Mapping sequence reads Calling variants NC000012 11 g














- Slides: 50

Mapping sequence reads & Calling variants NC_000012. 11: g. 21995261 C>T Martin Elferink Postdoc Bioinformatician, Genome Diagnostics, UMC Utrecht 23 -09 -2019

Genome ditiseendenkbeeldiggenoomvaneentetestensample ise ? ? ? fragmented DNA dit ? ? ? end ? ? ? bee ? ? ? gge Sequencing enk ? ? ? enk dit end sam bee ldi ? ? ? noo ? ? ? gge ? ? ? tes ise tes ? ? ? nee ? ? ? mva ? ? ? noo nte ? ? ? ten ? ? ? nte ten Hundred million reads vs billion position in the genome Mapping (using a reference genome) Variant Calling ditiseendenkbeeldiggenoomvaneentetestensample ditiseendenkbeeidiggenoomvaneentetestensample ditiseendenkbeeldiggenqomvaneentetestensample ditiveendenkbeeldiggenoomvan entetestensample ? ? ? nee ldi mva ? ? ? sam ple

Part 1: Short read illumina sequencing Part 2: Long read Oxford Nanopore sequencing

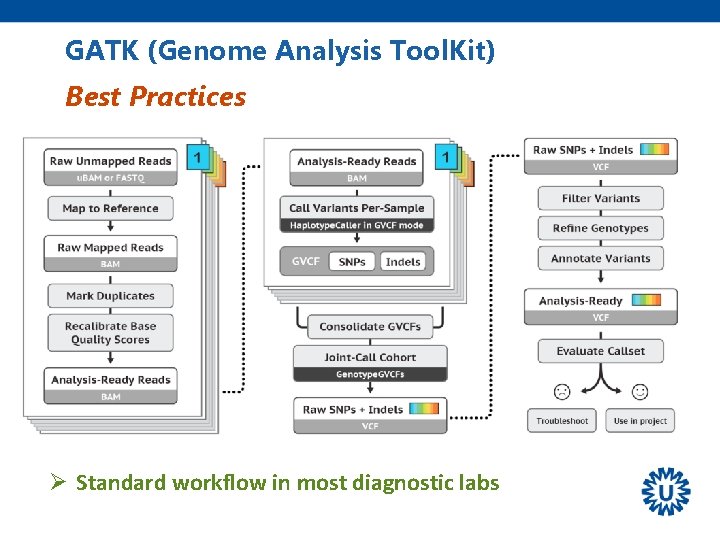
Part 1. GATK best practices for Illumina NGS • Introduction to GATK (best practice guidelines): – mapping and variant calling • File formats: – FASTQ format – BAM format – Variant Calling Format (VCF) Focus: • (Illumina) short-read sequencing • Reference based mapping and variant calling • Germline mutations in a diploid genome • Single nucleotide variants (SNV) and small insertions/deletions (INDEL)

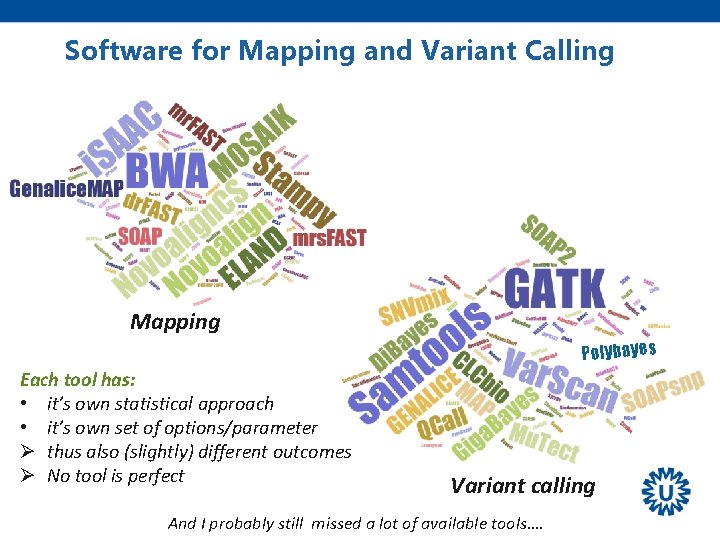
Software for Mapping and Variant Calling Mapping Polybayes Each tool has: • it’s own statistical approach • it’s own set of options/parameter Ø thus also (slightly) different outcomes Ø No tool is perfect Variant calling And I probably still missed a lot of available tools….

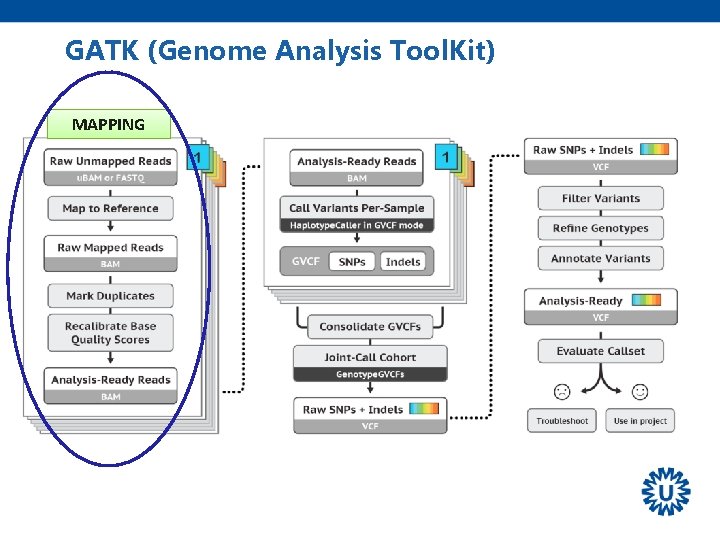
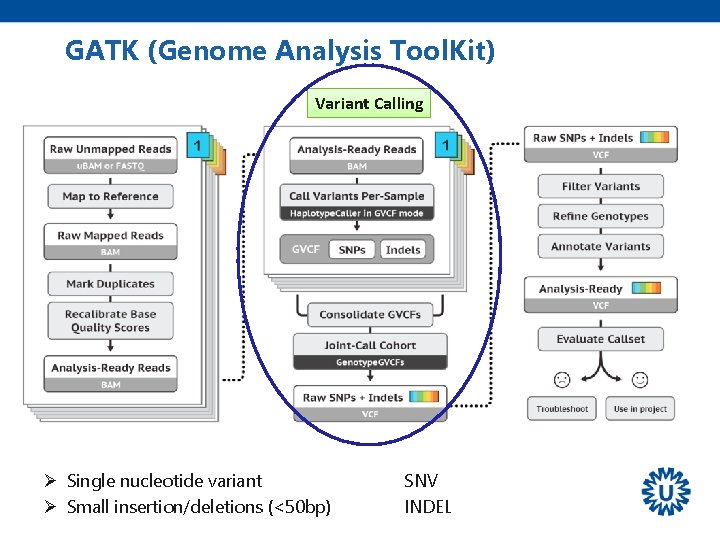
GATK (Genome Analysis Tool. Kit) Best Practices Ø Standard workflow in most diagnostic labs

GATK (Genome Analysis Tool. Kit) MAPPING

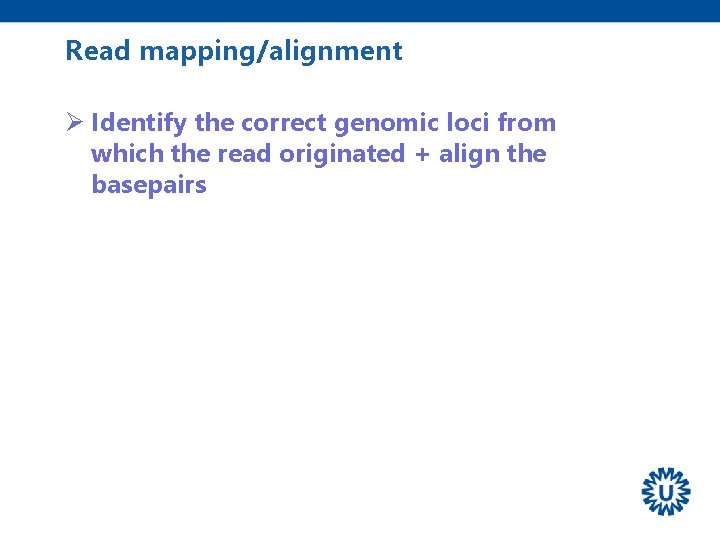
Read mapping/alignment Ø Identify the correct genomic loci from which the read originated + align the basepairs

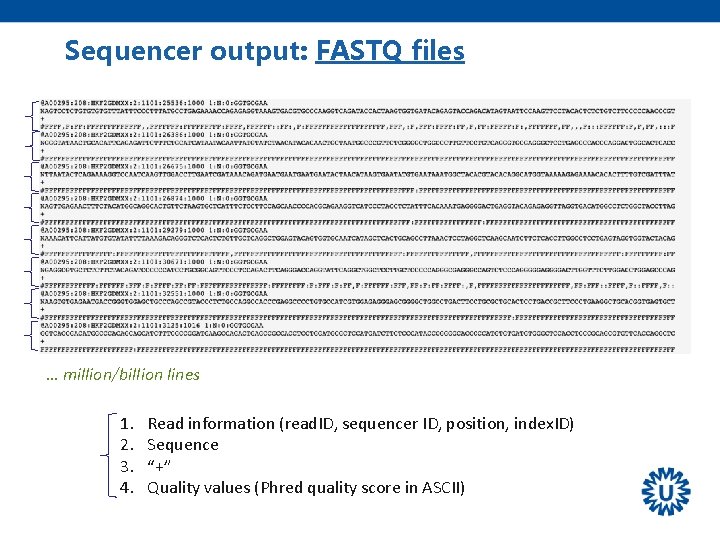
Sequencer output: FASTQ files … million/billion lines 1. 2. 3. 4. Read information (read. ID, sequencer ID, position, index. ID) Sequence “+” Quality values (Phred quality score in ASCII)

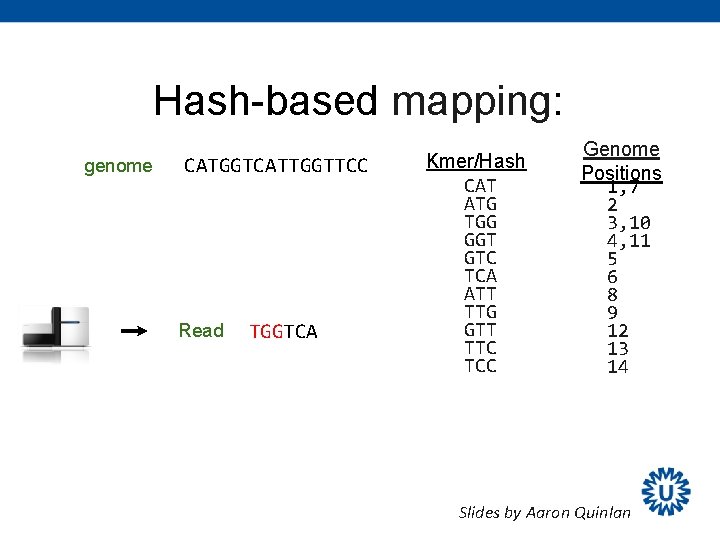
Mapping sequence reads Software: BWA (Burrow-Wheeler Aligner) Human Reference Sequence gtgccaggaccagatcgtgccaacggacaggtgctaagga Sequence read gtgccaagga Ø Mapping: determine the genomic location of the read Ø Alignment: alignment of the nucleotides between reference and read

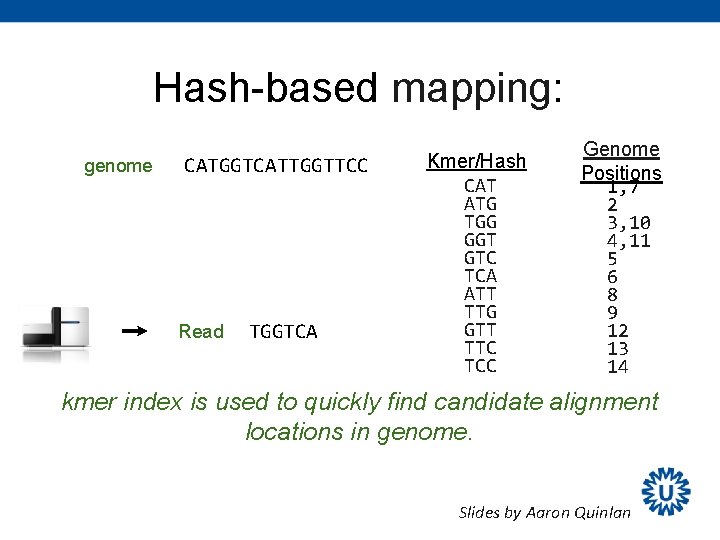
Hash-based mapping: genome CATGGTCATTGGTTCC Read TGGTCA Kmer/Hash CAT ATG TGG GGT GTC TCA ATT TTG GTT TTC TCC Genome Positions 1, 7 2 3, 10 4, 11 5 6 8 9 12 13 14 kmer index is used to quickly find candidate alignment locations in genome. Slides by Aaron Quinlan

Hash-based mapping: genome CATGGTCATTGGTTCC Read TGGTCA Kmer/Hash CAT ATG TGG GGT GTC TCA ATT TTG GTT TTC TCC Genome Positions 1, 7 2 3, 10 4, 11 5 6 8 9 12 13 14 Slides by Aaron Quinlan

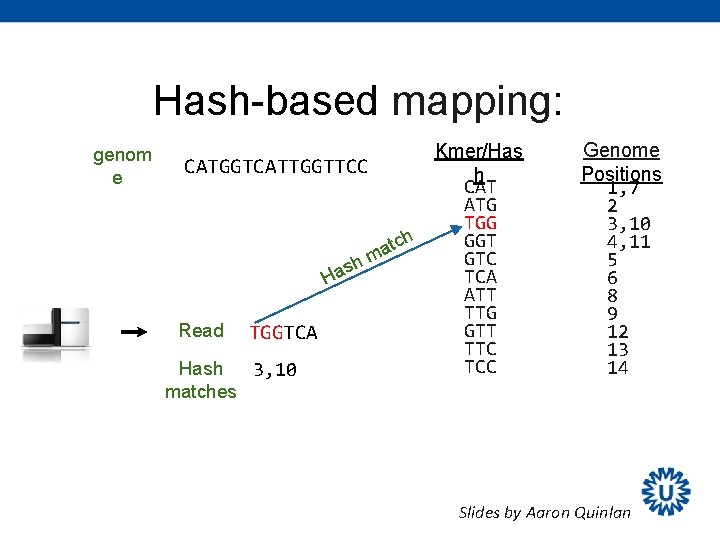
Hash-based mapping: genom e Kmer/Has CATGGTCATTGGTTCC h CAT ATG TGG GGT tch a m GTC h s a TCA H ATT TTG Read TGGTCA GTT TTC TCC Hash 3, 10 Genome Positions 1, 7 2 3, 10 4, 11 5 6 8 9 12 13 14 matches Slides by Aaron Quinlan

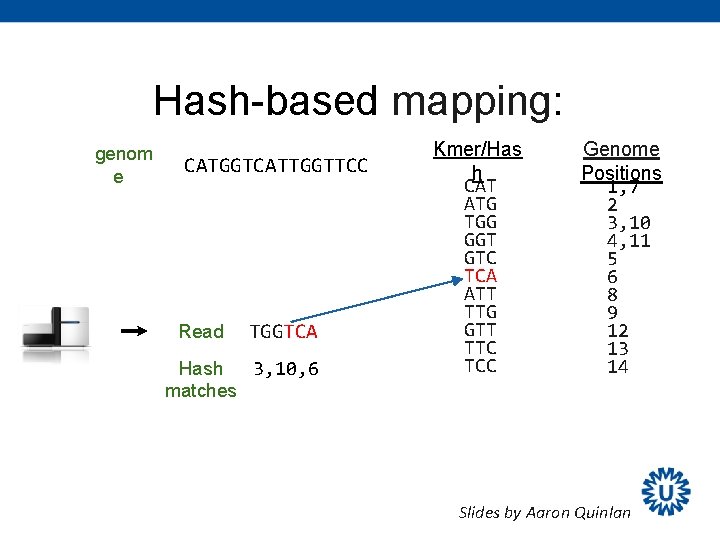
Hash-based mapping: genom e CATGGTCATTGGTTCC Read TGGTCA Hash 3, 10, 6 matches Kmer/Has h CAT ATG TGG GGT GTC TCA ATT TTG GTT TTC TCC Genome Positions 1, 7 2 3, 10 4, 11 5 6 8 9 12 13 14 Slides by Aaron Quinlan

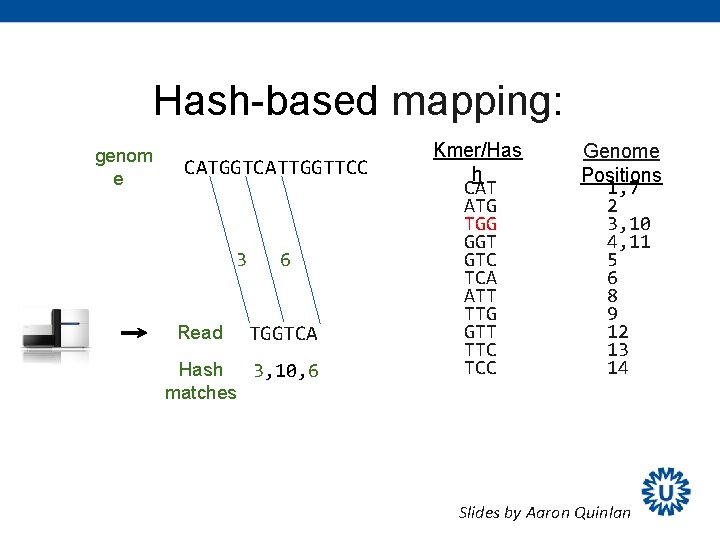
Hash-based mapping: genom e CATGGTCATTGGTTCC 3 Read 6 TGGTCA Hash 3, 10, 6 matches Kmer/Has h CAT ATG TGG GGT GTC TCA ATT TTG GTT TTC TCC Genome Positions 1, 7 2 3, 10 4, 11 5 6 8 9 12 13 14 Slides by Aaron Quinlan

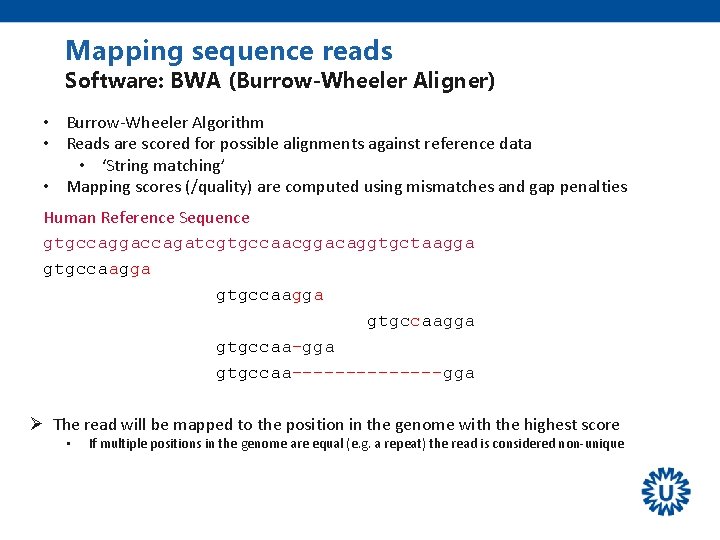
Mapping sequence reads Software: BWA (Burrow-Wheeler Aligner) • Burrow-Wheeler Algorithm • Reads are scored for possible alignments against reference data • ‘String matching’ • Mapping scores (/quality) are computed using mismatches and gap penalties Human Reference Sequence gtgccaggaccagatcgtgccaacggacaggtgctaagga gtgccaagga gtgccaa-gga gtgccaa-------gga Ø The read will be mapped to the position in the genome with the highest score • If multiple positions in the genome are equal (e. g. a repeat) the read is considered non-unique

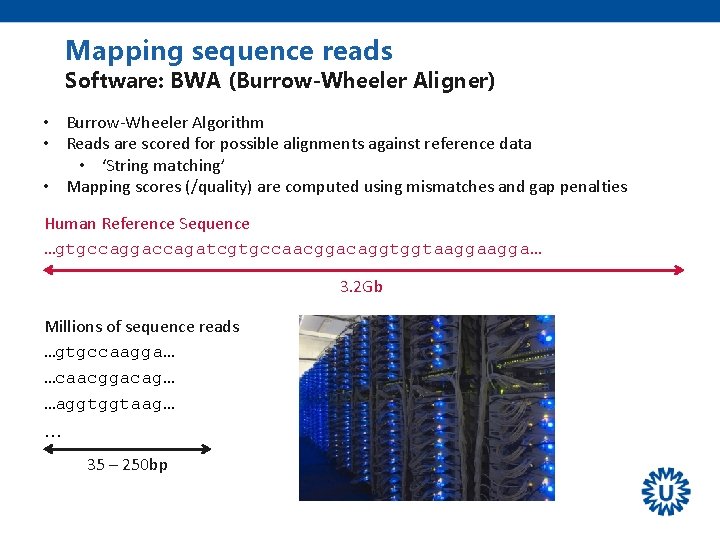
Mapping sequence reads Software: BWA (Burrow-Wheeler Aligner) • Burrow-Wheeler Algorithm • Reads are scored for possible alignments against reference data • ‘String matching’ • Mapping scores (/quality) are computed using mismatches and gap penalties Human Reference Sequence …gtgccaggaccagatcgtgccaacggacaggtggtaagga… 3. 2 Gb Millions of sequence reads …gtgccaagga… …caacggacag… …aggtggtaag… … 35 – 250 bp

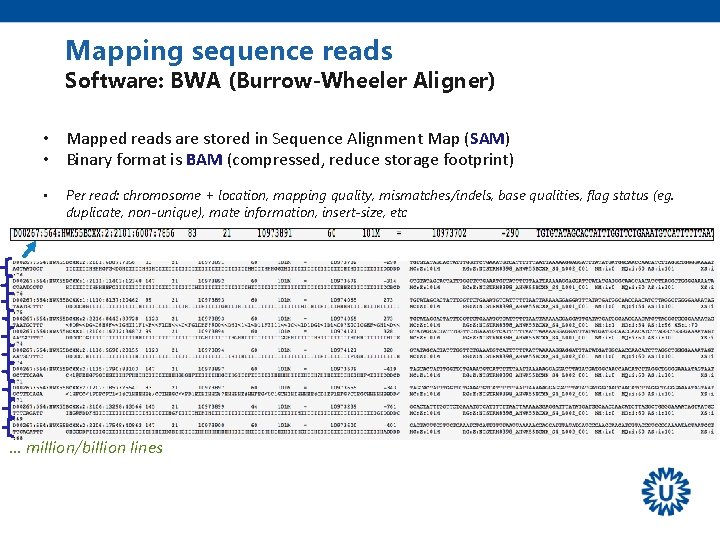
Mapping sequence reads Software: BWA (Burrow-Wheeler Aligner) • Mapped reads are stored in Sequence Alignment Map (SAM) • Binary format is BAM (compressed, reduce storage footprint) • Per read: chromosome + location, mapping quality, mismatches/indels, base qualities, flag status (eg. duplicate, non-unique), mate information, insert-size, etc … million/billion lines

GATK (Genome Analysis Tool. Kit) Variant Calling Ø Single nucleotide variant Ø Small insertion/deletions (<50 bp) SNV INDEL

Variant Calling Ø Variant calling is the process by which we identify variants from sequence data – Taking into account noise of sequencing and biology

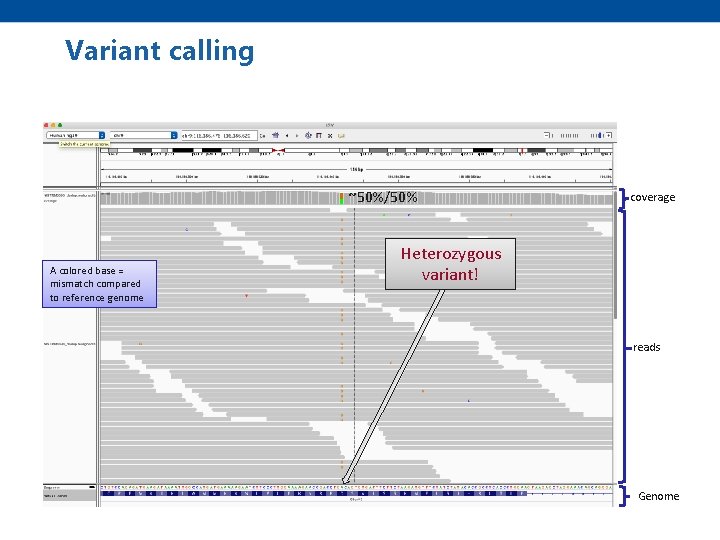
Variant calling ~50%/50% A colored base = mismatch compared to reference genome coverage Heterozygous variant! reads Genome

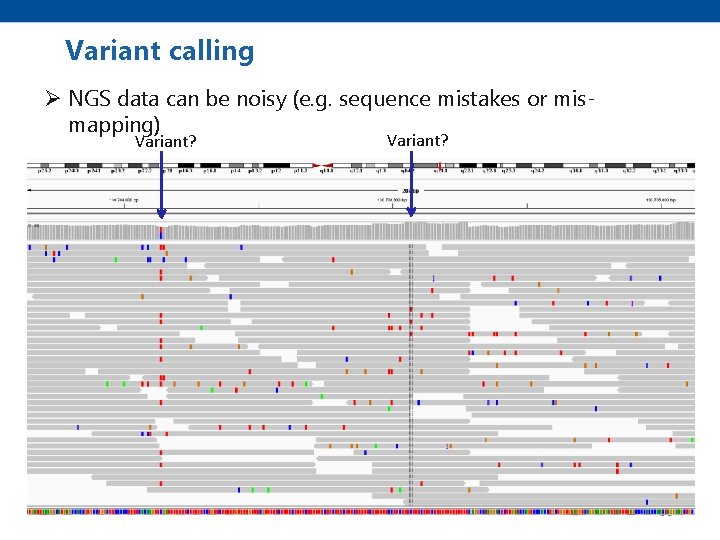
Variant calling Ø NGS data can be noisy (e. g. sequence mistakes or mismapping) Variant?

Variant calling Ø NGS data can be noisy – non-uniquely mapped reads (multiple locations in the reference genome) • e. g. duplications, repeats, pseudo/paralogous genes (in the reference genome) – flagged/filtered reads (e. g. read duplicates).

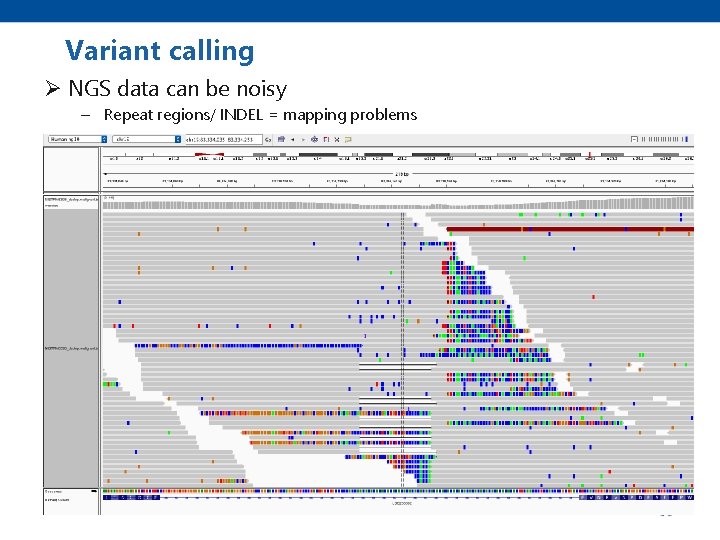
Variant calling Ø NGS data can be noisy – Repeat regions/ INDEL = mapping problems

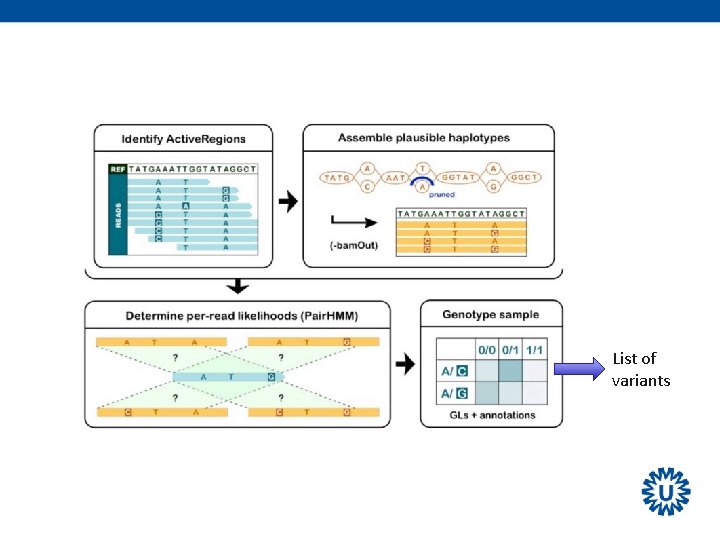
Variant calling GATK Haplotype. Caller algorithm • Calls SNVs and INDELs simultaneously • Uses all bases connected through local de novo assembly ØAccurate, especially for INDELs ØSupports “N+1” genotyping (Multisample calling) Øgenomic VCF (g. VCF) output

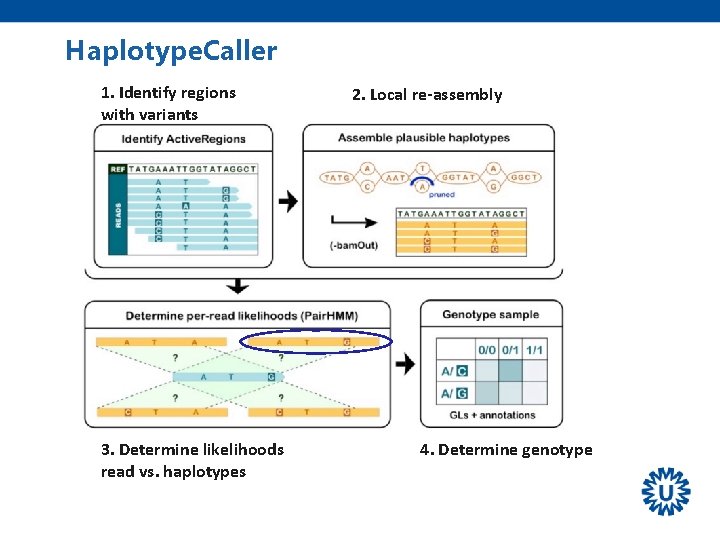
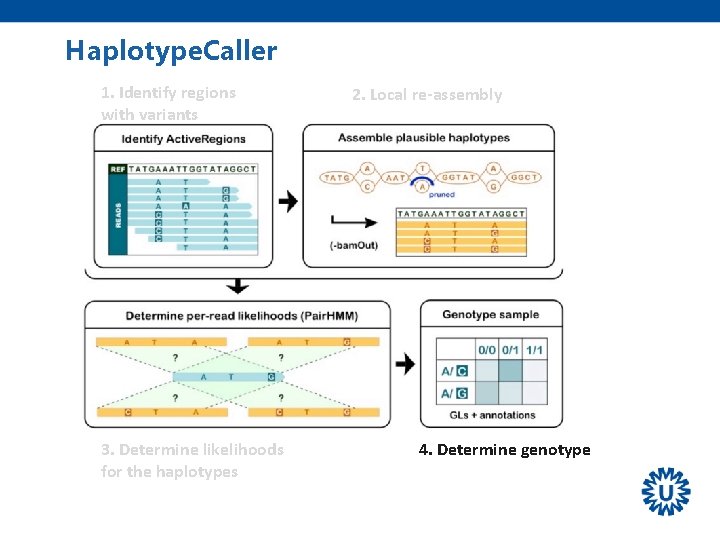
Haplotype. Caller 1. Identify regions with variants 3. Determine likelihoods read vs. haplotypes 2. Local re-assembly 4. Determine genotype

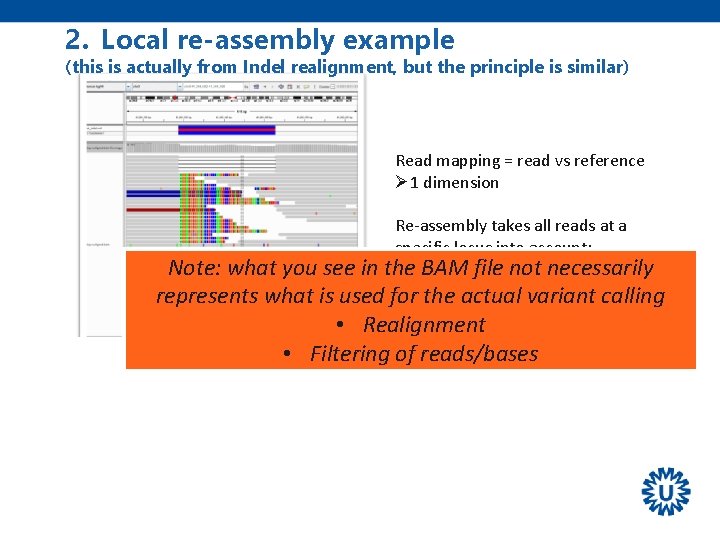
2. Local re-assembly example (this is actually from Indel realignment, but the principle is similar) Read mapping = read vs reference Ø 1 dimension Re-assembly takes all reads at a specific locus into account: Ø 2 BAM dimensions the file not necessarily Note: what you see in represents what is used for the actual variant calling • Realignment Increased accuracy, especially for detection! • Filtering INDEL of reads/bases

Haplotype. Caller 1. Identify regions with variants 3. Determine likelihoods for the haplotypes 2. Local re-assembly 4. Determine genotype

Variant calling Ø Coverage is important Random sampling Assuming a true heterozygous variant C/T (expect 50% C and 50% T) Human Reference Sequence …gtgccaggaccagatcg… gtgccaggacc @1 X gtgccaggacc @2 X gtgccaggacc @3 X gtgccaggacc @4 X gtgccaggacc @5 X 50% chance not to sample the T-allele 25% 12. 5% 6. 25% 3. 125% … @15 X 0, 003% Ø Increased coverage will increase detection of alleles Note: assuming an unbiased approach, which is not necessarily the case with sequence capture (such as WES), PCR amplifications, and reference based mapping.

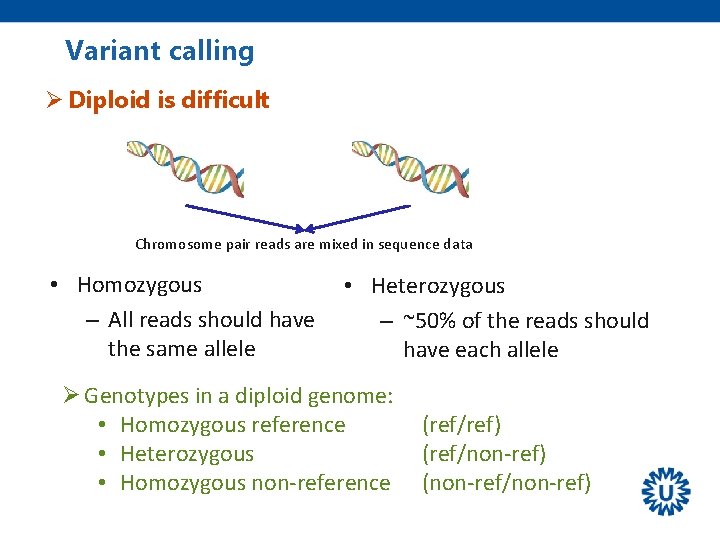
Variant calling Ø Diploid is difficult Chromosome pair reads are mixed in sequence data • Homozygous – All reads should have the same allele • Heterozygous – ~50% of the reads should have each allele Ø Genotypes in a diploid genome: • Homozygous reference • Heterozygous • Homozygous non-reference (ref/ref) (ref/non-ref) (non-ref/non-ref)

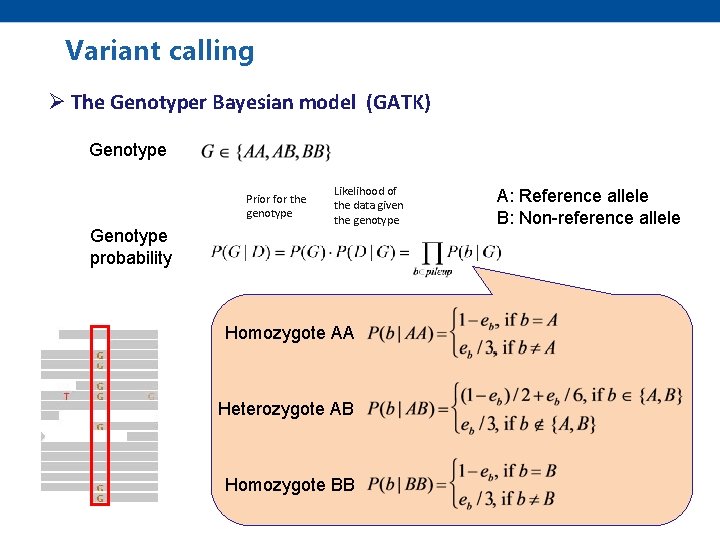
Variant calling Ø The Genotyper Bayesian model (GATK) Genotype Prior for the genotype Genotype probability Likelihood of the data given the genotype Homozygote AA Heterozygote AB Homozygote BB A: Reference allele B: Non-reference allele

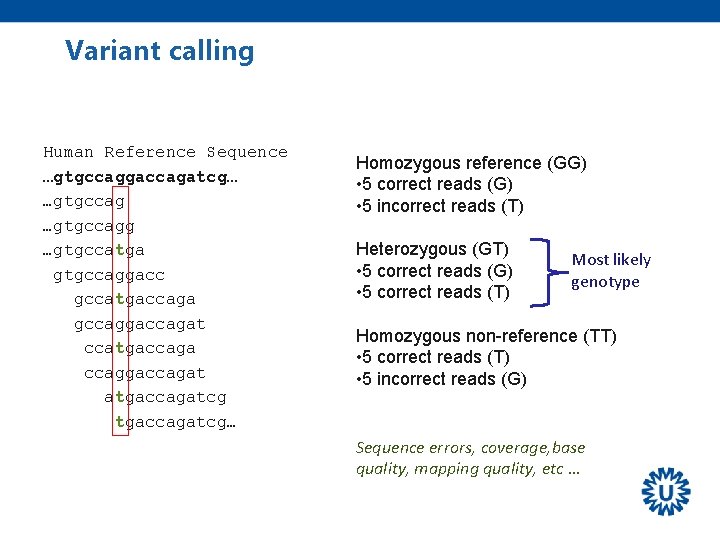
Variant calling Human Reference Sequence …gtgccaggaccagatcg… …gtgccagg …gtgccatga gtgccaggacc gccatgaccaga gccaggaccagat ccatgaccaga ccaggaccagat atgaccagatcg… Homozygous reference (GG) • 5 correct reads (G) • 5 incorrect reads (T) Heterozygous (GT) • 5 correct reads (G) • 5 correct reads (T) Most likely genotype Homozygous non-reference (TT) • 5 correct reads (T) • 5 incorrect reads (G) Sequence errors, coverage, base quality, mapping quality, etc …

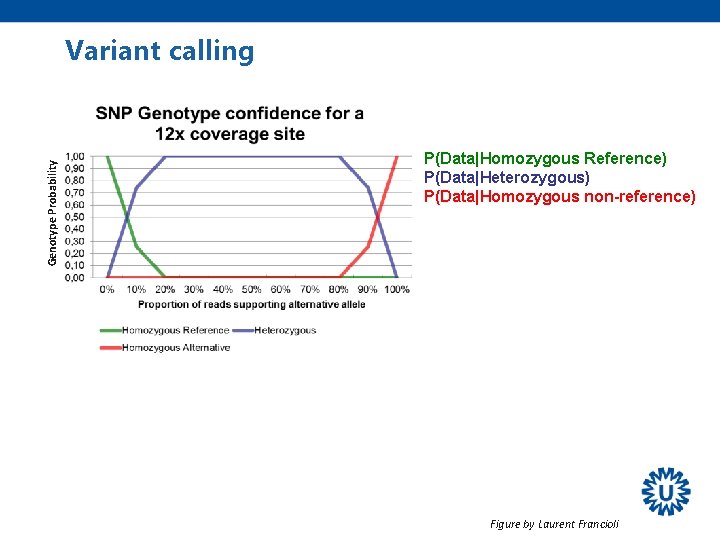
Genotype Probability Variant calling P(Data|Homozygous Reference) P(Data|Heterozygous) P(Data|Homozygous non-reference) Figure by Laurent Francioli

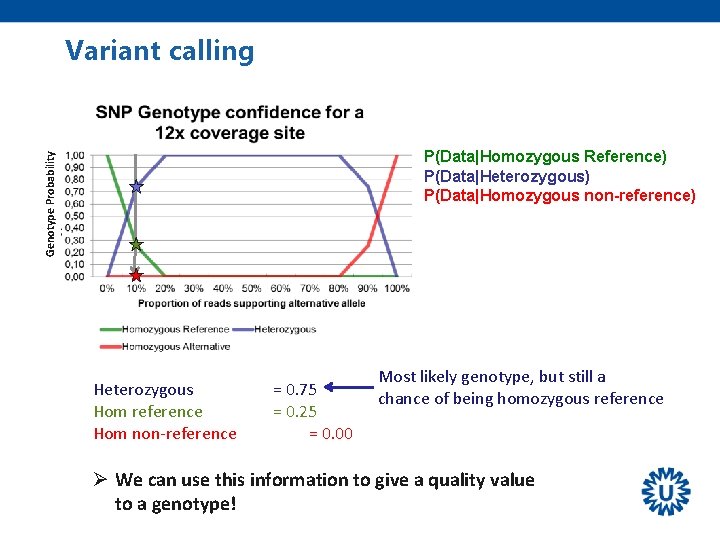
Variant calling Genotype Probability P(Data|Homozygous Reference) P(Data|Heterozygous) P(Data|Homozygous non-reference) Heterozygous Hom reference Hom non-reference = 0. 75 = 0. 25 = 0. 00 Most likely genotype, but still a chance of being homozygous reference Ø We can use this information to give a quality value to a genotype!

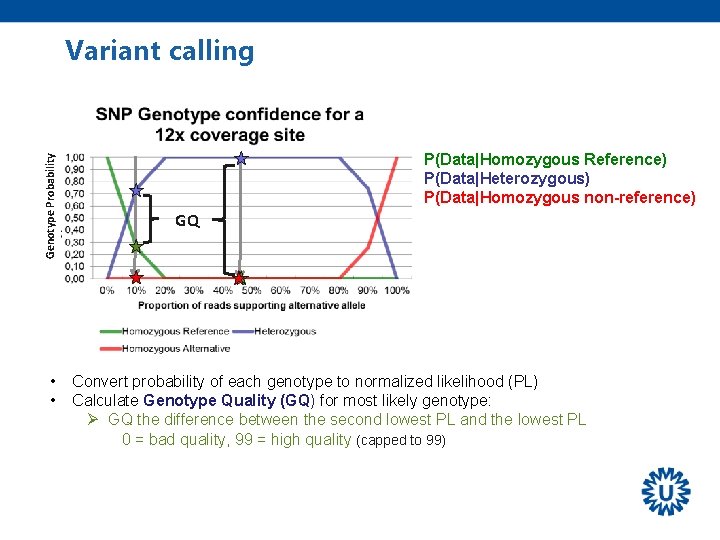
Genotype Probability Variant calling • • P(Data|Homozygous Reference) P(Data|Heterozygous) P(Data|Homozygous non-reference) GQ Convert probability of each genotype to normalized likelihood (PL) Calculate Genotype Quality (GQ) for most likely genotype: Ø GQ the difference between the second lowest PL and the lowest PL 0 = bad quality, 99 = high quality (capped to 99)

List of variants

Variant calling During variant calling many different parameters/values are calculated: • Position in the genome • The alleles involved • The actual genotype • Genotype Quality • Coverage information (total, for each allele) • Genotype likelihoods • And many more …. ØIdeally these should be stored in a single comprehensive file that can be used by many downstream tools (machine readable)

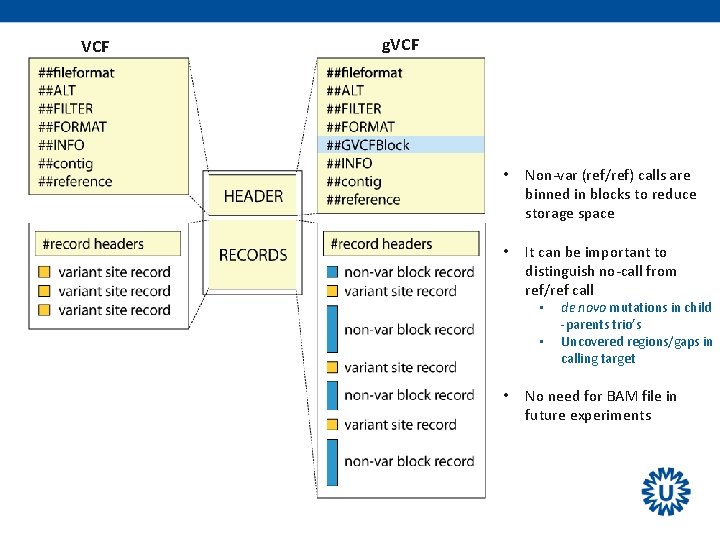
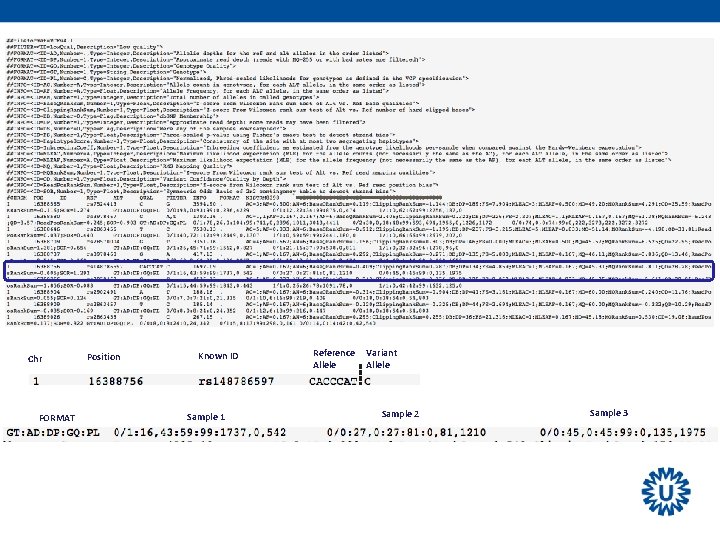
VCF-file • GATK variant calling output is stored in the Variant Call Format (VCF) • like most modern variant callers • Text file with comprehensive storage of all properties of variants – – – Variants: chromosome, position, reference/alternative bases Genotype, Genotype Quality, Coverage, Genotype Likelihoods Can contain information of one or more individuals. Can contain non-variant (ref/ref) information. Can contain annotations – filters: e. g. high or low quality variants, SNPs dense region, strand bias, etc. – predictions on protein effects, population frequencies, etc. Ø VCF’s are machine-readable (and less human-readable)

VCF g. VCF • Non-var (ref/ref) calls are binned in blocks to reduce storage space • It can be important to distinguish no-call from ref/ref call • • • de novo mutations in child -parents trio’s Uncovered regions/gaps in calling target No need for BAM file in future experiments

Chr FORMAT Position Known ID Sample 1 Reference Allele Variant Allele Sample 2 Sample 3

Part 2) Long read sequencing

Long read sequencing Illumina PACBIO ONT 300 bp (upto 500 bp) 10 -15 kb (upto ˜ 100 kb) Depends on DNA extraction, Standard DNA isolatie kits: 10 -30 kb New kits/other techniques >100 kb Full chromosomes possible?

Mapping using long reads 16

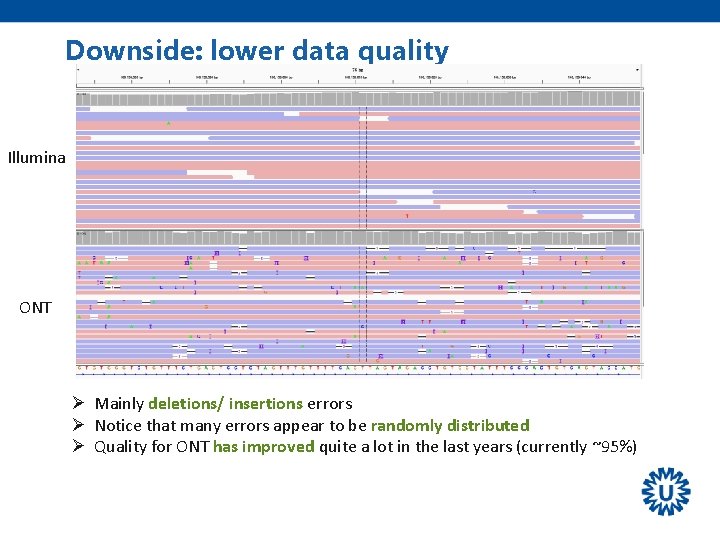
Downside: lower data quality Illumina ONT Ø Mainly deletions/ insertions errors Ø Notice that many errors appear to be randomly distributed Ø Quality for ONT has improved quite a lot in the last years (currently ~95%)

Mapping and variant calling • In theory, long reads are easier to map – Unique mapping due to length (remember hash/k-mers mapping) • But low quality add new complexities for mapping/alignment and variant calling due to mismatches, insertions, deletions – Many short read mappers/aligners and variant callers can not cope with this: e. g. Haplotype. Caller does not work As a result: new tools are (being) developed specially for long reads SNV/INDEL (<5 bp) sensitivity is still not comparable to Illumina (especially for diagnostic use)

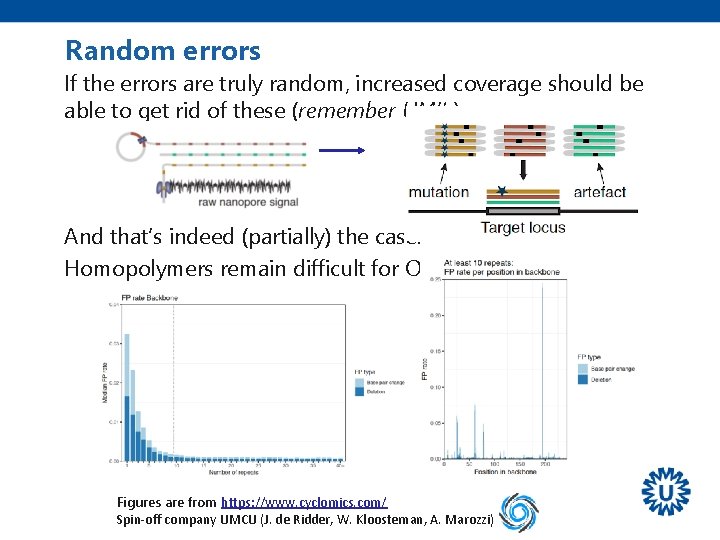
Random errors If the errors are truly random, increased coverage should be able to get rid of these (remember UMI’s). And that’s indeed (partially) the case. Homopolymers remain difficult for ONT. Figures are from https: //www. cyclomics. com/ Spin-off company UMCU (J. de Ridder, W. Kloosteman, A. Marozzi)

Short-term use of ONT within diagnostics Structural variants that do not require base pair resolution • Repeats (e. g. repeat expansions) • Structural variants (CNV, translocations, ánd inversions) • m. RNA sequencing (e. g. alternative splicing) • Phasing of variants (haplotyping)


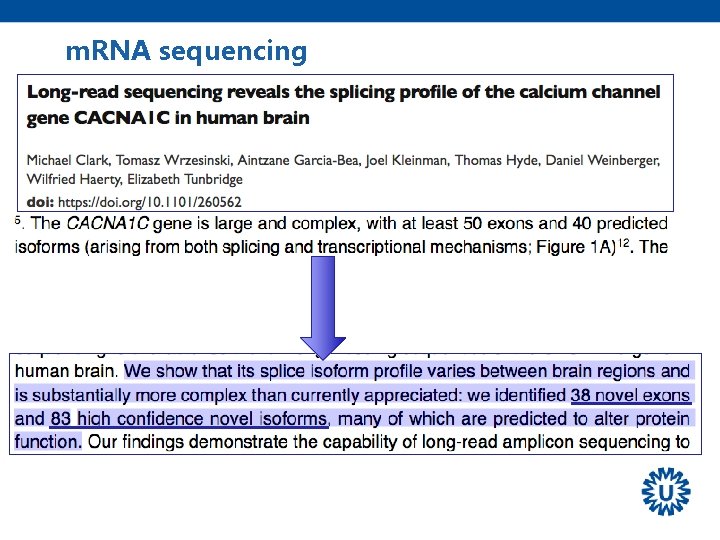
m. RNA sequencing

Fastq Read Mapping • Be critical to software when implementing: test and validate BAM Variant Calling VCF • Have trust in the outcome of the software – They might do a better job than humans • Long reads are here! – and will be in the clinic very soon Variant filtering/interpretation