Introduction to Gene Expression Analysis Phillip Lord Cedric








- Slides: 25

Introduction to Gene Expression Analysis Phillip Lord & Cedric Simillion

Resources http: //www. ebi. ac. uk/microarray/biology_intro. html http: //www. mged. org/ http: //www. tigr. org/tdb/microarray/ Microarray Bioinformatics Dov Stekel Product Details: Paperback 280 pages (September 8, 2003) Publisher: Cambridge University Press Language: English ISBN: 052152587 X

How do we measure gene expression? • Oldest technique is to look at a phenotype. • In this case, the ura 4+ gene from S. pombe • Most other techniques based on hybridisation. – Northern Blot – Quantative RT-PCR

Microarray analysis • Whole genome sequencing makes it possible to predict the entire gene complement • Various technologies have built on this knowledge to produce systems that will monitor the expression (usually transcription) at the whole genome level – Measurement of global transcription is called transcriptomics • Come by a variety of names – gene chips, arrays, DNA arrays. Can be somewhat confusing what is actually being described. • Not to be confused with Genotyping Microarrays

Generating Microarrays • There are many different systems for generating microarrays – spotting • original technology, now rather old • good for “one off” arrays – in-situ synthesis • newer, more reproducible • expensive first time around, then cheaper

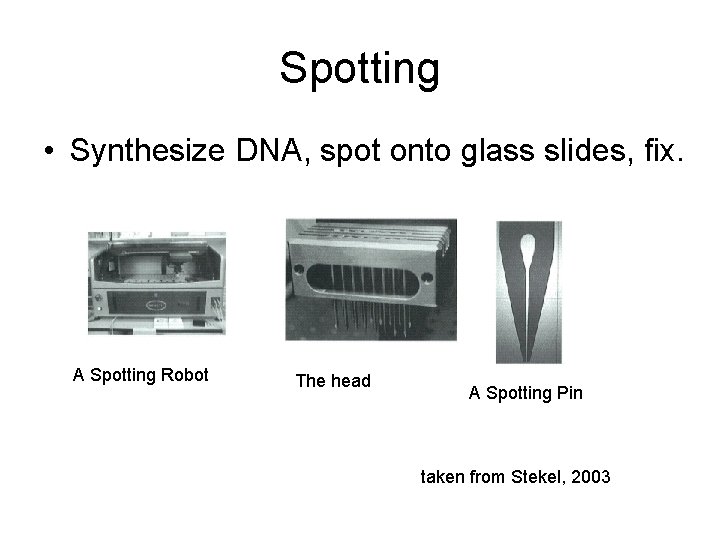
Spotting • Synthesize DNA, spot onto glass slides, fix. A Spotting Robot The head A Spotting Pin taken from Stekel, 2003

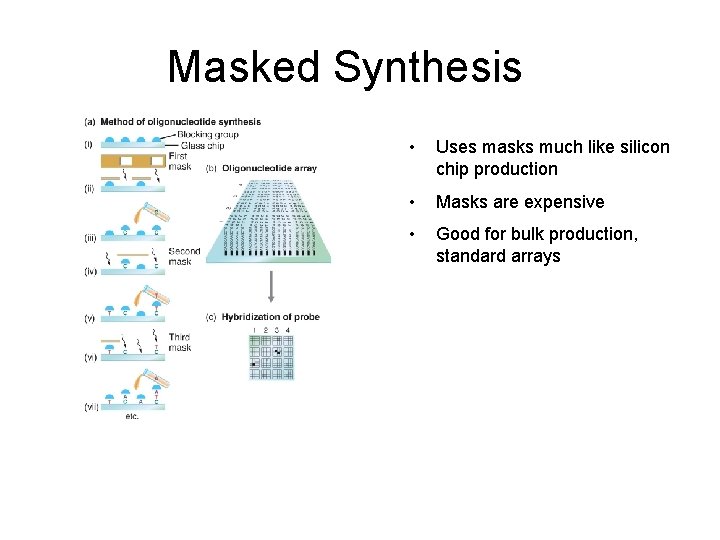
In-situ synthesis • • Uses chemically protected nucleotides Specific spots are “de-protected” Can then extend these oligos Different techniques for deprotection

Masked Synthesis • Uses masks much like silicon chip production • Masks are expensive • Good for bulk production, standard arrays

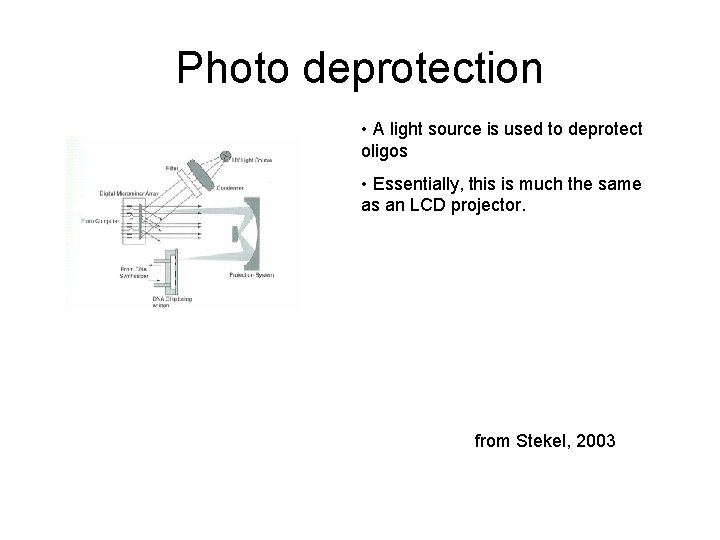
Photo deprotection • A light source is used to deprotect oligos • Essentially, this is much the same as an LCD projector. from Stekel, 2003

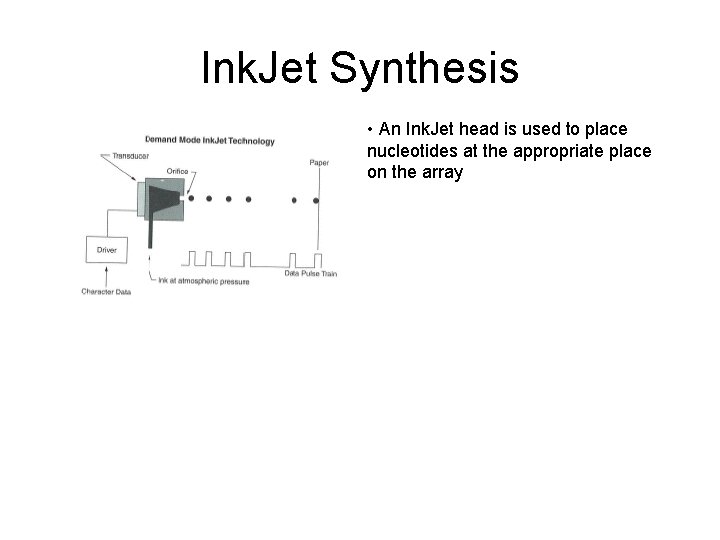
Ink. Jet Synthesis • An Ink. Jet head is used to place nucleotides at the appropriate place on the array

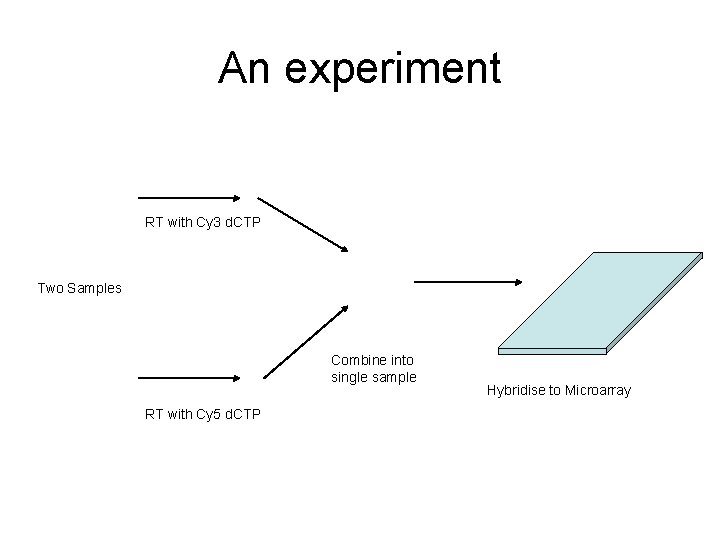
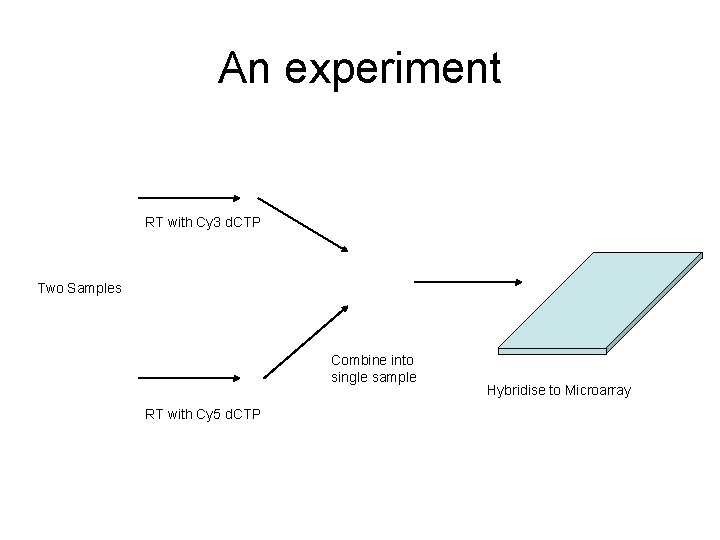
An experiment RT with Cy 3 d. CTP Two Samples Combine into single sample RT with Cy 5 d. CTP Hybridise to Microarray

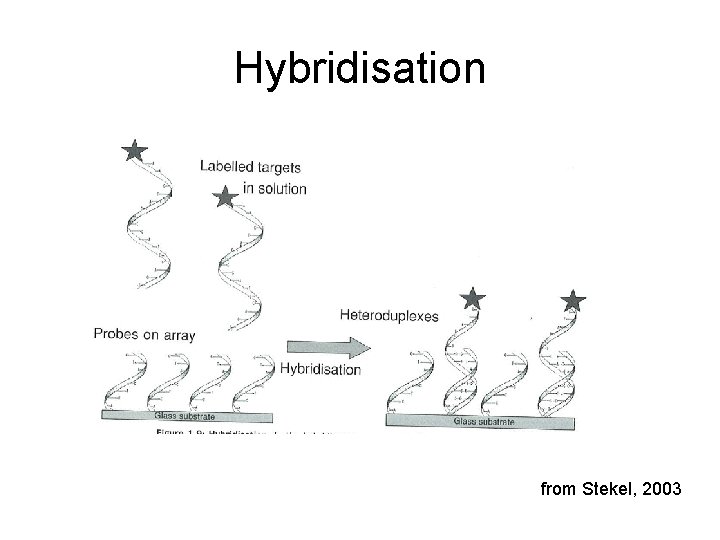
Hybridisation from Stekel, 2003

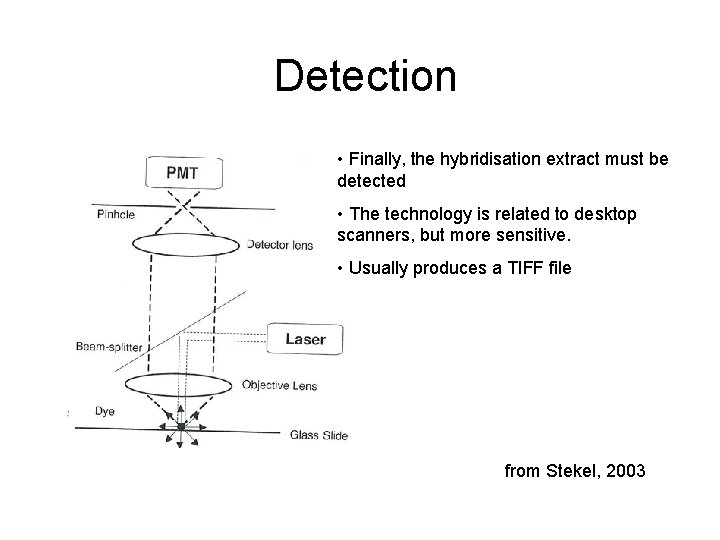
Detection • Finally, the hybridisation extract must be detected • The technology is related to desktop scanners, but more sensitive. • Usually produces a TIFF file from Stekel, 2003

The end result from Stekel, 2003

Problems • We are looking for variability between the expression of different genes. • There are many (many!) other sources of variability • Most microarray analysis is about trying to normalise these sources of variability, leaving biological variability

Artifacts The Jolly Green Giant The Yellow Splodge Peril Space Invaders

Solutions • Removing Sections • Background Subtraction • Start Again

Feature Recognition • Not all spots are equal – different sizes, different shapes. • Identifying the exact scope of the spot on an array can therefore be hard. • Often solved in the initial detection of spots.

Spot Detection The Doughnut A general disaster The basic solution to this is to not use circular spots for detection. There a variety of edge detection algorithms, or manual tools which work.

An experiment RT with Cy 3 d. CTP Two Samples Combine into single sample RT with Cy 5 d. CTP Hybridise to Microarray

Channel Variability • Cy 3/Cy 5 dyes have different properties. • So do the lasers at different frequencies. • So do the photomultipliers which detect them.

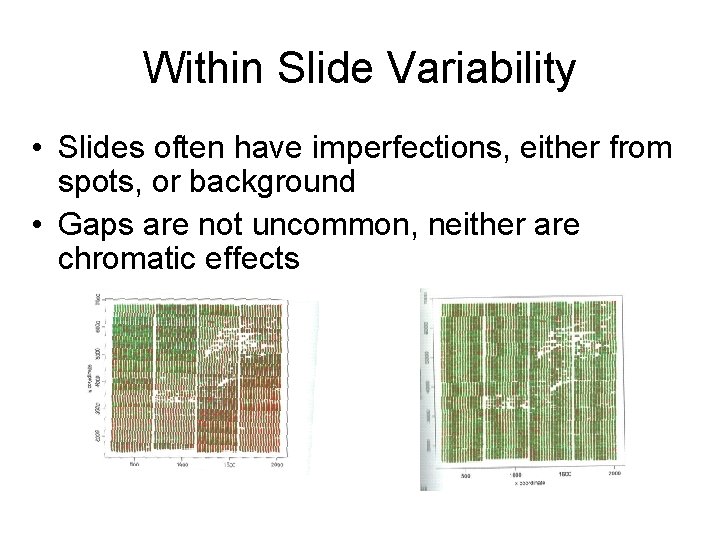
Within Slide Variability • Slides often have imperfections, either from spots, or background • Gaps are not uncommon, neither are chromatic effects

Inslide Normalisation

Between slide variability • Results between different slides are not directly comparable. • Results between different experiments are not directly comparable.

Further work • Please read • Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW "A genome-wide transcriptional analysis of the mitotic cell cycle. " Mol Cell. 1998; 2: 1: 65 -73 .