • v. RNA translated to form a large polyprotein which is then cleaved to yield 3 major segments – P 1 - the capsid proteins are at the 5' end – P 2 - regulatory protein in the centre – P 3 -growth functional proteins at the 3' end • Polyprotein is cleaved in a cascade process with early cleavage by protein 2 A (a nascent protease), intermediate cleavage by 3 C, maturation cleavage of V 0 by VP 2 to yield VP 2 and VP 4. 27

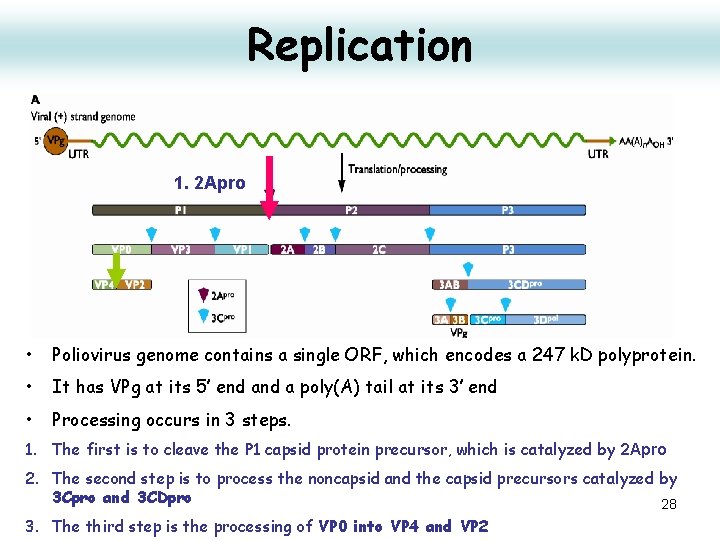
Replication 1. 2 Apro • Poliovirus genome contains a single ORF, which encodes a 247 k. D polyprotein. • It has VPg at its 5’ end a poly(A) tail at its 3’ end • Processing occurs in 3 steps. 1. The first is to cleave the P 1 capsid protein precursor, which is catalyzed by 2 Apro 2. The second step is to process the noncapsid and the capsid precursors catalyzed by 3 Cpro and 3 CDpro 28 3. The third step is the processing of VP 0 into VP 4 and VP 2

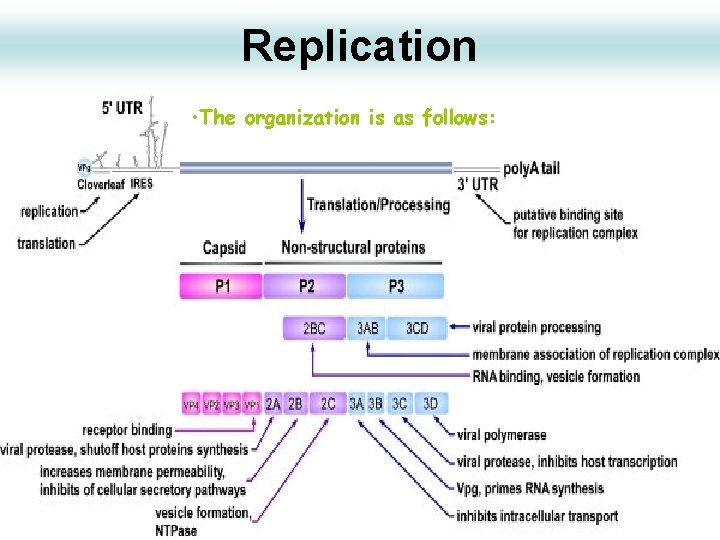
Replication • The organization is as follows: 29

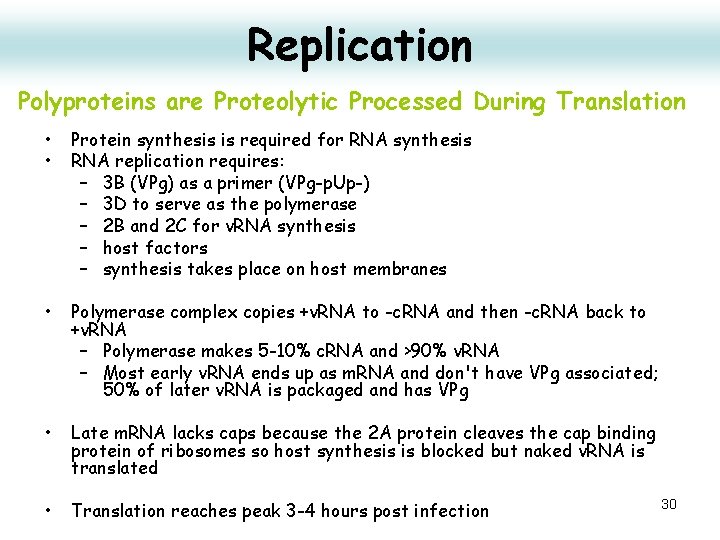
Replication Polyproteins are Proteolytic Processed During Translation • • Protein synthesis is required for RNA synthesis RNA replication requires: – 3 B (VPg) as a primer (VPg-p. Up-) – 3 D to serve as the polymerase – 2 B and 2 C for v. RNA synthesis – host factors – synthesis takes place on host membranes • Polymerase complex copies +v. RNA to -c. RNA and then -c. RNA back to +v. RNA – Polymerase makes 5 -10% c. RNA and >90% v. RNA – Most early v. RNA ends up as m. RNA and don't have VPg associated; 50% of later v. RNA is packaged and has VPg • Late m. RNA lacks caps because the 2 A protein cleaves the cap binding protein of ribosomes so host synthesis is blocked but naked v. RNA is translated • Translation reaches peak 3 -4 hours post infection 30

Polyproteins are Proteolytic Processed During Translation 31

Replication 1 Attachment. Capsid RNA Nucleus Host cell Cytoplasm 2 Entry and uncoating. 4 Translation and syn- thesis of viral proteins. 3 RNA replication by viral RNA- dependent RNA polymerase. – strand is transcribed from + viral genome. Capsid protein + strand m. RNA is transcribed from the – strand. Uncoating releases viral RNA and proteins. Viral genome (RNA) ss. RNA; + or sense strand; Picornaviridae Viral protein

Replication 1 Attachment. Capsid RNA Nucleus Host cell Cytoplasm 2 Entry 5 Maturation and uncoating. and release. 4 Translation and syn- thesis of viral proteins. 3 RNA replication by viral RNA- dependent RNA polymerase. – strand is transcribed from + viral genome. Capsid protein Uncoating releases viral RNA and proteins. Viral genome (RNA) + strand m. RNA is transcribed from the – strand. ss. RNA; + or sense strand; Picornaviridae Viral protein

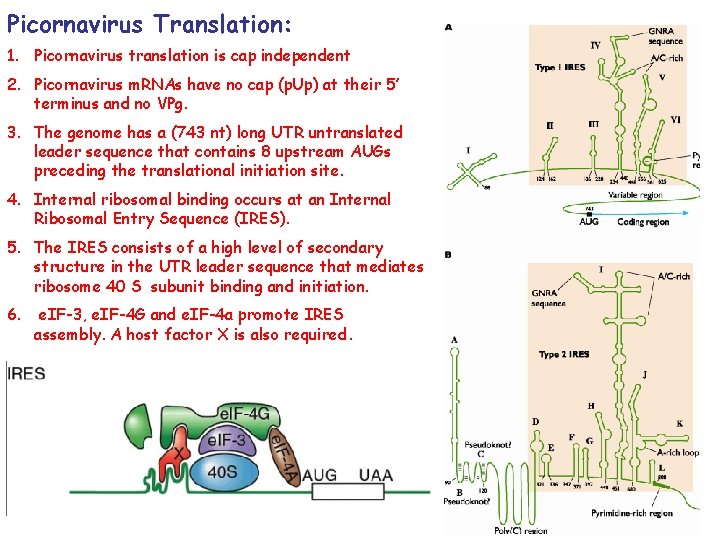
Picornavirus Translation: 1. Picornavirus translation is cap independent 2. Picornavirus m. RNAs have no cap (p. Up) at their 5’ terminus and no VPg. 3. The genome has a (743 nt) long UTR untranslated leader sequence that contains 8 upstream AUGs preceding the translational initiation site. 4. Internal ribosomal binding occurs at an Internal Ribosomal Entry Sequence (IRES). 5. The IRES consists of a high level of secondary structure in the UTR leader sequence that mediates ribosome 40 S subunit binding and initiation. 6. e. IF-3, e. IF-4 G and e. IF-4 a promote IRES assembly. A host factor X is also required. 34

A Host Protein is Required for Synthesis of Polio Virus RNA A highly based paired structure formed from the 108 nucleotides at the 5’ end of the (+) strand RNA of poliovirus, which forms a cloverleaf like structure. The viral protein 3 CD and a PCBP a host Cellular protein, (Polyr. C Binding Protein) interact with different loops of the cloverleaf. This interaction brings the 5’ end close to the 3’ end. At this point VPg priming can occur. Formation of this ribonucleoprotein complex is required for synthesis of both plus and minus strand RNA. 35

Steps in the Assembly of Poliovirus Ø After Translation and release by the 2 A protease, P 1 folds and is cleaved by the 3 CD protease to form the 5 S protomers. Ø assemble into a 14 S complex. Ø The 14 S complex may then form a 75 S empty capsid that assembles with the RNA to form a noninfectious 150 S provirion. Ø Some researchers argue that the 14 S subunits assemble in a series of steps with the viral RNA to form provirions. Ø The provirion is converted to an infectous form by internal cleavage of VP 0 molecules to VP 4 plus VP 2. P 1 Peptide Assembly steps are not reversible Folded P 1 5 S Unit 36

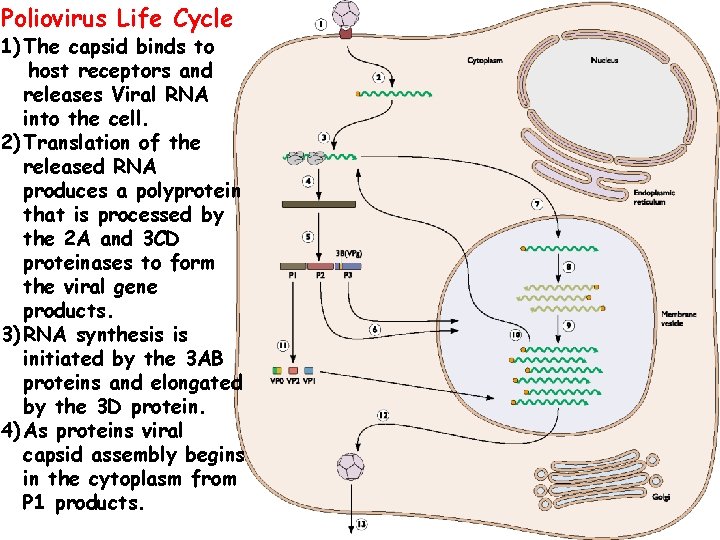
Poliovirus Life Cycle 1) The capsid binds to host receptors and releases Viral RNA into the cell. 2) Translation of the released RNA produces a polyprotein that is processed by the 2 A and 3 CD proteinases to form the viral gene products. 3) RNA synthesis is initiated by the 3 AB proteins and elongated by the 3 D protein. 4) As proteins viral capsid assembly begins in the cytoplasm from P 1 products. 37

Laboratory Diagnosis of Infections • Most common method: isolate virus from stool samples or throat samples from the mouth – Rarely isolated from CSF • Grows well in any human or monkey kidney cell lines (causes good CPEs). • Identify serotype with neutralization assays • Nucleic acid methods: genomic sequencing to determine if the infection is caused by vaccine or wild-type virus. 38

39

40
 Picornaviridae adalah
Picornaviridae adalah Estrutura do virus
Estrutura do virus Fields virology
Fields virology Introduction to medical virology
Introduction to medical virology Fields virology
Fields virology Virology
Virology Dispersive model
Dispersive model Watson strand crick strand
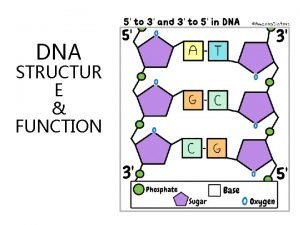
Watson strand crick strand Mrna strand that is complementary to the dna strand aattgc
Mrna strand that is complementary to the dna strand aattgc Template strand, new strand, base pair, and dna polymerase.
Template strand, new strand, base pair, and dna polymerase. Watson strand crick strand
Watson strand crick strand Strand of virus
Strand of virus Virusmax
Virusmax Corona virus dna or rna
Corona virus dna or rna Polyproteins
Polyproteins Rna virus
Rna virus Familia picornaviridae
Familia picornaviridae Big families vs small families
Big families vs small families Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Zinc oxide + nitric acid → zinc nitrate + water
Zinc oxide + nitric acid → zinc nitrate + water Je contiens du sucre sans être sucré que suis-je
Je contiens du sucre sans être sucré que suis-je Trias jura kreda
Trias jura kreda Plus j'apprends plus je me rends compte de mon ignorance
Plus j'apprends plus je me rends compte de mon ignorance Jerusalem cite de dieu chant
Jerusalem cite de dieu chant Vakantie op het strand
Vakantie op het strand Why is fiber class evidence
Why is fiber class evidence Strangers strand van sint anneke
Strangers strand van sint anneke Vakantie op het strand
Vakantie op het strand Pc steel wire
Pc steel wire Leading and lagging strand
Leading and lagging strand Strand in lesson plan
Strand in lesson plan Strand in lesson plan
Strand in lesson plan Sanaysay tungkol sa isports
Sanaysay tungkol sa isports Chargaff rule
Chargaff rule Ability of a substance to be pulled into a thin strand
Ability of a substance to be pulled into a thin strand Track and strand
Track and strand Foley manometer
Foley manometer Dna template strand to mrna
Dna template strand to mrna