Parallelizing the graph isomorphism portion of an automatic




- Slides: 21

Parallelizing the graph isomorphism portion of an automatic reaction mechanism generation algorithm Geoff Oxberry 18. 337 Project, Spring 2009

Automatic reaction mechanism generation yields models quickly Reaction mechanisms are used to model chemistry in a wide range of applications p Generating first principles reaction mechanisms can take years, requires lots of expertise p The Bill Green group developed software (RMG) that automatically generates these models based on rules p

For some problems, RMG takes days to generate a mechanism We want it to take a day or less on a cluster p Big bottleneck for us is that we have to repeatedly solve a colored graph isomorphism (GI) problem p If we can speed it up, we can solve many more interesting chemistry problems p Parallelism is one option p

We want to see if parallelism can be used to speed up RMG Want to see if a parallel version of RMG is faster than a serial version p Due to time constraints, I chose to implement skeletal prototypes of serial and parallel versions of RMG in Python p Idea is to use results for the prototypes to see if it is worth parallelizing the production-scale code p

Parallelism does speed up RMG on intermediate-sized case studies When searching for graph isomorphisms in collections of 20 or fewer graphs, serial code is faster p When searching for graph isomorphisms in collections of ~100 graphs, parallel code is faster p When searching for graph isomorphisms in collections of ~2000 graphs, serial code is faster again p

Outline Brief overview of graph isomorphism p Discussion of existing RMG algorithm and how to parallelize p Python prototypes of serial and parallel versions of RMG algorithm p Results p Discussion of obstacles p Conclusions p

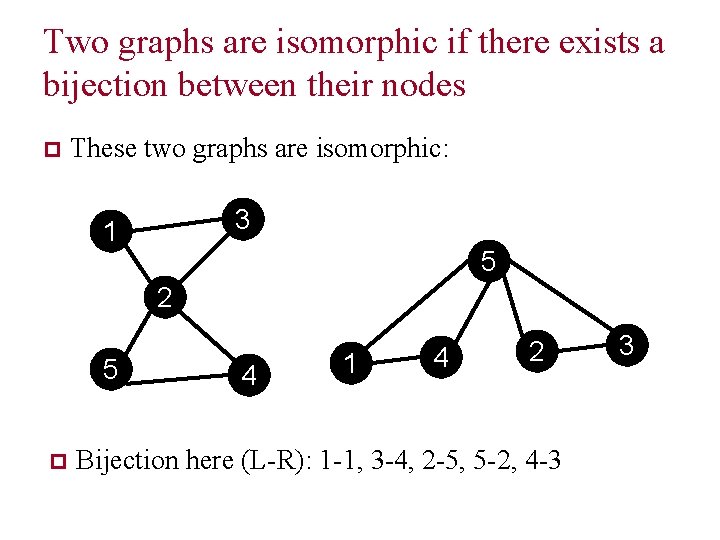
Two graphs are isomorphic if there exists a bijection between their nodes p These two graphs are isomorphic: 3 1 5 2 5 p 4 1 4 2 Bijection here (L-R): 1 -1, 3 -4, 2 -5, 5 -2, 4 -3 3

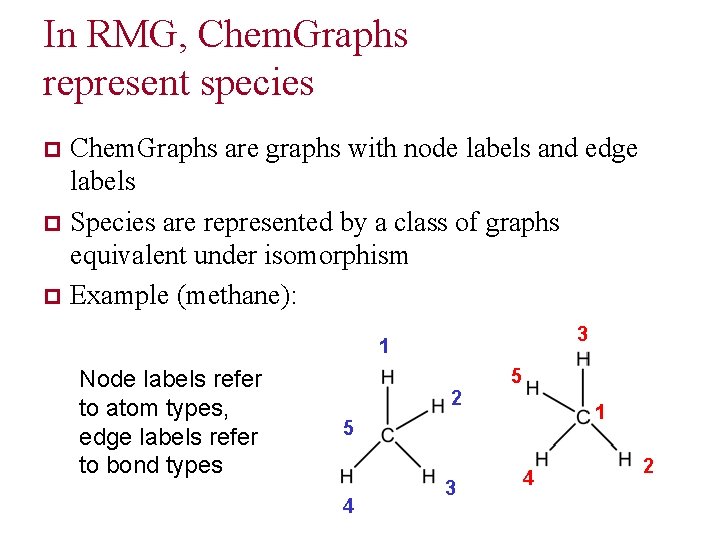
In RMG, Chem. Graphs represent species Chem. Graphs are graphs with node labels and edge labels p Species are represented by a class of graphs equivalent under isomorphism p Example (methane): p 3 1 Node labels refer to atom types, edge labels refer to bond types 2 5 1 5 4 3 4 2

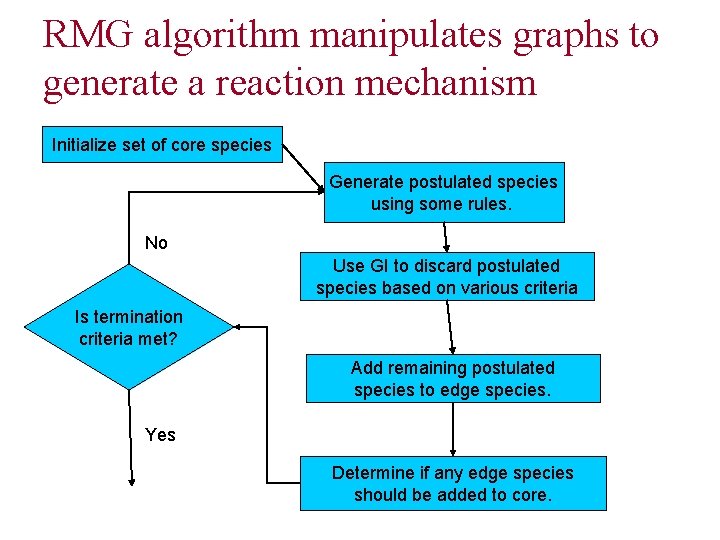
RMG classifies species as one of three types Core species make up all of the reactants of the reaction mechanism p Edge species are products of the reaction mechanism not included in the core; they may be added to the core over the course of the algorithm p Postulated species are proposed species that may be added to the edge over the course of the algorithm p

RMG algorithm manipulates graphs to generate a reaction mechanism Initialize set of core species Generate postulated species using some rules. No Use GI to discard postulated species based on various criteria Is termination criteria met? Add remaining postulated species to edge species. Yes Determine if any edge species should be added to core.

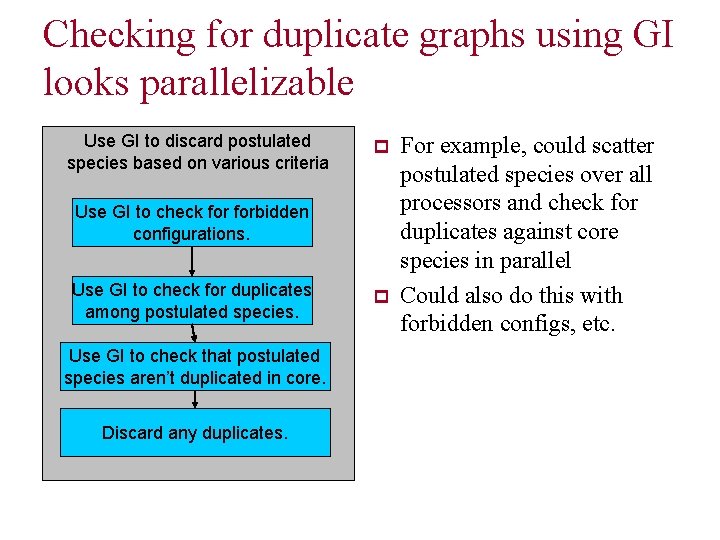
Checking for duplicate graphs using GI looks parallelizable Use GI to discard postulated species based on various criteria p Use GI to check forbidden configurations. Use GI to check for duplicates among postulated species. Use GI to check that postulated species aren’t duplicated in core. Discard any duplicates. p For example, could scatter postulated species over all processors and check for duplicates against core species in parallel Could also do this with forbidden configs, etc.

Instead of working with RMG directly, I created a prototype RMG takes 18 mos. for a developer to get up to speed; this project was ~6 wks. p To save time, I built a prototype in Python because its syntax and available libraries enable rapid development p Also enabled me to focus on the parts of the code that matter (GI algorithms) and ignore the rest p

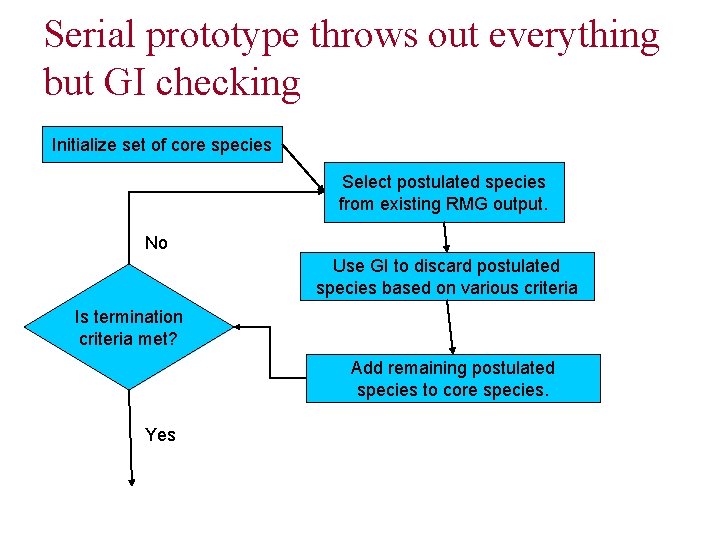
Serial prototype throws out everything but GI checking Initialize set of core species Select postulated species from existing RMG output. No Use GI to discard postulated species based on various criteria Is termination criteria met? Add remaining postulated species to core species. Yes

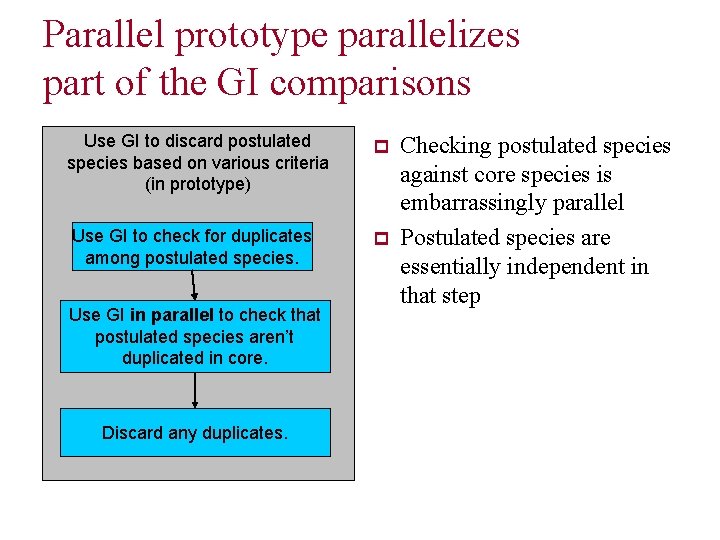
Parallel prototype parallelizes part of the GI comparisons Use GI to discard postulated species based on various criteria (in prototype) p Use GI to check for duplicates among postulated species. p Use GI in parallel to check that postulated species aren’t duplicated in core. Discard any duplicates. Checking postulated species against core species is embarrassingly parallel Postulated species are essentially independent in that step

Prototypes were implemented in Python/MPI on a cluster p Software: n n n Python 2. 5 (w/ C extensions) igraph module (graph data structure, GI algorithms) mpi 4 py module (MPI bindings for Python) p Hardware: n n n 64 -node cluster (pharos. mit. edu) 8 GB RAM per node Each node has 2 quadcore Xeon processors (either 2. 33 GHz or 2. 66 GHz)

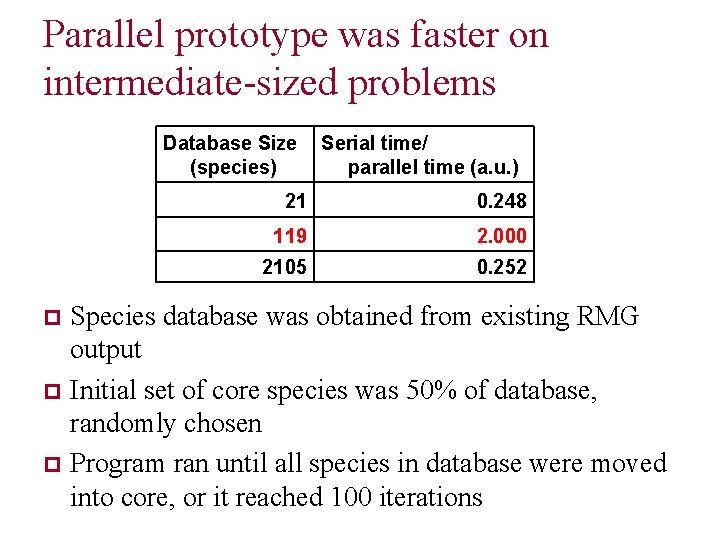
Parallel prototype was faster on intermediate-sized problems Database Size (species) Serial time/ parallel time (a. u. ) 21 0. 248 119 2. 000 2105 0. 252 Species database was obtained from existing RMG output p Initial set of core species was 50% of database, randomly chosen p Program ran until all species in database were moved into core, or it reached 100 iterations p

Communication is slow in large test cases due to passing graph objects Graphs are implemented using a class in the igraph library p mpi 4 py converts non-native Python objects using c. Pickle, which is compute-intensive p c. Pickle is probably why the serial code is faster in large test cases p Alternative approach would use Num. Py and define an MPI derived data type; would be faster p

Many technical problems occurred during the project Laptop experienced hardware failures p Difficulties installing igraph and mpi 4 py on pharos p n n System libraries had to be recompiled Environment variables were reset so igraph and mpi 4 py could be recognized on all nodes Incomplete mpi 4 py documentation p Python extended debugger not installed; no graphical front-end p

Parallelism can be used to speed up RMG for some case studies Saw speed up for intermediate-sized case studies on parallel prototype p Additional opportunities for parallelism within RMG algorithm p Can also decrease MPI communication costs w/ additional development, use of debugger/profiler p

Future Work Install extended Python debugger/profiler p Use Num. Py and MPI derived data type to reduced communication overhead p Try alternative strategies for parallelization: p n n Reorganize algorithm (check core species, then postulated species) Parallelize checks of postulated species against themselves

Acknowledgments p RMG team: n n n Franklin Goldsmith Sandeep Sharma Josh Allen Richard West Michael Harper Greg Magoon Ray Speth p Kushal Kedia p Prof. Bill Green p DOE CSGF for funding p