Opera Reconstructing optimal genomic scaffolds with highthroughput pairedend


![ü Scaffolding Problem[Huson et al, 2002] Discordant Read Contig 1 k Paired-end Read 3 ü Scaffolding Problem[Huson et al, 2002] Discordant Read Contig 1 k Paired-end Read 3](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-5.jpg)
![Statistics of Assembled Genomes[Schatz et al, 2010] Organism Genome Size # of Contigs N Statistics of Assembled Genomes[Schatz et al, 2010] Organism Genome Size # of Contigs N](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-6.jpg)
![NP-Complete [Huson et al, 2002] * Huson, D. H. , Reinert, K. , Myers NP-Complete [Huson et al, 2002] * Huson, D. H. , Reinert, K. , Myers](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-7.jpg)
![Heuristic Methods - Celera Assembler[Myers et al, 2000] - Euler[Pevzner et al, 2001] - Heuristic Methods - Celera Assembler[Myers et al, 2000] - Euler[Pevzner et al, 2001] -](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-8.jpg)

![1. Pre-Processing Paired-end Reads -> Clusters [Huson et al, 2002] 3 Chimeric Noise Filtered 1. Pre-Processing Paired-end Reads -> Clusters [Huson et al, 2002] 3 Chimeric Noise Filtered](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-10.jpg)
![Dynamic Programming üScaffold Tail is Sufficient Upper Bound width(w) üAnalogous to Bandwidth Problem[Saxe, 1980] Dynamic Programming üScaffold Tail is Sufficient Upper Bound width(w) üAnalogous to Bandwidth Problem[Saxe, 1980]](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-12.jpg)


- Slides: 26

Opera: Reconstructing optimal genomic scaffolds with highthroughput paired-end sequences Song Gao, Niranjan Nagarajan, Wing-Kin Sung National University of Singapore Genome Institute of Singapore

Outline ØOverview • Methods - 1. Pre-Processing - 2. A Special Case - 3. Full Algorithm - 4. Graph Contraction - 5. Gap Estimation • Results • Ongoing Work 2

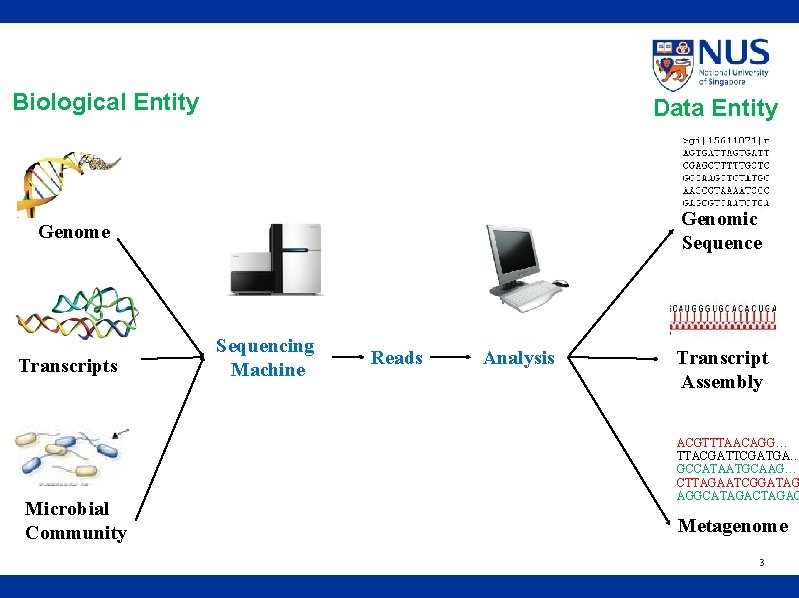
Biological Entity Data Entity Genomic Sequence Genome Transcripts Microbial Community Sequencing Machine Reads Analysis Transcript Assembly ACGTTTAACAGG… TTACGATTCGATGA… GCCATAATGCAAG… CTTAGAATCGGATAG AGGCATAGACTAGAG Metagenome 3

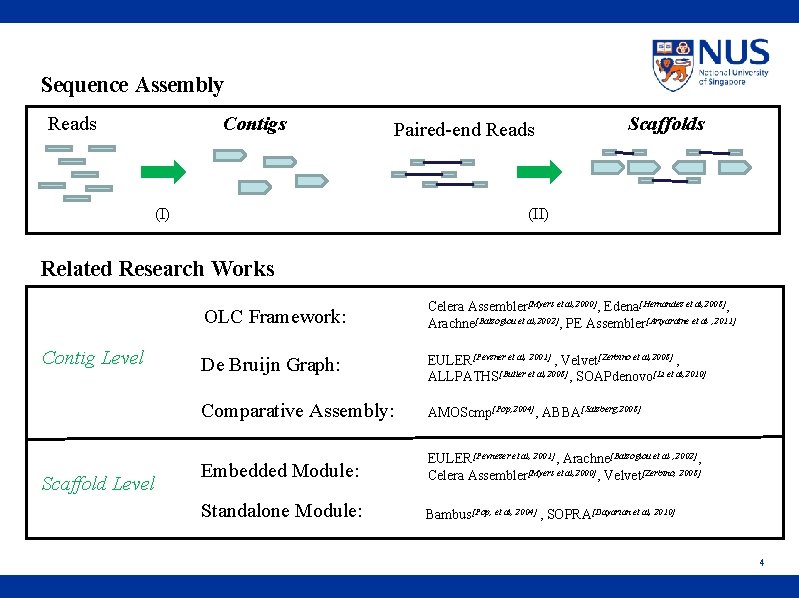
Sequence Assembly Reads Contigs Paired-end Reads (I) Scaffolds (II) Related Research Works Contig Level Scaffold Level OLC Framework: Celera Assembler[Myers et al, 2000], Edena[Hernandez et al, 2008], Arachne[Batzoglou et al, 2002], PE Assembler[Ariyaratne et al , 2011] De Bruijn Graph: EULER[Pevzner et al, 2001] , Velvet[Zerbino et al, 2008] , ALLPATHS[Butler et al, 2008], SOAPdenovo[Li et al, 2010] Comparative Assembly: AMOScmp[Pop, 2004], ABBA[Salzberg, 2008] Embedded Module: EULER[Pevnezer et al, 2001], Arachne[Batzoglou et al , 2002], Celera Assembler[Myers et al, 2000], Velvet[Zerbino, 2008] Standalone Module: Bambus[Pop, et al, 2004] , SOPRA[Dayarian et al, 2010] 4
![ü Scaffolding ProblemHuson et al 2002 Discordant Read Contig 1 k Pairedend Read 3 ü Scaffolding Problem[Huson et al, 2002] Discordant Read Contig 1 k Paired-end Read 3](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-5.jpg)
ü Scaffolding Problem[Huson et al, 2002] Discordant Read Contig 1 k Paired-end Read 3 k 2. 5 k Scaffold ü Value Addition Gap Filling: Gap. Closer Module of SOAPdenovo Repeat Resolution Long-Range Genomic Structure * Huson, D. H. , Reinert, K. , Myers E. W. : The greedy path-merging algorithm for contig scaffolding. Journal of the ACM 49(5), 603– 615 (2002) 5
![Statistics of Assembled GenomesSchatz et al 2010 Organism Genome Size of Contigs N Statistics of Assembled Genomes[Schatz et al, 2010] Organism Genome Size # of Contigs N](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-6.jpg)
Statistics of Assembled Genomes[Schatz et al, 2010] Organism Genome Size # of Contigs N 50 # of Scaffold N 50 Grapevine 500 Mb 58, 611 18. 2 kb 2, 093 1. 33 Mb Panda 2. 4 Gb 200, 604 36. 7 kb 81, 469 1. 22 Mb Strawberry 220 Mb 16, 487 28. 1 kb 3, 263 1. 44 Mb Turkey 1. 1 Gb 128, 271 12. 6 kb 26, 917 1. 5 Mb * N 50: Given a set of sequences of varying lengths, the N 50 length is defined as the length N for which 50% of all bases in the sequences are in a sequence of length L >= N. Data ØSequencing Errors ØRead Length ØCoverage Analysis Long Insert vs. Long Read[Chaisson, 2009; Zerbino, 2009] * Schatz M. C. , Arthur L. D. , Steven L. S. : Assembly of large genomes using second-generation sequencing. Genome Research, 20 -9, 1165 -1173 (2010) * Zerbino, D. R. : Pebble and rock band: heuristic resolution of repeats and scaolding in the velvet short-read de novo assembler. PLo. S ONE, 4(12) (2009) * Chaisson, M. J. , Brinza, D. , Pevzner, P. A. : De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Research 19, 336 -346 (2009) 6
![NPComplete Huson et al 2002 Huson D H Reinert K Myers NP-Complete [Huson et al, 2002] * Huson, D. H. , Reinert, K. , Myers](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-7.jpg)
NP-Complete [Huson et al, 2002] * Huson, D. H. , Reinert, K. , Myers E. W. : The greedy path-merging algorithm for contig scaffolding. Journal of the ACM 49(5), 603– 615 (2002) 7
![Heuristic Methods Celera AssemblerMyers et al 2000 EulerPevzner et al 2001 Heuristic Methods - Celera Assembler[Myers et al, 2000] - Euler[Pevzner et al, 2001] -](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-8.jpg)
Heuristic Methods - Celera Assembler[Myers et al, 2000] - Euler[Pevzner et al, 2001] - Jazz[Chapman et al, 2002] - Velvet[Zerbino et al, 2008] - Arachne[Batzoglou et al , 2002] - Bambus[Pop, et al, 2004] “True Complexity” Phase transition based on parameters[Hayes, 1996] 3 -SAT Problem Parametric Complexity[Rodney et al, 1999] Vertex Cover Problem * Hayes, B. Can't get no satisfaction. American. Scientist. 85, 108 -112 (1996). * Rodney G. D. , et al. Parameterized Complexity: A Framework for Systematically Confronting Computational Intractability. DIMACS. Vol 49. 1999 8

Outline • Overview Ø Methods - 1. Pre-Processing - 2. A Special Case - 3. Full Algorithm - 4. Graph Contraction - 5. Gap Estimation • Results • Ongoing Work 9
![1 PreProcessing Pairedend Reads Clusters Huson et al 2002 3 Chimeric Noise Filtered 1. Pre-Processing Paired-end Reads -> Clusters [Huson et al, 2002] 3 Chimeric Noise Filtered](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-10.jpg)
1. Pre-Processing Paired-end Reads -> Clusters [Huson et al, 2002] 3 Chimeric Noise Filtered by simulation Chimera * Upper Bound of Paired-end Reads * Huson, D. H. , Reinert, K. , Myers E. W. : The greedy path-merging algorithm for contig scaffolding. Journal of the ACM 49(5), 603– 615 (2002) 10

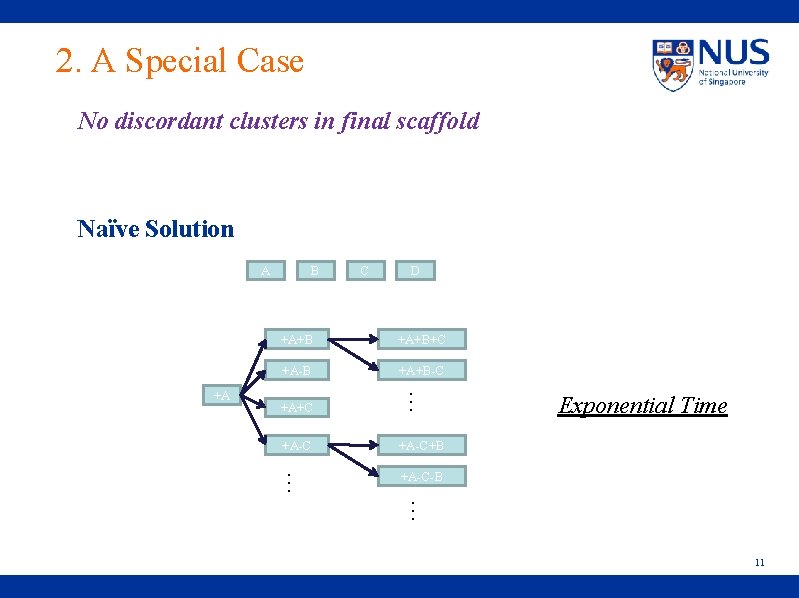
2. A Special Case No discordant clusters in final scaffold Naïve Solution A C D +A+B+C +A-B +A+B-C +A+C … +A B +A-C+B … +A-C-B Exponential Time … 11
![Dynamic Programming üScaffold Tail is Sufficient Upper Bound widthw üAnalogous to Bandwidth ProblemSaxe 1980 Dynamic Programming üScaffold Tail is Sufficient Upper Bound width(w) üAnalogous to Bandwidth Problem[Saxe, 1980]](https://slidetodoc.com/presentation_image_h2/21b849382e816d582dc3090f69783138/image-12.jpg)
Dynamic Programming üScaffold Tail is Sufficient Upper Bound width(w) üAnalogous to Bandwidth Problem[Saxe, 1980] Orientation of Nodes Direction of Edges Discordant Edges … * J. Saxe: Dynamic programming algorithms for recognizing small-bandwidth graphs in polynomial time SIAM J. on Algebraic and Discrete Methodd, 1(4), 363369 (1980) 12

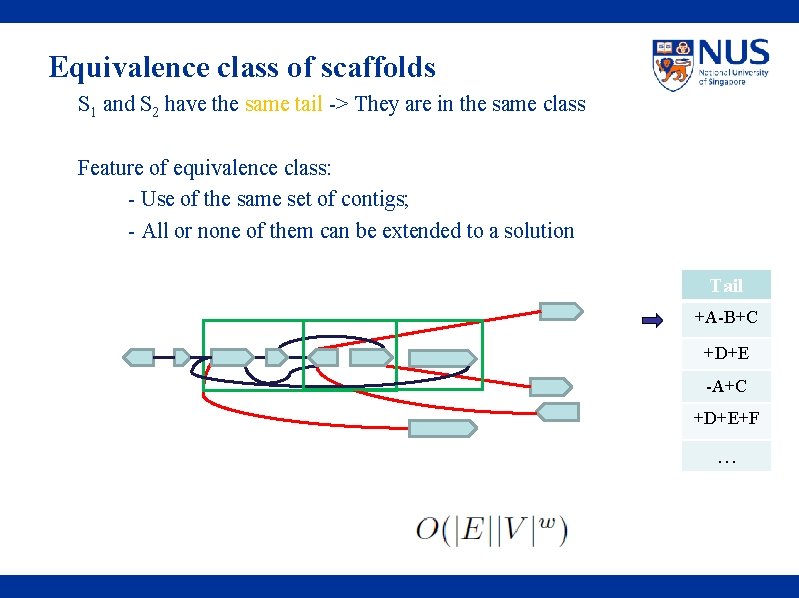
Equivalence class of scaffolds S 1 and S 2 have the same tail -> They are in the same class Feature of equivalence class: - Use of the same set of contigs; - All or none of them can be extended to a solution Tail +A-B+C +D+E -A+C +D+E+F …

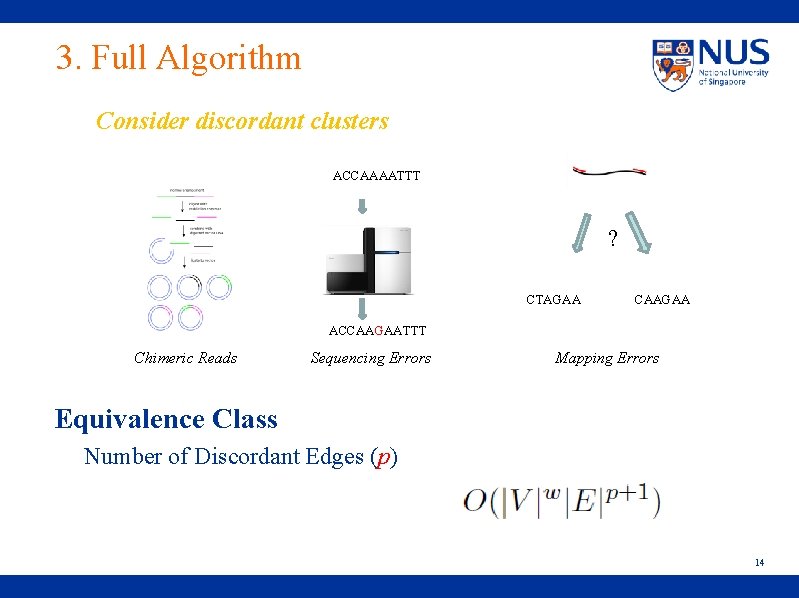
3. Full Algorithm Consider discordant clusters ACCAAAATTT ? CTAGAA CAAGAA ACCAAGAATTT Chimeric Reads Sequencing Errors Mapping Errors Equivalence Class Number of Discordant Edges (p) 14

4. Graph Contraction 20 k

4. Graph Contraction

4. Graph Contraction

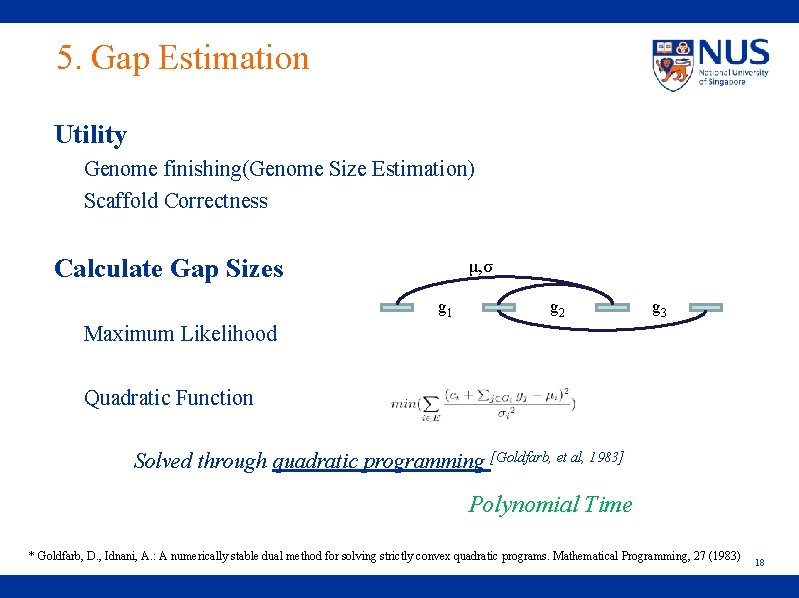
5. Gap Estimation Utility Genome finishing(Genome Size Estimation) Scaffold Correctness Calculate Gap Sizes μ, σ g 1 g 2 g 3 Maximum Likelihood Quadratic Function Solved through quadratic programming [Goldfarb, et al, 1983] Polynomial Time * Goldfarb, D. , Idnani, A. : A numerically stable dual method for solving strictly convex quadratic programs. Mathematical Programming, 27 (1983) 18

Outline • Overview • Methods - 1. Pre-Processing - 2. A Special Case - 3. Full Algorithm - 4. Graph Contraction - 5. Gap Estimation Ø Results • Ongoing Work 19

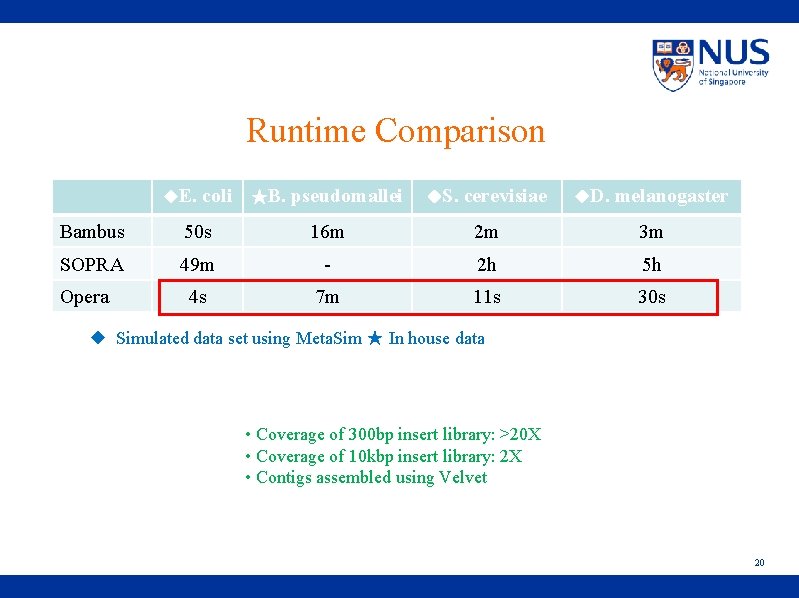
Runtime Comparison ◆E. coli ★B. pseudomallei ◆S. cerevisiae ◆D. melanogaster Bambus 50 s 16 m 2 m 3 m SOPRA 49 m - 2 h 5 h 4 s 7 m 11 s 30 s Opera ◆ Simulated data set using Meta. Sim ★ In house data • Coverage of 300 bp insert library: >20 X • Coverage of 10 kbp insert library: 2 X • Contigs assembled using Velvet 20

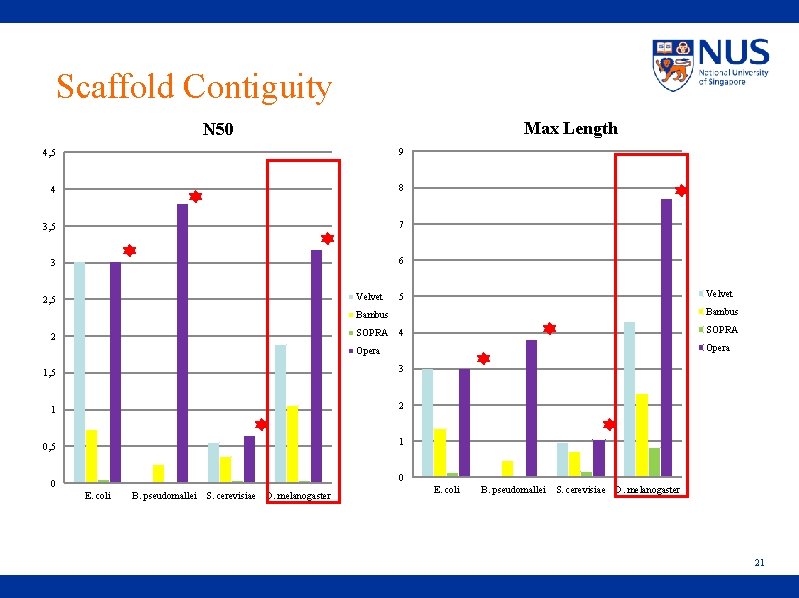
Scaffold Contiguity Max Length N 50 4, 5 9 4 8 3, 5 7 3 6 Velvet 2, 5 Velvet 5 Bambus SOPRA 2 SOPRA 4 Opera 1, 5 3 1 2 1 0, 5 0 0 E. coli B. pseudomallei S. cerevisiae D. melanogaster 21

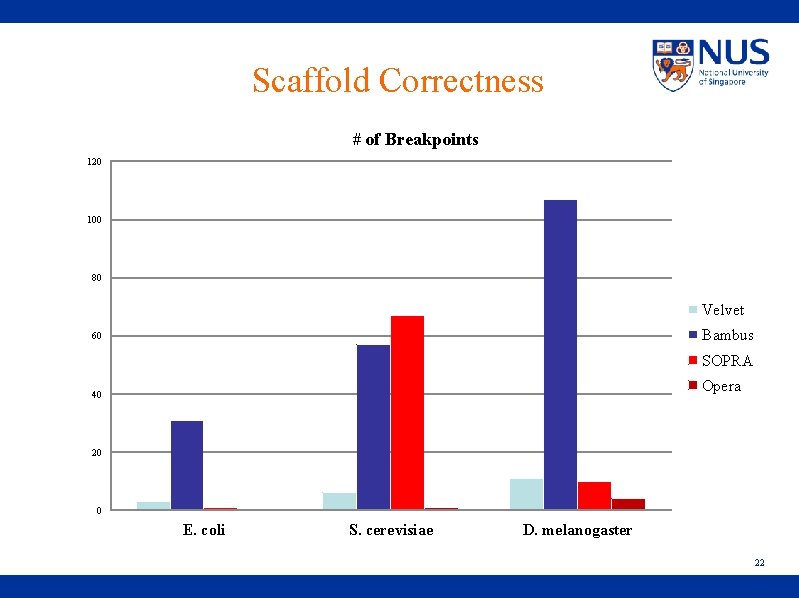
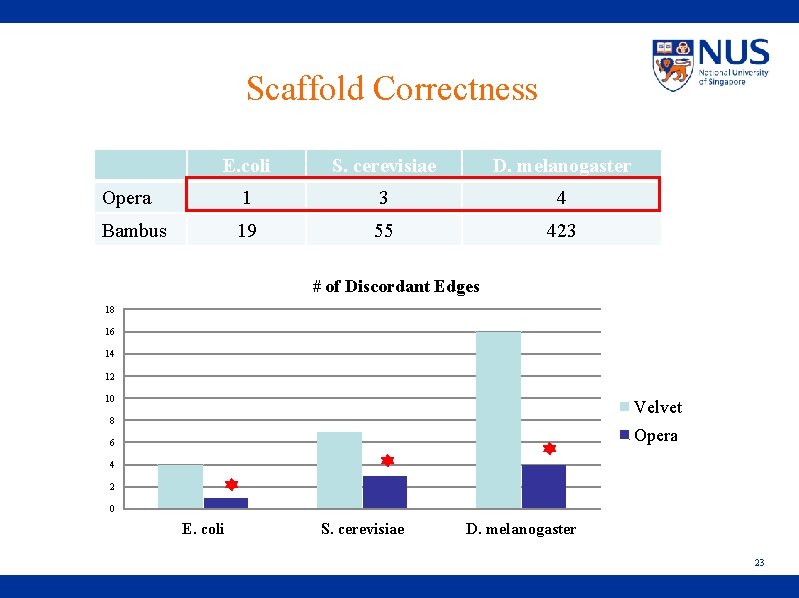
Scaffold Correctness # of Breakpoints 120 100 80 Velvet Bambus 60 SOPRA Opera 40 20 0 E. coli S. cerevisiae D. melanogaster 22

Scaffold Correctness E. coli S. cerevisiae D. melanogaster Opera 1 3 4 Bambus 19 55 423 # of Discordant Edges 18 16 14 12 10 Velvet 8 Opera 6 4 2 0 E. coli S. cerevisiae D. melanogaster 23

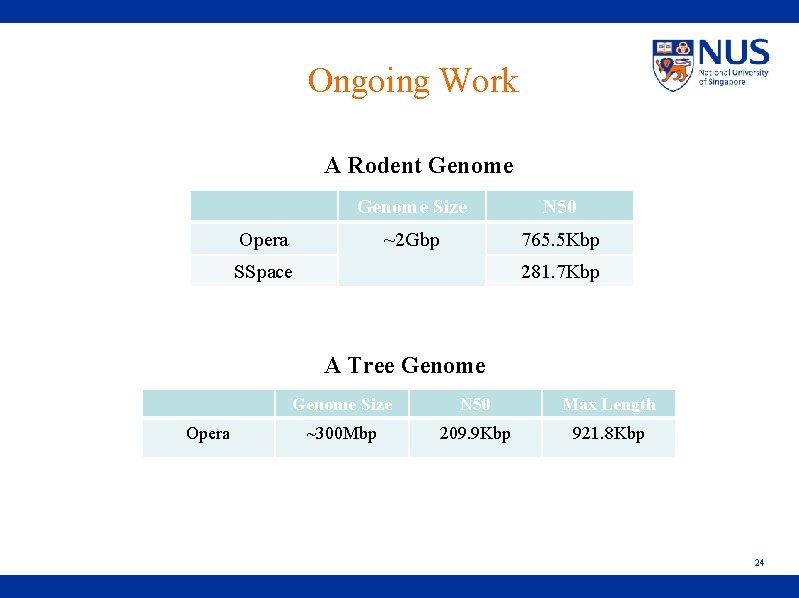
Ongoing Work A Rodent Genome Size N 50 ~2 Gbp 765. 5 Kbp Opera SSpace 281. 7 Kbp A Tree Genome Opera Genome Size N 50 Max Length ~300 Mbp 209. 9 Kbp 921. 8 Kbp 24

Ongoing Work Repeats Lower bounds and better scaffold Multiple Libraries Other applications Metagenomics Cancer Genomics Link: https: //sourceforge. net/projects/operasf/ 25

Acknowledgement Niranjan Nagarajan Wing-Kin Sung Pramila N. Ariyaratne Fundings: NUS Graduate School for Integrative Sciences and Engineering (NGS) A*STAR of Singapore Ministry of Education, Singapore Questions? 26