Introduction to Enzymes Michaelis Menten Kinetics Nicole Catterlin


![Are You Catching a Name Trend? [-ase = enzyme] Remember it! Are You Catching a Name Trend? [-ase = enzyme] Remember it!](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-11.jpg)
![How do Enzymes work? [Substrate + Enzyme] ➡ S/E Complex ➡ Product ● binds How do Enzymes work? [Substrate + Enzyme] ➡ S/E Complex ➡ Product ● binds](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-12.jpg)



![Substrate and Km ● [S] = concentration of Substrate o usually experiments are done Substrate and Km ● [S] = concentration of Substrate o usually experiments are done](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-19.jpg)






- Slides: 33

Introduction to Enzymes, Michaelis Menten Kinetics Nicole Catterlin, P 3 Howard University ACSEP 2015: Biochemistry

Enzyme Introduction? Enzyme Review. ● We already know about enzymes! Aren’t you smart? ● Let’s go over some of the stuff we’ve learned.

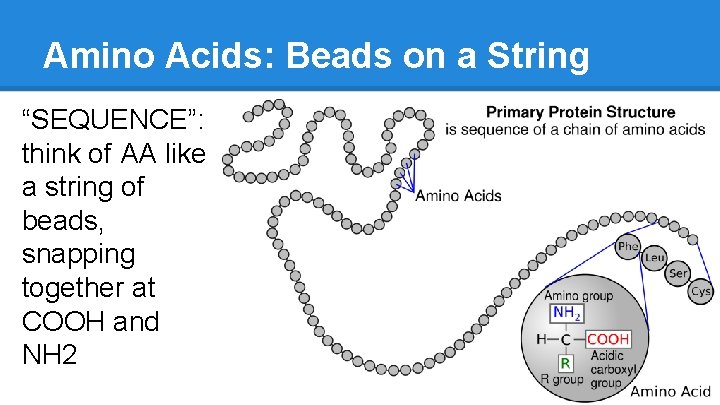
Amino Acids: Beads on a String “SEQUENCE”: think of AA like a string of beads, snapping together at COOH and NH 2



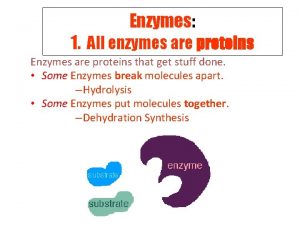
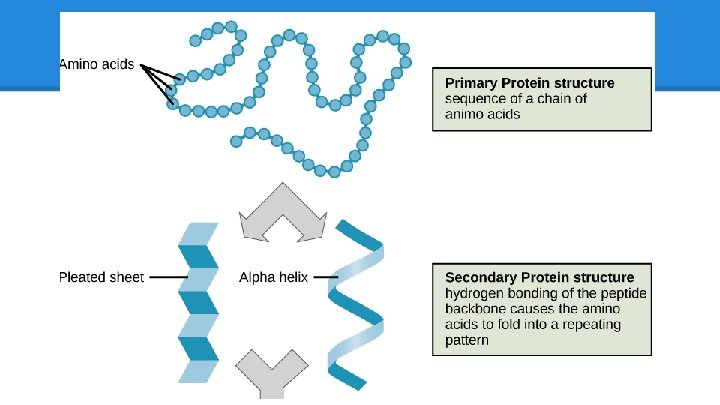
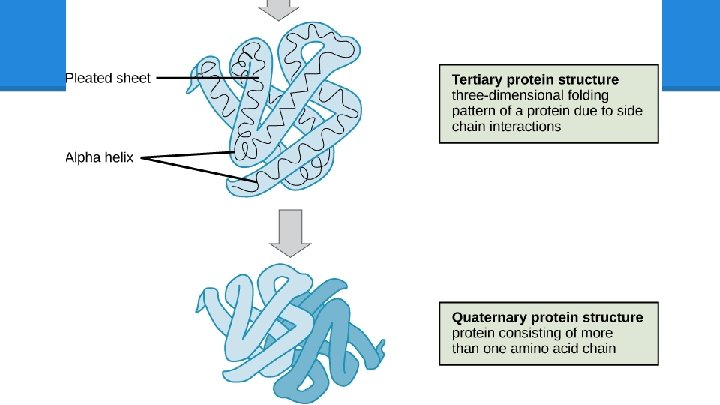
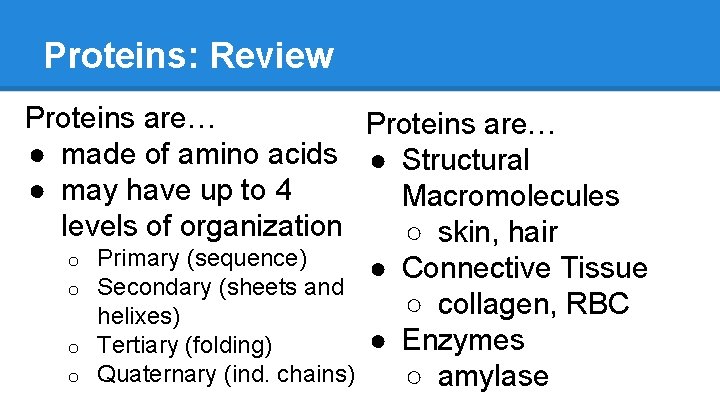
Proteins: Review Proteins are… ● made of amino acids ● Structural ● may have up to 4 Macromolecules levels of organization ○ skin, hair o Primary (sequence) ● Connective Tissue o Secondary (sheets and ○ collagen, RBC helixes) ● Enzymes o Tertiary (folding) o Quaternary (ind. chains) ○ amylase

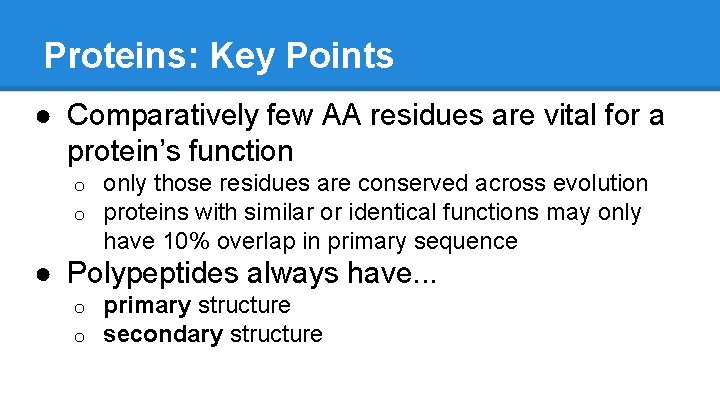
Proteins: Key Points ● Comparatively few AA residues are vital for a protein’s function o o only those residues are conserved across evolution proteins with similar or identical functions may only have 10% overlap in primary sequence ● Polypeptides always have. . . o o primary structure secondary structure

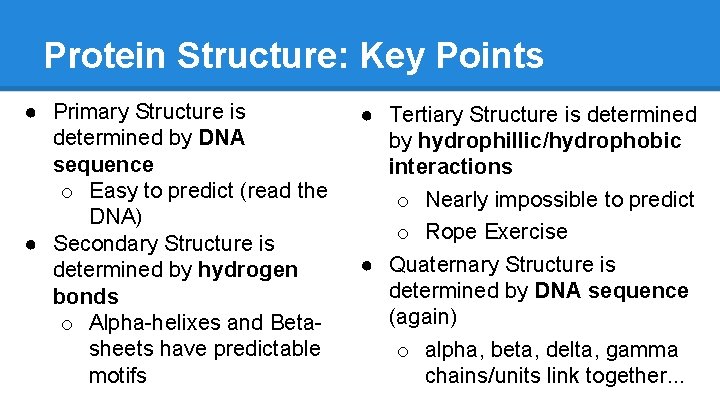
Protein Structure: Key Points ● Primary Structure is determined by DNA sequence o Easy to predict (read the DNA) ● Secondary Structure is determined by hydrogen bonds o Alpha-helixes and Betasheets have predictable motifs ● Tertiary Structure is determined by hydrophillic/hydrophobic interactions o Nearly impossible to predict o Rope Exercise ● Quaternary Structure is determined by DNA sequence (again) o alpha, beta, delta, gamma chains/units link together. . .

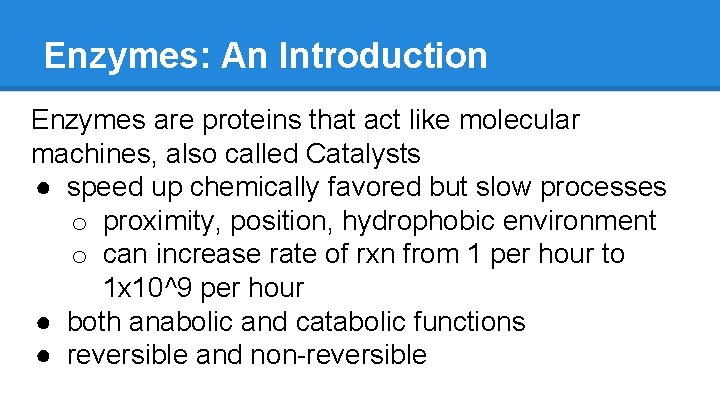
Enzymes: An Introduction Enzymes are proteins that act like molecular machines, also called Catalysts ● speed up chemically favored but slow processes o proximity, position, hydrophobic environment o can increase rate of rxn from 1 per hour to 1 x 10^9 per hour ● both anabolic and catabolic functions ● reversible and non-reversible

Enzymes: Examples ● Amylase o converts starch (polysaccharide) into simple sugars o ana/cata? ● Lipase o breaks down triglycerides into fatty acids o ana/cata? ● Aminotransferase o transfers an amino group from one molecule to another o ana/cata? ● DNA ligase o mends “nicks” in DNA strands after repair o ana/cata?
![Are You Catching a Name Trend ase enzyme Remember it Are You Catching a Name Trend? [-ase = enzyme] Remember it!](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-11.jpg)
Are You Catching a Name Trend? [-ase = enzyme] Remember it!
![How do Enzymes work Substrate Enzyme SE Complex Product binds How do Enzymes work? [Substrate + Enzyme] ➡ S/E Complex ➡ Product ● binds](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-12.jpg)
How do Enzymes work? [Substrate + Enzyme] ➡ S/E Complex ➡ Product ● binds SPECIFIC substrates o brings them into proximity o brings them into proper position o hydrophobic (water exclusionary) environment ● self-regenerating o any altered AA residues are restored by the end by acid-base interactions ● transcribed and translated from genes o DNA > RNA > translation via ribosomes o translation can be “up-regulated” or “down-regulated” according to body’s needs

Enzymes, As Seen Through Math ● Enzymes can be examined theoretically and mathematically o Theoretically: structure, inhibitors, activators § Why they work o Mathematically: how fast and what slows it down § How they work ● Because we use enzymes for many processes in our world, it is important to know how fast they go ● This is known as “enzyme kinetics”

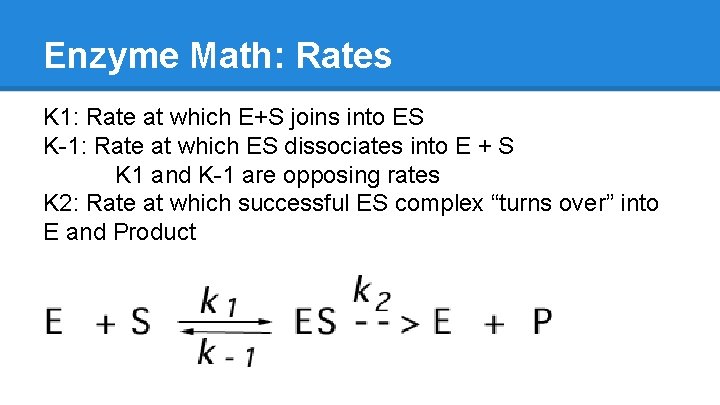
Enzyme Math: Rates K 1: Rate at which E+S joins into ES K-1: Rate at which ES dissociates into E + S K 1 and K-1 are opposing rates K 2: Rate at which successful ES complex “turns over” into E and Product

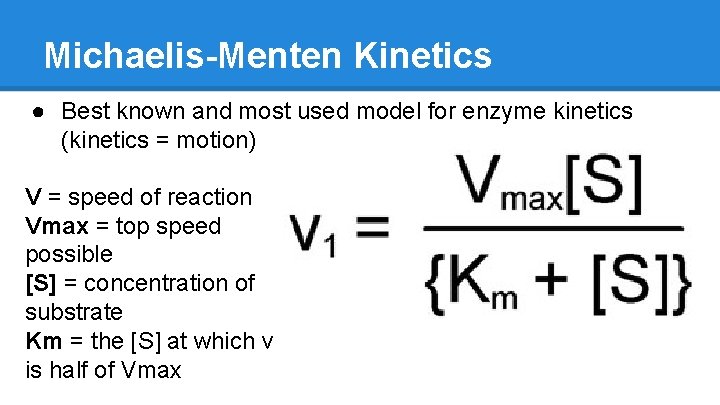
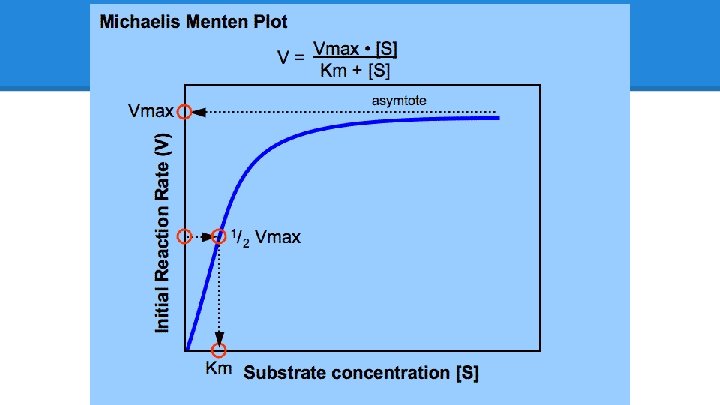
Michaelis-Menten Kinetics ● Best known and most used model for enzyme kinetics (kinetics = motion) V = speed of reaction Vmax = top speed possible [S] = concentration of substrate Km = the [S] at which v is half of Vmax


Vmax: Otters and Oysters ● Imagine a pool with 10 otters floating happily ● Throw the otter closest to you one (1) oyster o He de-shells it, scoops it out and eats it as the other otters watch jealously, hungry. o You start throwing two at a time. Three at a time. The oysters disappear almost instantly, leaving only shells, because there are so many hungry otters and not enough oysters. ● But what if you unloaded a truck full of oysters onto this pool of otters?

Vmax: Pedal to the Metal ● Vmax “represents the maximum rate achieved by the system, at maximum (saturating) substrate concentrations" ● Units: umol/min (or mol/second) ● So, how many mols of substrate are turned into product per second o if you know the [E], you can take Vmax/[E] and get “turnover rate” (per second), or how much work each enzyme is doing ● If given all the oysters they could possibly shell, how many oysters would ten otters eat in a minute? A second? o Enzymes like carbonic anhydrase are otters that can shell 400, 000 oysters per second!
![Substrate and Km S concentration of Substrate o usually experiments are done Substrate and Km ● [S] = concentration of Substrate o usually experiments are done](https://slidetodoc.com/presentation_image/281e88fd6ac75b3b7c131410181c4e17/image-19.jpg)
Substrate and Km ● [S] = concentration of Substrate o usually experiments are done with increasing levels of substrate: 100 mg substate, 200 mg substrate, 500 mg, 1000 mg, etc, to see at what point Vmax is reached o How many oysters does it take to truly overwhelm the hungry otters?

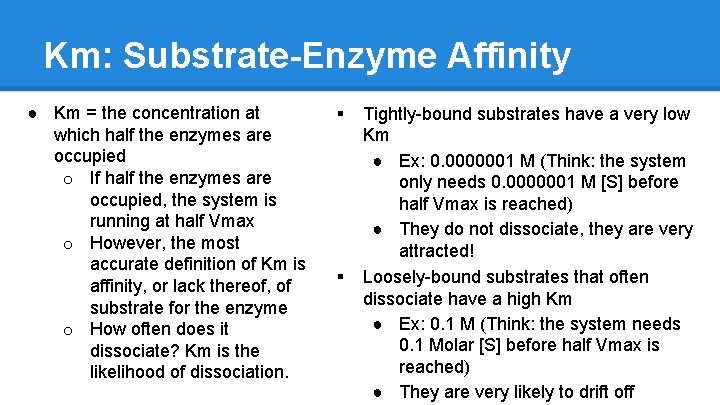
Km: Substrate-Enzyme Affinity ● Km = the concentration at which half the enzymes are occupied o If half the enzymes are occupied, the system is running at half Vmax o However, the most accurate definition of Km is affinity, or lack thereof, of substrate for the enzyme o How often does it dissociate? Km is the likelihood of dissociation. § § Tightly-bound substrates have a very low Km ● Ex: 0. 0000001 M (Think: the system only needs 0. 0000001 M [S] before half Vmax is reached) ● They do not dissociate, they are very attracted! Loosely-bound substrates that often dissociate have a high Km ● Ex: 0. 1 M (Think: the system needs 0. 1 Molar [S] before half Vmax is reached) ● They are very likely to drift off

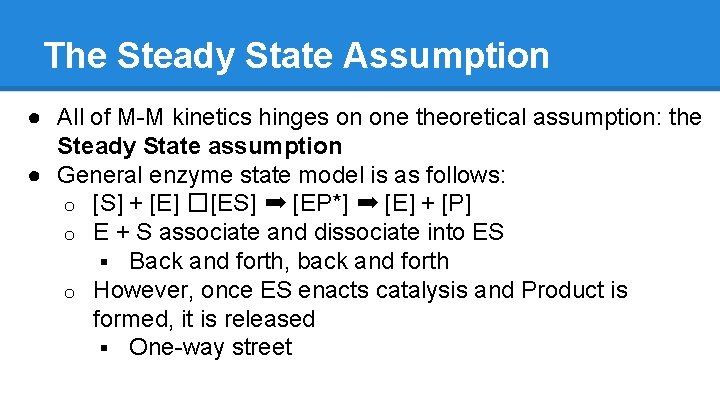
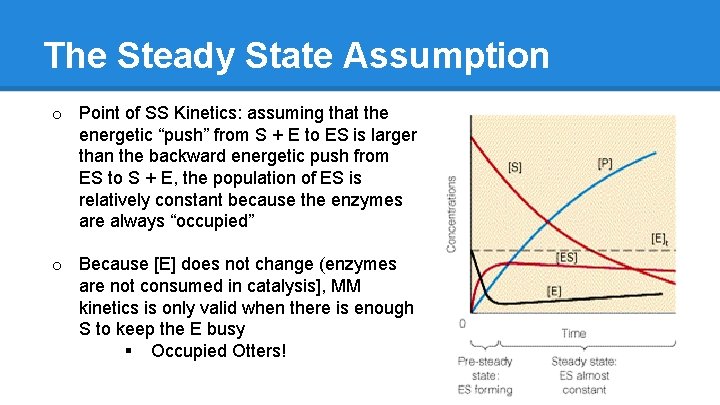
The Steady State Assumption ● All of M-M kinetics hinges on one theoretical assumption: the Steady State assumption ● General enzyme state model is as follows: o [S] + [E] � [ES] ➡ [EP*] ➡ [E] + [P] o E + S associate and dissociate into ES § Back and forth, back and forth o However, once ES enacts catalysis and Product is formed, it is released § One-way street

The Steady State Assumption o Point of SS Kinetics: assuming that the energetic “push” from S + E to ES is larger than the backward energetic push from ES to S + E, the population of ES is relatively constant because the enzymes are always “occupied” o Because [E] does not change (enzymes are not consumed in catalysis], MM kinetics is only valid when there is enough S to keep the E busy § Occupied Otters!

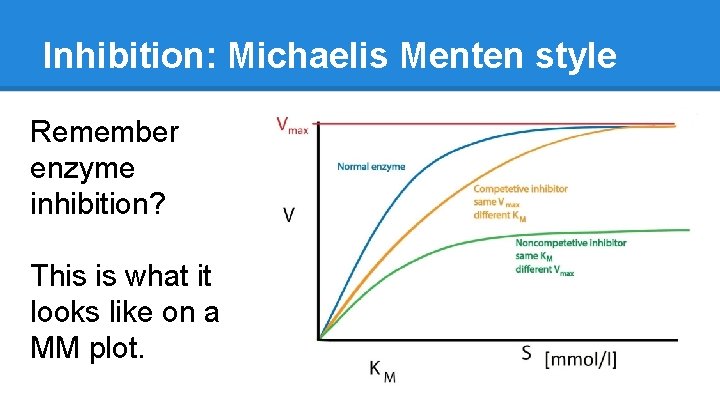
Inhibition: Michaelis Menten style Remember enzyme inhibition? This is what it looks like on a MM plot.

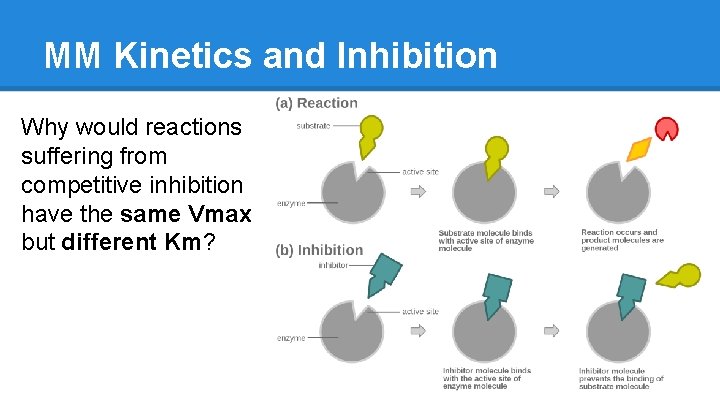
MM Kinetics and Inhibition Why would reactions suffering from competitive inhibition have the same Vmax but different Km?

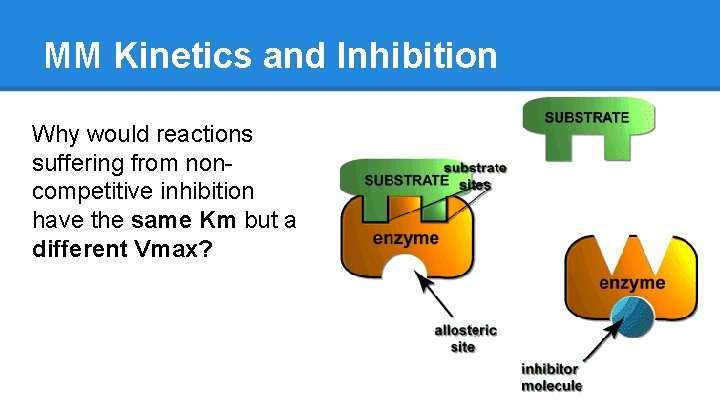
MM Kinetics and Inhibition Why would reactions suffering from noncompetitive inhibition have the same Km but a different Vmax?

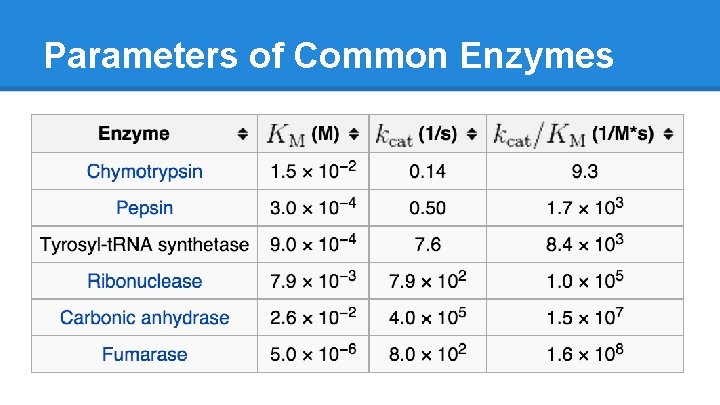
Parameters of Common Enzymes

Parameter Paradox ● Just because it binds tightly doesn’t mean it works quickly o ex: Pepsin has a Km of 0. 0003 but only produces 1 product every 2 seconds ● Just because it binds loosely doesn’t mean it works slowly o ex: Carbonic anhydrase has a Km of 0. 026 but has a turnover rate of 400, 000 per second

Let’s Look at a Problem. . .

Solution

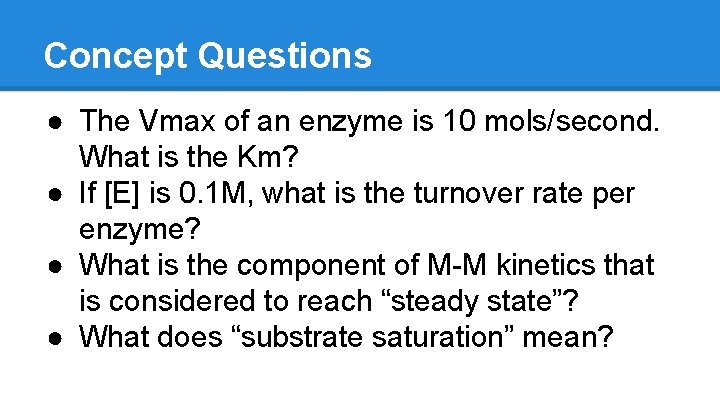
Concept Questions ● The Vmax of an enzyme is 10 mols/second. What is the Km? ● If [E] is 0. 1 M, what is the turnover rate per enzyme? ● What is the component of M-M kinetics that is considered to reach “steady state”? ● What does “substrate saturation” mean?

Thanks for Your Attention! We’re done! Quiz in an hour, review tomorrow.

What I Want You to Know. . . ● Refer to Lecture 3… ● What does a basic Michaelis Menten graph look like and why? ● What are the components of the Michaelis-Menten kinetics equation? o What is Vmax? o What is Km? (two definitions) o What is [S]? o What is V? ● What is the Steady State Assumption? ● If provided with the M-M equation, can you do a simple Vmax calculation?

Sources Enzyme k rates: http: //faculty. smu. edu/svik/5310 lectures/ES. gif M-M equation: http: //themedicalbiochemistrypage. org/images/michaelismentenvelocityequation. jpg M-M blue graph: http: //plantphys. info/plant_physiology/images/enzmichaelis. gif M-M SS kinetics: https: //classconnection. s 3. amazonaws. com/1613/flashcards/883732/png/picture 10. png Oyster and Otter: http: //www. oceanlight. com/stock-photo/sea-otter-eating-shellfish-picture-21609 -557678. jpg Parameters of Common Enzymes: http: //en. wikipedia. org/wiki/Michaelis%E 2%80%93 Menten_kinetics#Quasi-steady-state_approximation MM Enzyme Problems: http: //202. 114. 65. 51/fzjx/wsw/website/mit/eb/kinetics/solvingkinetics. html
 Vmax and km
Vmax and km Significance of michaelis menten equation
Significance of michaelis menten equation Michaelis menten steady state
Michaelis menten steady state Pseudo 1st order reaction
Pseudo 1st order reaction Equação de michaelis menten
Equação de michaelis menten Alp dıraman
Alp dıraman Michaelis menten steady state
Michaelis menten steady state Grafica de eadie hofstee
Grafica de eadie hofstee Vmax and km
Vmax and km Inhibición mixta km y vmax
Inhibición mixta km y vmax Homotrof
Homotrof Enzim
Enzim Substrat artinya
Substrat artinya Vmax equation
Vmax equation Limitations of michaelis menten equation
Limitations of michaelis menten equation Haloenzim
Haloenzim Kanzlei michaelis
Kanzlei michaelis Tipos de pelvis femenina
Tipos de pelvis femenina петитов треугольник
петитов треугольник Molecularity of reaction
Molecularity of reaction Kinetics and equilibrium
Kinetics and equilibrium Zero order kinetics
Zero order kinetics Planar kinetics of a rigid body
Planar kinetics of a rigid body Difference between zero and first order kinetics
Difference between zero and first order kinetics Difference between 1st order and zero order kinetics
Difference between 1st order and zero order kinetics Collision theory of kinetics
Collision theory of kinetics Types of reactions grade 11
Types of reactions grade 11 Kinetics flotation chemicals
Kinetics flotation chemicals Half life chemistry kinetics
Half life chemistry kinetics Chemical kinetics definition
Chemical kinetics definition Planar kinetics of a rigid body: force and acceleration
Planar kinetics of a rigid body: force and acceleration Kinetic of rigid body
Kinetic of rigid body Kinetics of a particle force and acceleration
Kinetics of a particle force and acceleration Ap chemistry kinetics
Ap chemistry kinetics