Reconstructing and Using Phylogenies Reconstructing and Using Phylogenies























- Slides: 60

Reconstructing and Using Phylogenies

Reconstructing and Using Phylogenies Key Concepts • Phylogeny Is the Basis of Biological Classification • All of Life Is Connected through Its Evolutionary History • Phylogeny Can Be Reconstructed from Traits of Organisms • Phylogeny Makes Biology Comparative and Predictive

Phylogeny Is the Basis of Biological Classification Phylogeny—the evolutionary history of evolutionary relationships Phylogenetic tree—a diagrammatic reconstruction of that history

Phylogeny Is the Basis of Biological Classification The biological classification system was started by Swedish biologist Carolus Linnaeus in the 1700 s. Binomial nomenclature gives every species a unique name consisting of two parts: the genus to which it belongs, and the species name. Example: • Homo sapiens Linnaeus (Linnaeus is the person who first proposed the name)

Phylogeny Is the Basis of Biological Classification Species and genera are further grouped into a hierarchical system of higher categories such as family—the taxon (classification group) above genus. Examples: • The family Hominidae contains humans, plus our recent fossil relatives, plus our closest living relatives, the chimpanzees and gorillas. • Rosaceae is the family that includes the genus Rosa (roses) and its relatives.

Phylogeny Is the Basis of Biological Classification Families are grouped into orders Orders into classes Classes into phyla (singular phylum) Phyla into kingdoms The ranking of taxa within the Linnaean classification is subjective.

Phylogeny Is the Basis of Biological Classification Linnaeus recognized the hierarchy of life, but he developed his system before evolutionary thought had become widespread. Today, biological classifications express the evolutionary relationships of organisms.

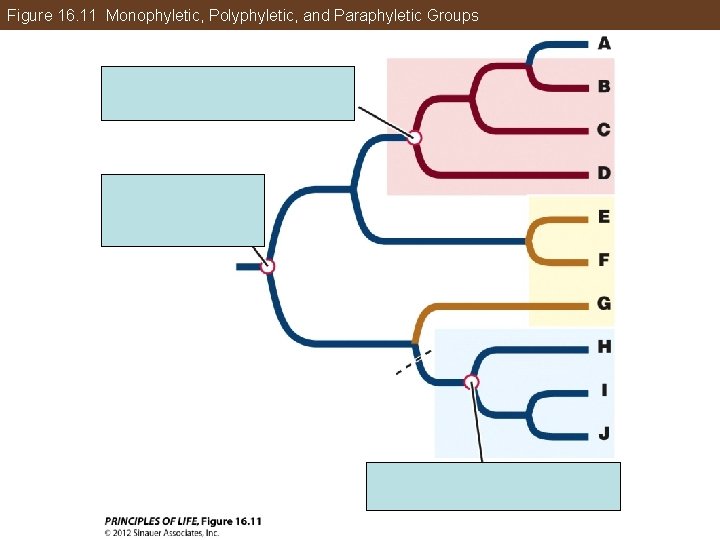
Phylogeny Is the Basis of Biological Classification But detailed phylogenetic information is not always available. Taxa are monophyletic—they contain an ancestor and all descendants of that ancestor, and no other organisms. Polyphyletic—a group that does not include its common ancestor Paraphyletic—a group that does not include all the descendants of a common ancestor

Figure 16. 11 Monophyletic, Polyphyletic, and Paraphyletic Groups

Phylogeny Is the Basis of Biological Classification Codes of biological nomenclature: Biologists around the world follow rules for the use of scientific names, to facilitate communication and dialogue. There may be many common names for one organism, or the same common name may refer to several species. But there is only one correct scientific name.

Figure 16. 12 Same Common Name, Not the Same Species

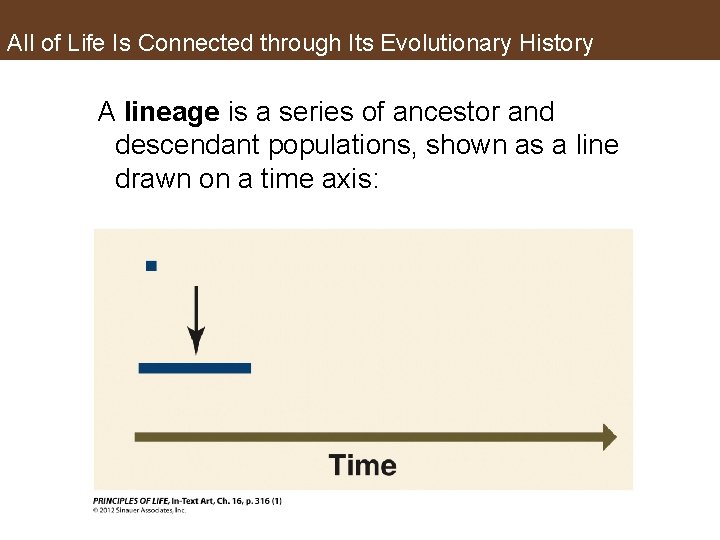
All of Life Is Connected through Its Evolutionary History A lineage is a series of ancestor and descendant populations, shown as a line drawn on a time axis:

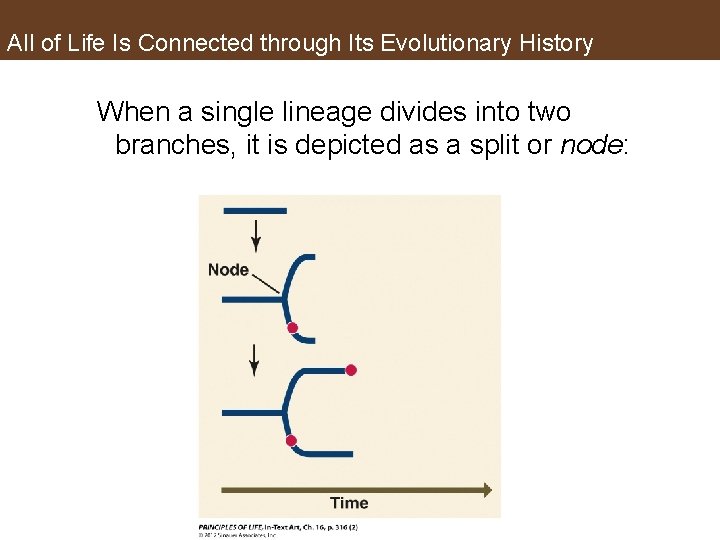
All of Life Is Connected through Its Evolutionary History When a single lineage divides into two branches, it is depicted as a split or node:

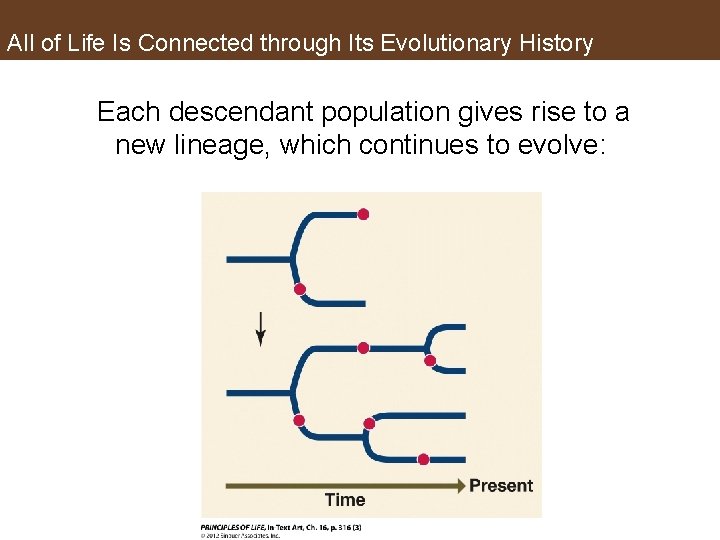
All of Life Is Connected through Its Evolutionary History Each descendant population gives rise to a new lineage, which continues to evolve:

All of Life Is Connected through Its Evolutionary History A phylogenetic tree may portray the evolutionary history of: • All life forms • Major evolutionary groups • Small groups of closely related species • Individuals • Populations • Genes

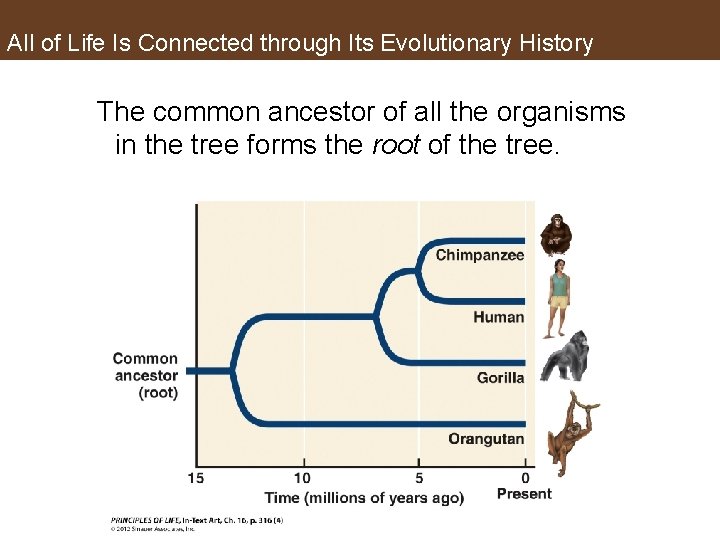
All of Life Is Connected through Its Evolutionary History The common ancestor of all the organisms in the tree forms the root of the tree.

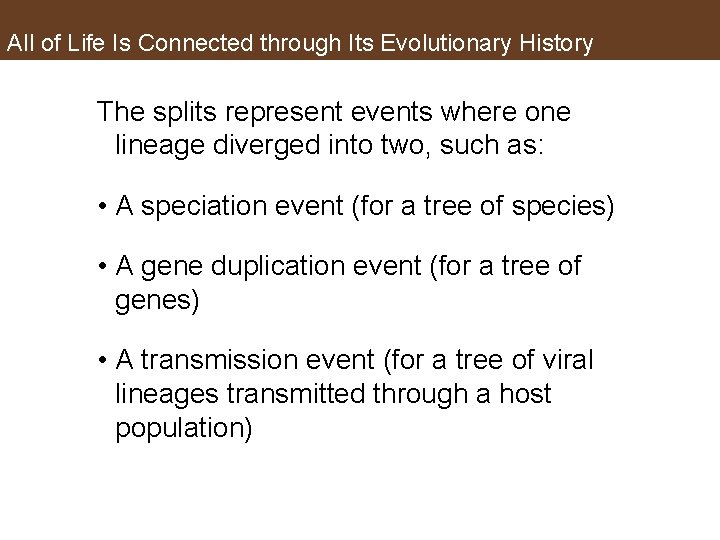
All of Life Is Connected through Its Evolutionary History The splits represent events where one lineage diverged into two, such as: • A speciation event (for a tree of species) • A gene duplication event (for a tree of genes) • A transmission event (for a tree of viral lineages transmitted through a host population)

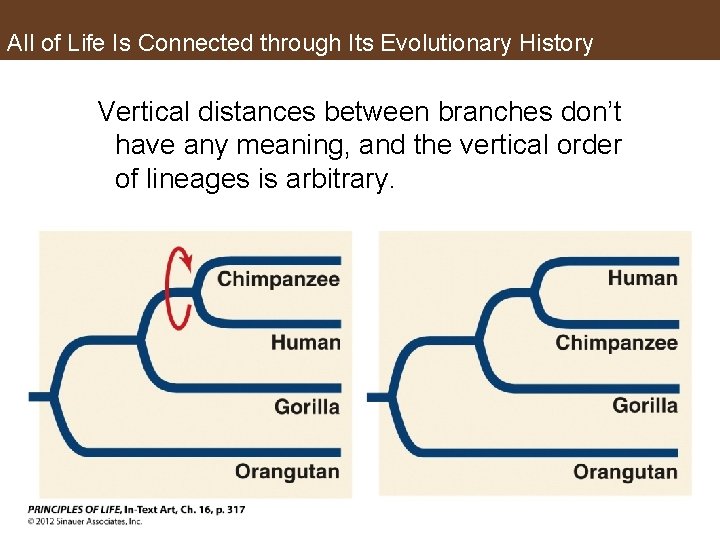
All of Life Is Connected through Its Evolutionary History Vertical distances between branches don’t have any meaning, and the vertical order of lineages is arbitrary.

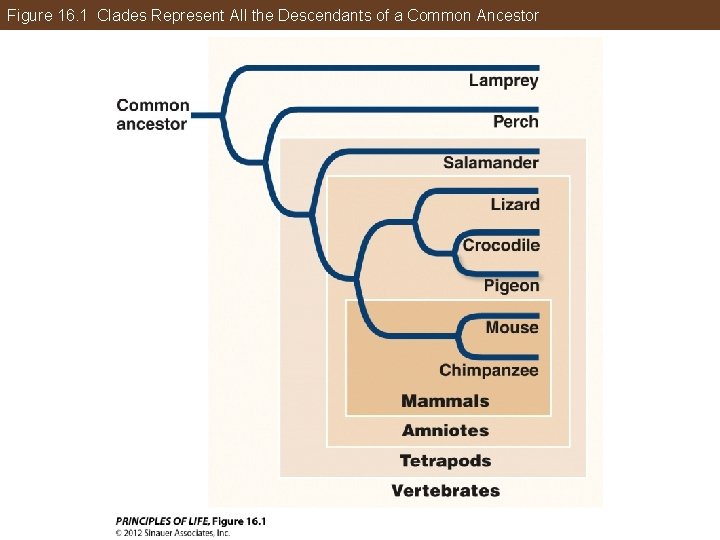
All of Life Is Connected through Its Evolutionary History Taxon—any group of species that we designate with a name Clade—taxon that consists of all the evolutionary descendants of a common ancestor Identify a clade by picking any point on the tree and tracing all the descendant lineages.

Figure 16. 1 Clades Represent All the Descendants of a Common Ancestor

All of Life Is Connected through Its Evolutionary History Sister species: Two species that are each other’s closest relatives Sister clades: Any two clades that are each other’s closest relatives Refer back to Figure 16. 1 and identify the sister species and clades

All of Life Is Connected through Its Evolutionary History Before the 1980 s, phylogenetic trees were used mostly in evolutionary biology, and in systematics—the study and classification of biodiversity. Today trees are widely used in molecular biology, biomedicine, physiology, behavior, ecology, and virtually all other fields of biology.

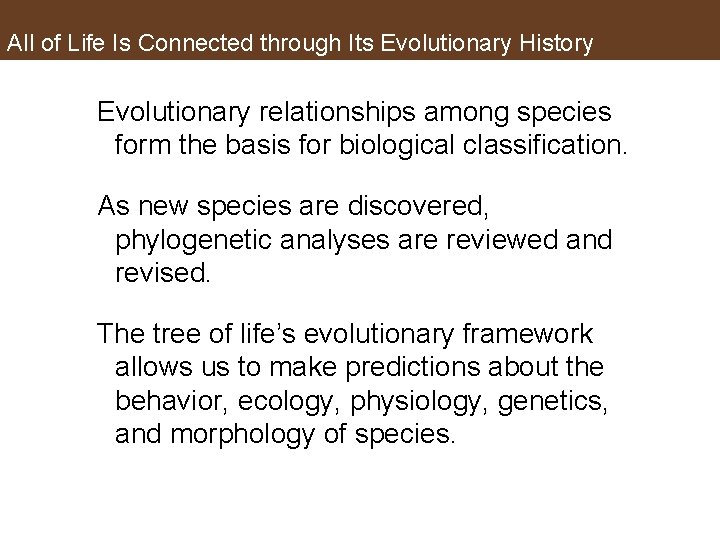
All of Life Is Connected through Its Evolutionary History Evolutionary relationships among species form the basis for biological classification. As new species are discovered, phylogenetic analyses are reviewed and revised. The tree of life’s evolutionary framework allows us to make predictions about the behavior, ecology, physiology, genetics, and morphology of species.

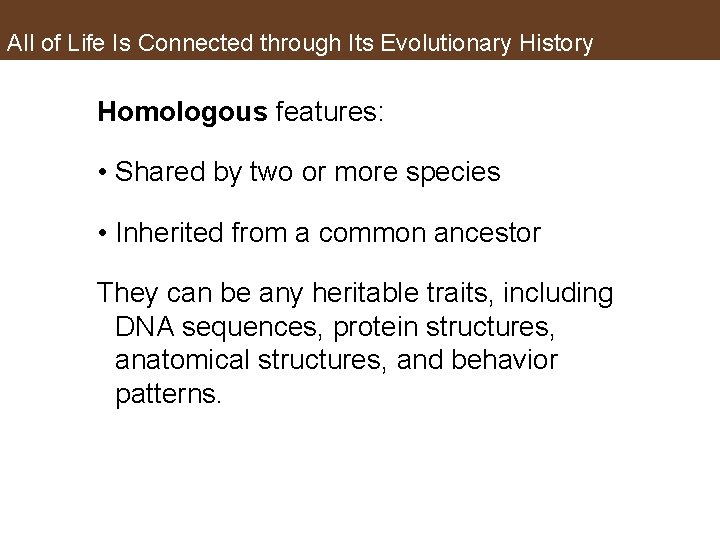
All of Life Is Connected through Its Evolutionary History Homologous features: • Shared by two or more species • Inherited from a common ancestor They can be any heritable traits, including DNA sequences, protein structures, anatomical structures, and behavior patterns.

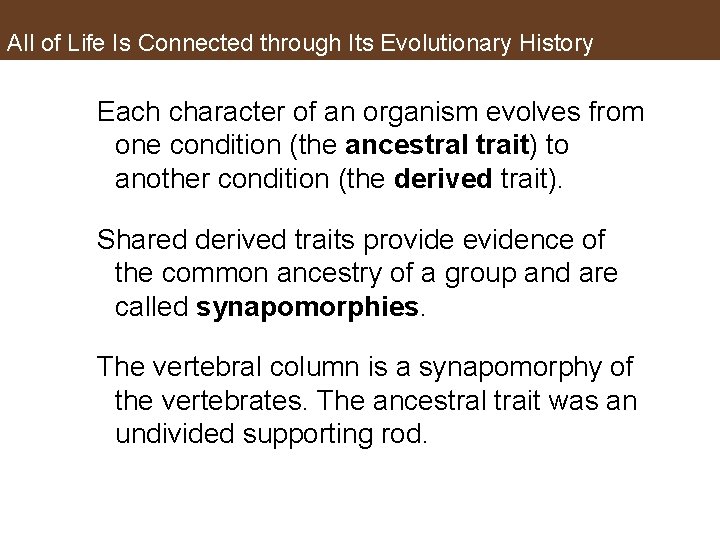
All of Life Is Connected through Its Evolutionary History Each character of an organism evolves from one condition (the ancestral trait) to another condition (the derived trait). Shared derived traits provide evidence of the common ancestry of a group and are called synapomorphies. The vertebral column is a synapomorphy of the vertebrates. The ancestral trait was an undivided supporting rod.

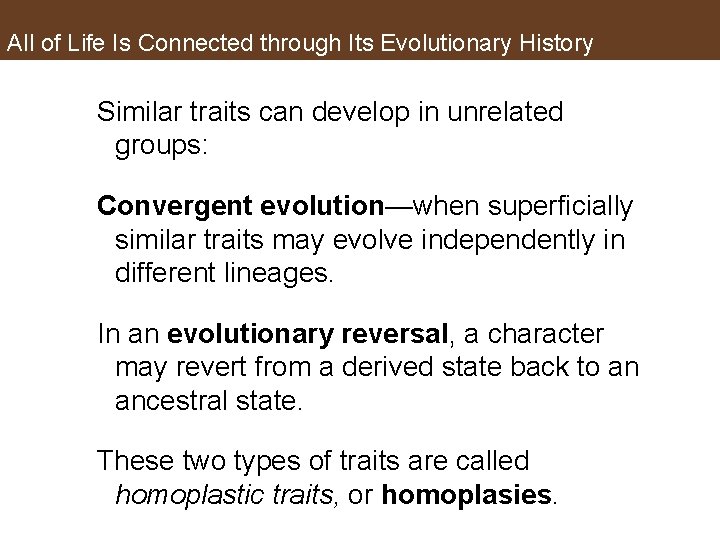
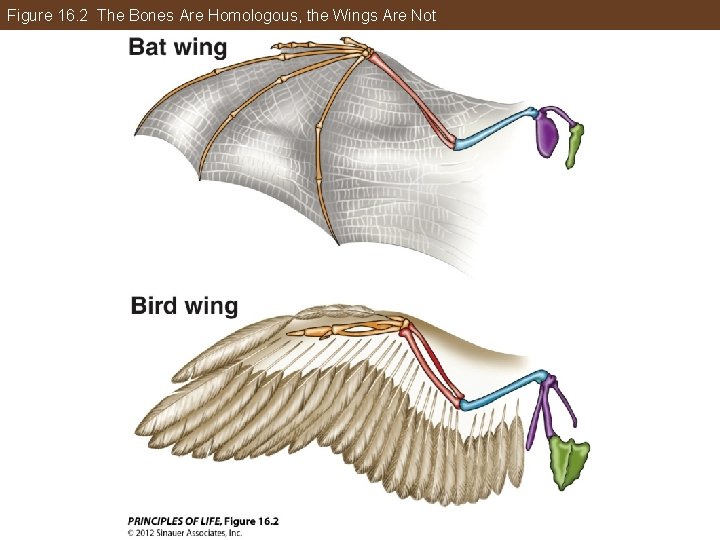
All of Life Is Connected through Its Evolutionary History Similar traits can develop in unrelated groups: Convergent evolution—when superficially similar traits may evolve independently in different lineages. In an evolutionary reversal, a character may revert from a derived state back to an ancestral state. These two types of traits are called homoplastic traits, or homoplasies.

Figure 16. 2 The Bones Are Homologous, the Wings Are Not

All of Life Is Connected through Its Evolutionary History A trait may be ancestral or derived, depending on the point of reference. Example: • Feathers are an ancestral trait for modern birds. But in a phylogeny of all living vertebrates, they are a derived trait found only in birds.

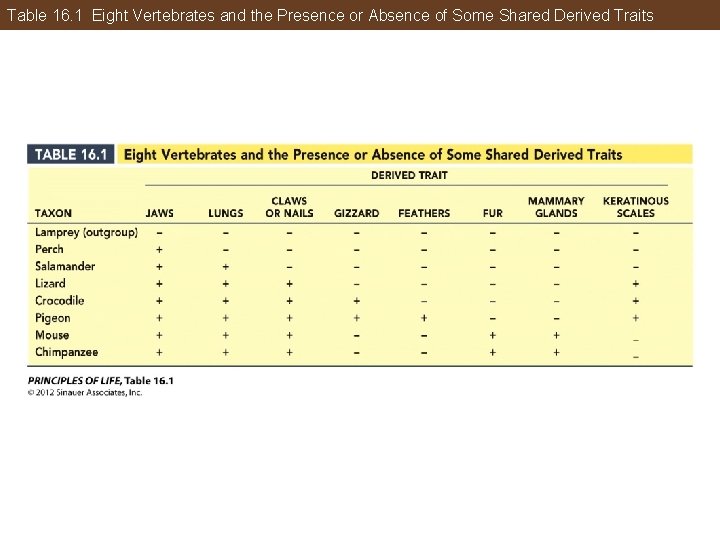
Phylogeny Can Be Reconstructed from Traits of Organisms Ingroup—the group of organisms of primary interest Outgroup—species or group known to be closely related to, but phylogenetically outside, the group of interest

Table 16. 1 Eight Vertebrates and the Presence or Absence of Some Shared Derived Traits

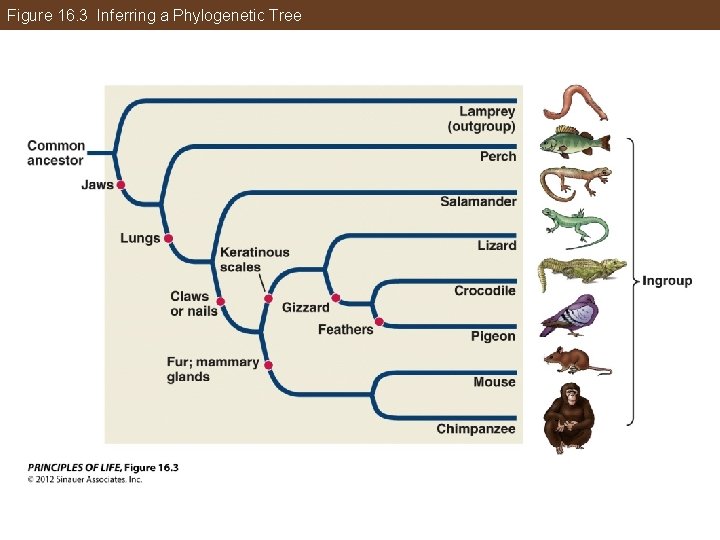
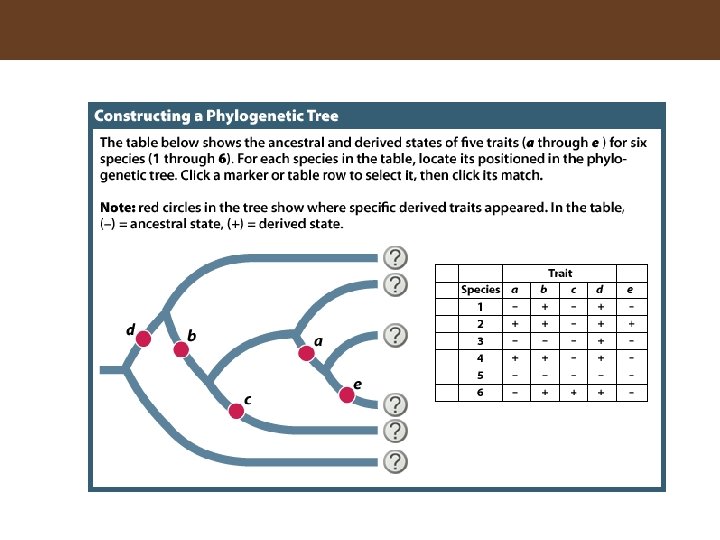
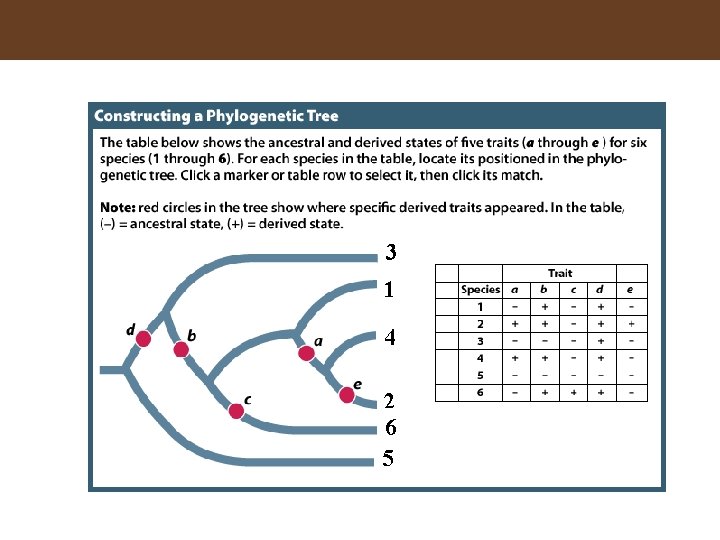
Figure 16. 3 Inferring a Phylogenetic Tree

Phylogeny Can Be Reconstructed from Traits of Organisms Parsimony principle—the preferred explanation of observed data is the simplest explanation In phylogenies, this entails minimizing the number of evolutionary changes that need to be assumed over all characters in all groups. The best hypothesis is one that requires the fewest homoplasies.

Phylogeny Can Be Reconstructed from Traits of Organisms Any trait that is genetically determined can be used in a phylogenetic analysis. Morphology—presence, size, shape, or other attributes of body parts Phylogenies of most extinct species depend almost exclusively on morphology. Fossils provide evidence that helps distinguish ancestral from derived traits. The fossil record can also reveal when lineages diverged.

Phylogeny Can Be Reconstructed from Traits of Organisms Limitations of using morphology: • Some taxa show few morphological differences • It is difficult to compare distantly related species • Some morphological variation is caused by environment

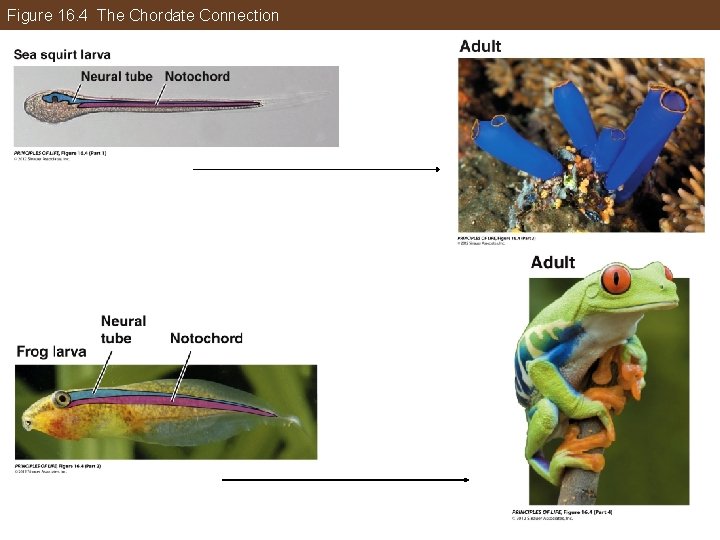
Phylogeny Can Be Reconstructed from Traits of Organisms Development: Similarities in developmental patterns may reveal evolutionary relationships. Example: • The larvae of sea squirts has a notochord, which is also present in all vertebrates.

Figure 16. 4 The Chordate Connection

Phylogeny Can Be Reconstructed from Traits of Organisms Behavior: Some traits are cultural or learned, and may not reflect evolutionary relationships (e. g. bird songs). Other traits have a genetic basis and can be used in phylogenies (e. g. frog calls).

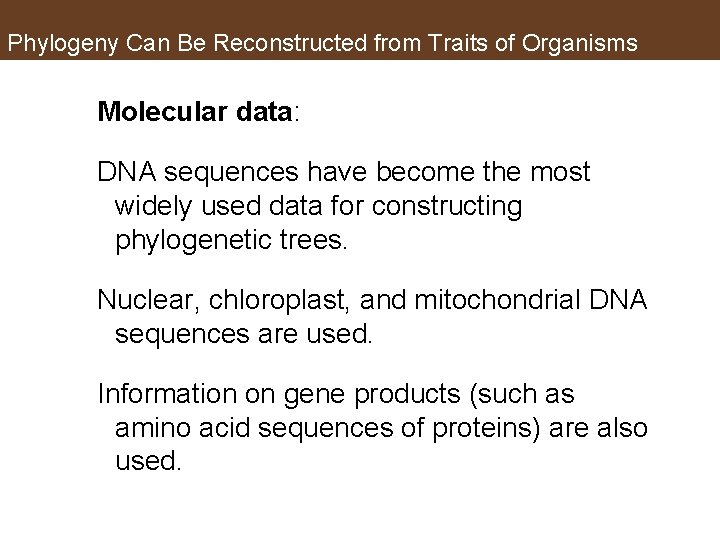
Phylogeny Can Be Reconstructed from Traits of Organisms Molecular data: DNA sequences have become the most widely used data for constructing phylogenetic trees. Nuclear, chloroplast, and mitochondrial DNA sequences are used. Information on gene products (such as amino acid sequences of proteins) are also used.

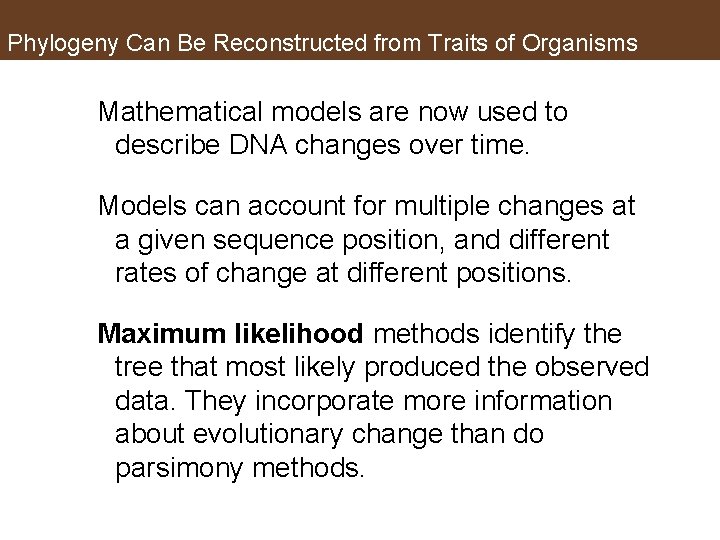
Phylogeny Can Be Reconstructed from Traits of Organisms Mathematical models are now used to describe DNA changes over time. Models can account for multiple changes at a given sequence position, and different rates of change at different positions. Maximum likelihood methods identify the tree that most likely produced the observed data. They incorporate more information about evolutionary change than do parsimony methods.

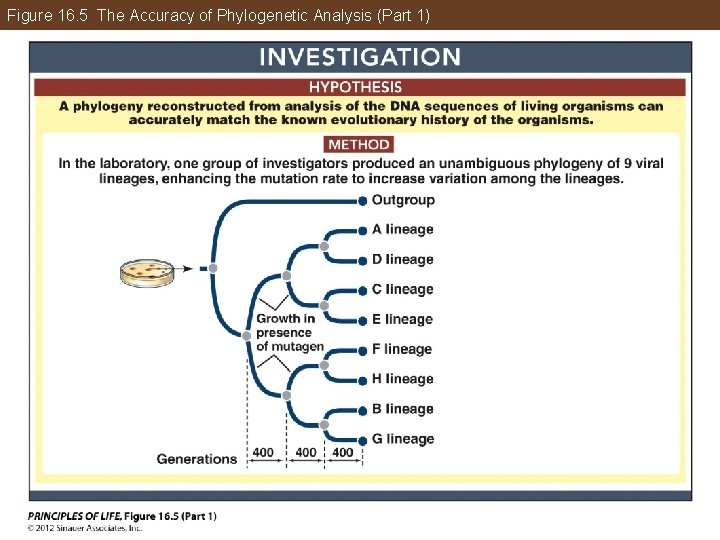
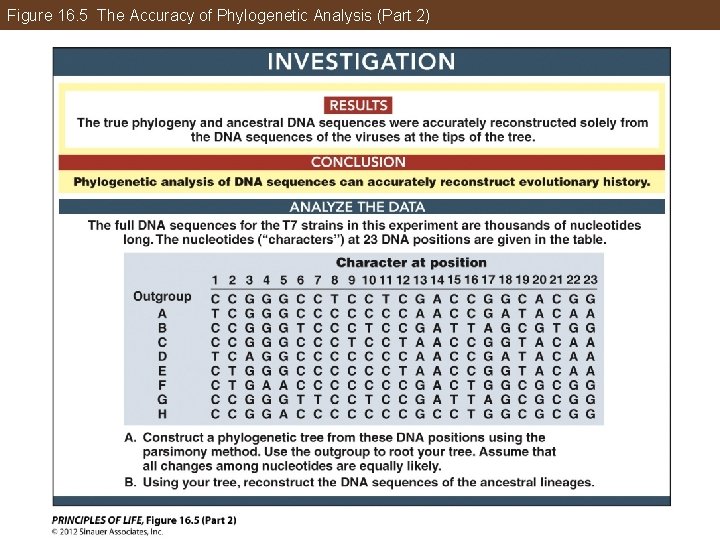
Phylogeny Can Be Reconstructed from Traits of Organisms Phylogenetic trees can be tested with computer simulations and by experiments on living organisms. These studies have confirmed the accuracy of phylogenetic methods and have been used to refine those methods and extend them to new applications.


3 1 4 2 6 5

Figure 16. 5 The Accuracy of Phylogenetic Analysis (Part 1)

Figure 16. 5 The Accuracy of Phylogenetic Analysis (Part 2)

Phylogeny Makes Biology Comparative and Predictive Applications of phylogenetic trees • Phylogeny can clarify the origin and evolution of traits that help in understanding fundamental biological processes. This information is then widely applied in life sciences fields, including agriculture and medicine.

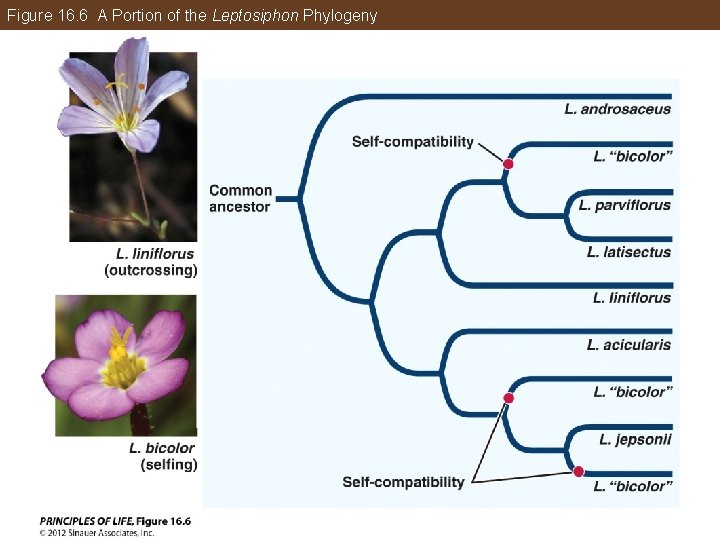
Phylogeny Makes Biology Comparative and Predictive Self-compatibility: • Most flowering plants reproduce by mating with another individual (outcrossing) Self-incompatible species have mechanisms to prevent self-fertilization. Other plants are selfing, which requires that they be self-compatible. The evolution of angiosperm fertilization mechanisms was examined in the genus Leptosiphon.

Figure 16. 6 A Portion of the Leptosiphon Phylogeny

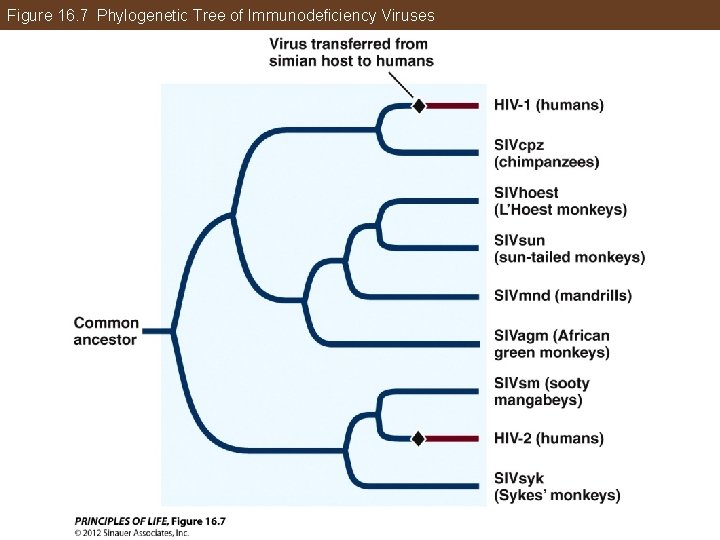
Phylogeny Makes Biology Comparative and Predictive Zoonotic diseases: • Caused by infectious organisms transmitted from an animal of a different species (e. g. rabies, AIDS) Phylogenetic analyses help determine when, where, and how a disease first entered a human population. One example is Human Immunodeficiency Virus (HIV).

Figure 16. 7 Phylogenetic Tree of Immunodeficiency Viruses

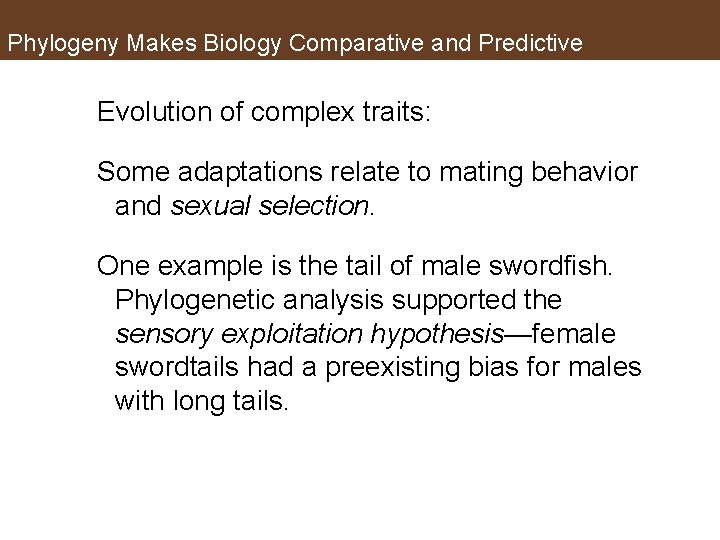
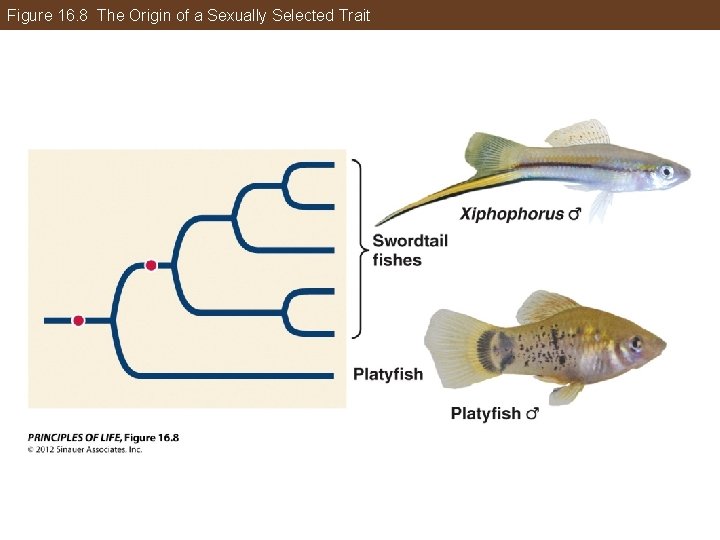
Phylogeny Makes Biology Comparative and Predictive Evolution of complex traits: Some adaptations relate to mating behavior and sexual selection. One example is the tail of male swordfish. Phylogenetic analysis supported the sensory exploitation hypothesis—female swordtails had a preexisting bias for males with long tails.

Figure 16. 8 The Origin of a Sexually Selected Trait

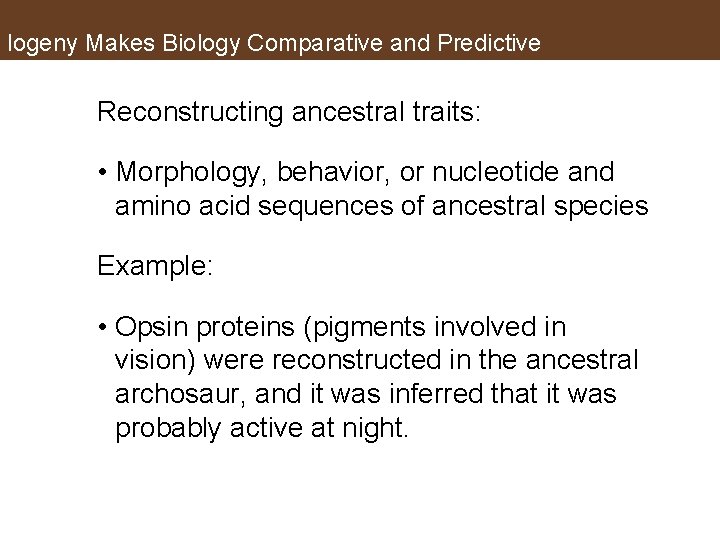
logeny Makes Biology Comparative and Predictive Reconstructing ancestral traits: • Morphology, behavior, or nucleotide and amino acid sequences of ancestral species Example: • Opsin proteins (pigments involved in vision) were reconstructed in the ancestral archosaur, and it was inferred that it was probably active at night.

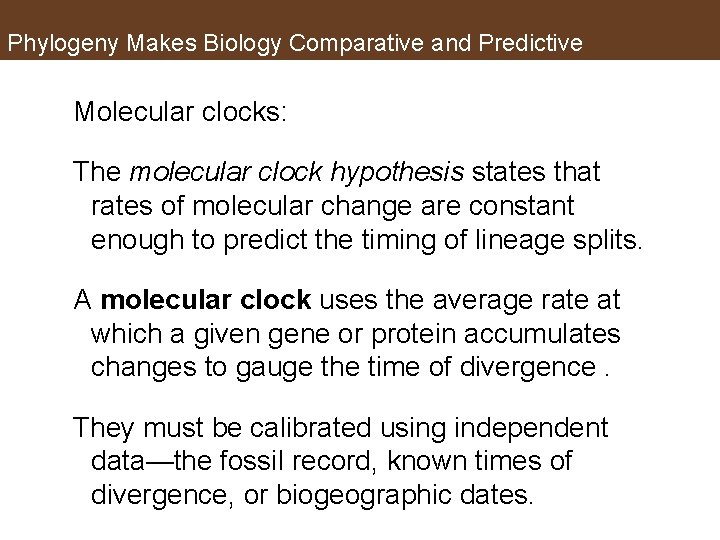
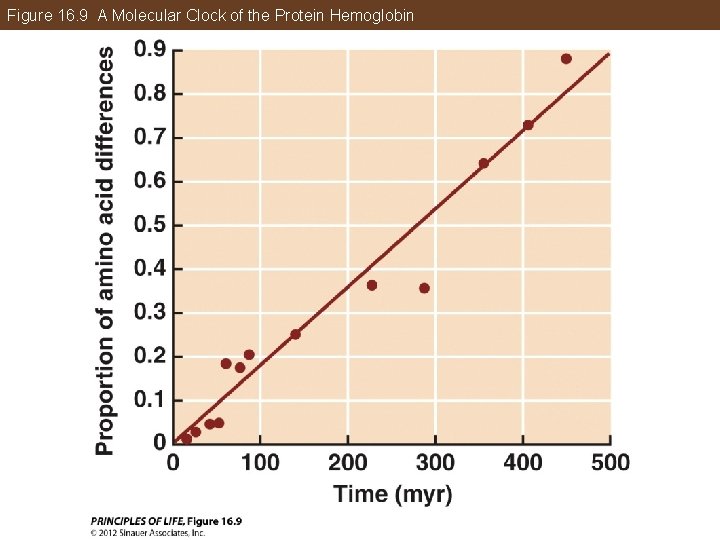
Phylogeny Makes Biology Comparative and Predictive Molecular clocks: The molecular clock hypothesis states that rates of molecular change are constant enough to predict the timing of lineage splits. A molecular clock uses the average rate at which a given gene or protein accumulates changes to gauge the time of divergence. They must be calibrated using independent data—the fossil record, known times of divergence, or biogeographic dates.

Figure 16. 9 A Molecular Clock of the Protein Hemoglobin

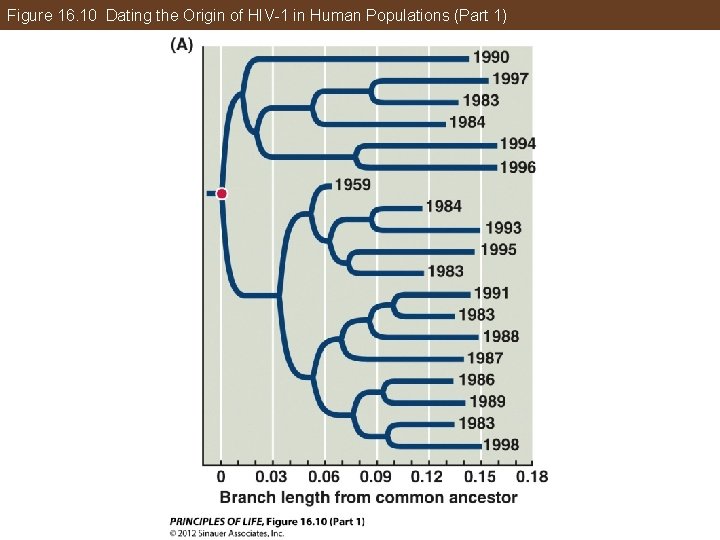
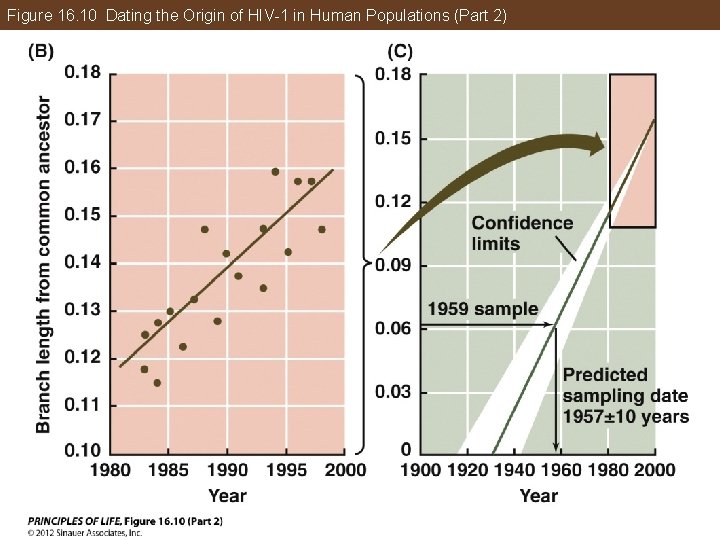
Phylogeny Makes Biology Comparative and Predictive A molecular clock was used to estimate the time when HIV-1 first entered human populations from chimpanzees. The clock was calibrated using the samples from the 1980 s and 1990 s, then tested using the samples from the 1950 s. The common ancestor of this group of HIV-1 viruses can also be determined, with an estimated date of origin of about 1930.

Figure 16. 10 Dating the Origin of HIV-1 in Human Populations (Part 1)

Figure 16. 10 Dating the Origin of HIV-1 in Human Populations (Part 2)

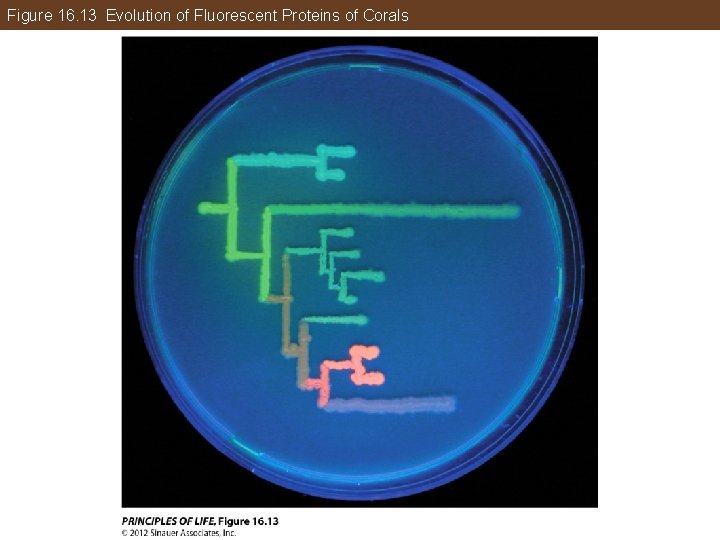
Figure 16. 13 Evolution of Fluorescent Proteins of Corals

Chapter 16 Opening Question How are phylogenetic methods used to resurrect protein sequences from extinct organisms?

Answer to Opening Question Biologists can reconstruct DNA and protein sequences of a clade’s ancestors if there is enough information about the genomes of their descendants. Real proteins that correspond to proteins in longextinct species can be reconstructed. Mathematical models that incorporates of replacement among different amino acid residues, substitution rates among nucleotides, and changes in the rate of molecular evolution among different lineages, are used.