Genome Rearrangement Phylogeny Tandy Warnow The University of




![Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999] Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999]](https://slidetodoc.com/presentation_image_h2/a68ccc29b906da2402ef93f46bd45e1e/image-15.jpg)
![Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999] Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999]](https://slidetodoc.com/presentation_image_h2/a68ccc29b906da2402ef93f46bd45e1e/image-16.jpg)





- Slides: 34

Genome Rearrangement Phylogeny Tandy Warnow The University of Illinois at Urbana-Champaign

In 1999, Bob Jansen needed help with 13 Campanulaceae genomes

In 1999, Bob Jansen needed help with 13 Campanulaceae genomes

In 1999, Bob Jansen needed help with 13 Campanulaceae genomes

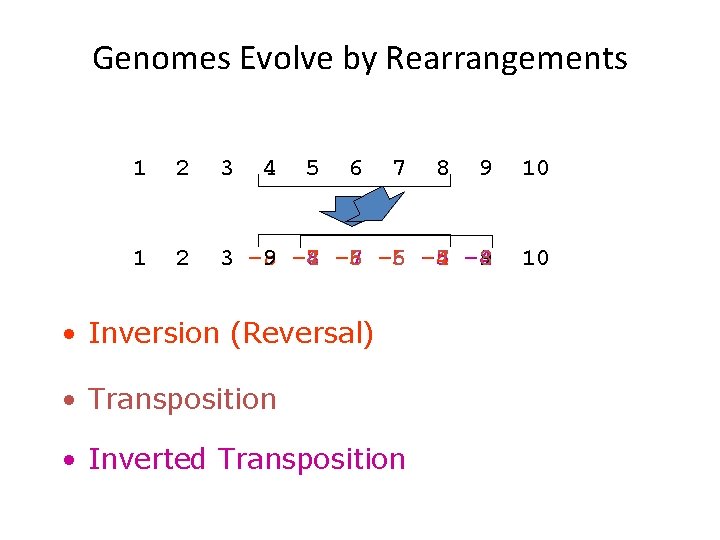
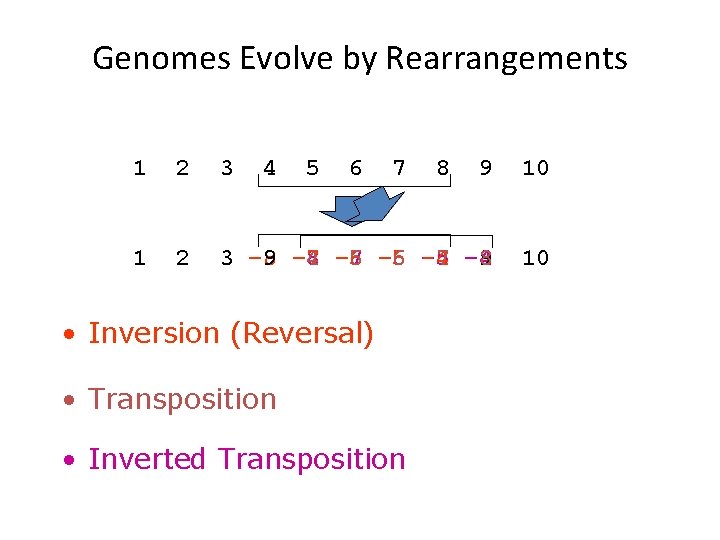
Genomes Evolve by Rearrangements 1 2 3 4 5 6 7 9 10 1 2 3 – 8 9 – 7 -8 4 – 6 – 7 5 – 6 6 -4 – 5 7 – 4 8 9 10 • Inversion (Reversal) • Transposition • Inverted Transposition 8

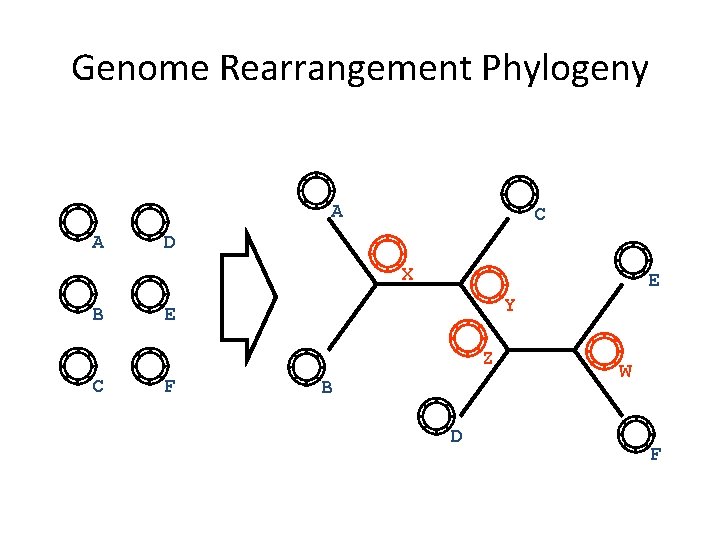
Genome Rearrangement Phylogeny A A C D X B E Y E Z C F B D W F

Breakpoint Phylogeny Proposed by Sankoff and Blanchette in J Comp. Biol. 1998 • Input: Chromosomes given as signed gene orders, one copy of each gene in each chromosome • Output: Tree with the minimum number of breakpoints

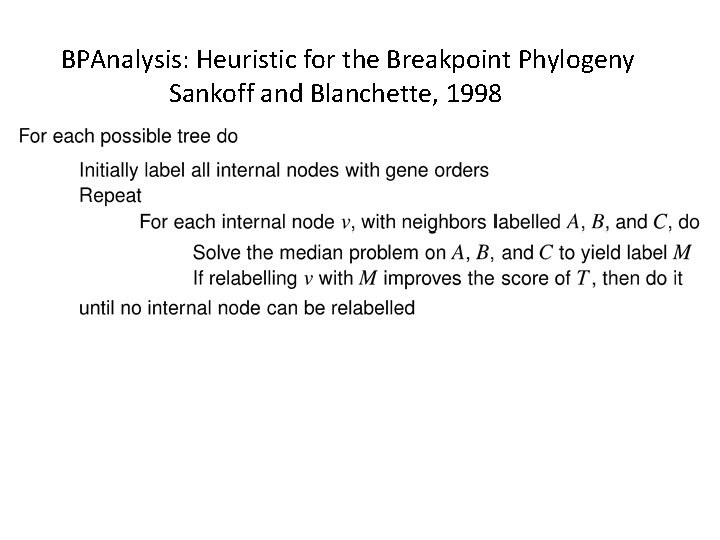
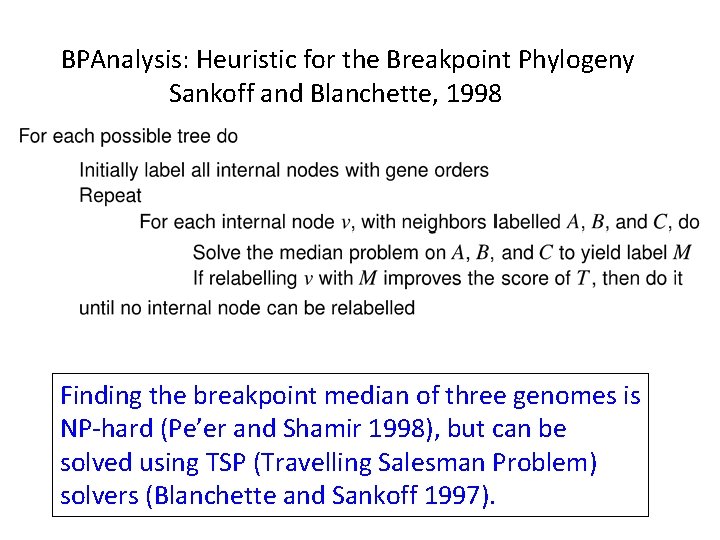
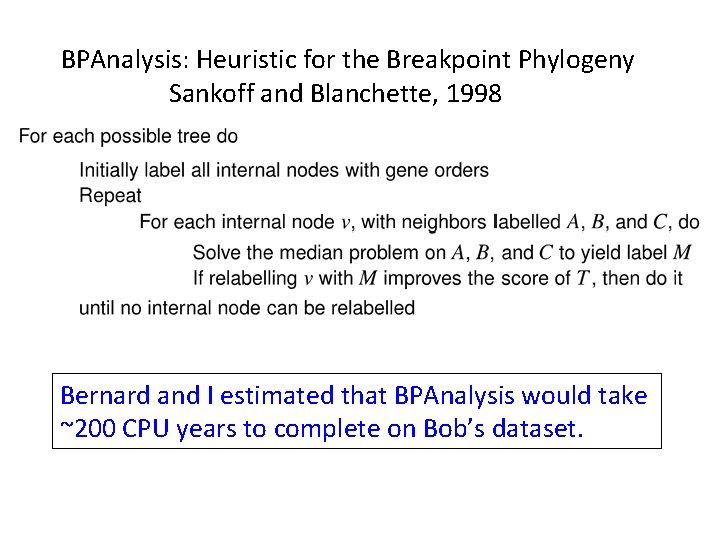
BPAnalysis: Heuristic for the Breakpoint Phylogeny Sankoff and Blanchette, 1998

BPAnalysis: Heuristic for the Breakpoint Phylogeny Sankoff and Blanchette, 1998 Finding the breakpoint median of three genomes is NP-hard (Pe’er and Shamir 1998), but can be solved using TSP (Travelling Salesman Problem) solvers (Blanchette and Sankoff 1997).

BPAnalysis: Heuristic for the Breakpoint Phylogeny Sankoff and Blanchette, 1998 Bernard and I estimated that BPAnalysis would take ~200 CPU years to complete on Bob’s dataset.

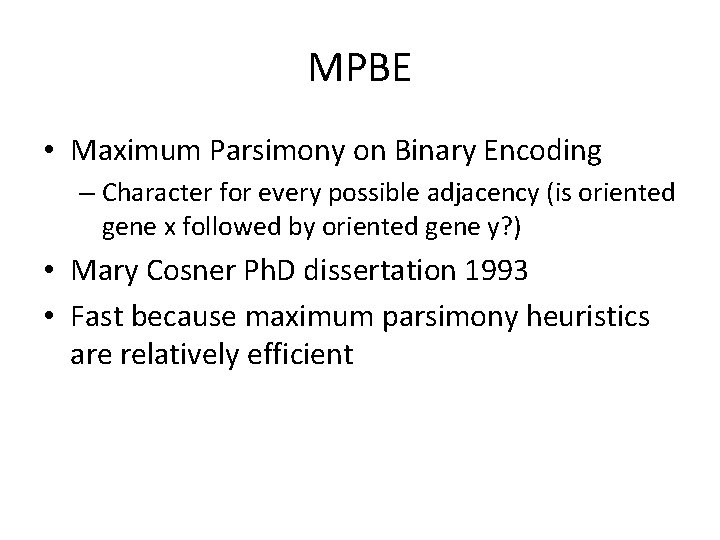
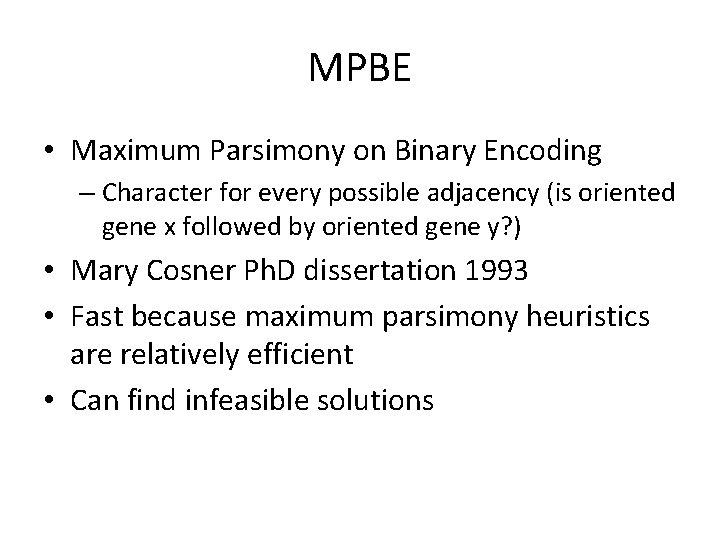
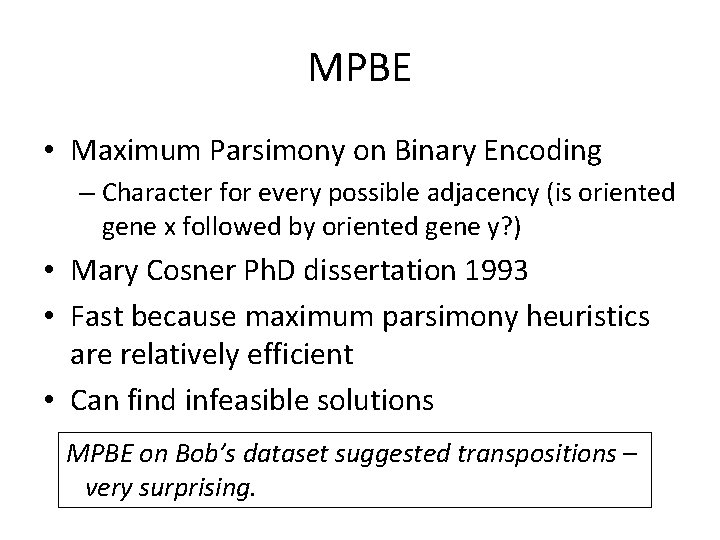
MPBE • Maximum Parsimony on Binary Encoding – Character for every possible adjacency (is oriented gene x followed by oriented gene y? ) • Mary Cosner Ph. D dissertation 1993 • Fast because maximum parsimony heuristics are relatively efficient • Can find infeasible solutions MPBE on Bob’s dataset suggested

MPBE • Maximum Parsimony on Binary Encoding – Character for every possible adjacency (is oriented gene x followed by oriented gene y? ) • Mary Cosner Ph. D dissertation 1993 • Fast because maximum parsimony heuristics are relatively efficient • Can find infeasible solutions MPBE on Bob’s dataset suggested

MPBE • Maximum Parsimony on Binary Encoding – Character for every possible adjacency (is oriented gene x followed by oriented gene y? ) • Mary Cosner Ph. D dissertation 1993 • Fast because maximum parsimony heuristics are relatively efficient • Can find infeasible solutions MPBE on Bob’s dataset suggested transpositions – very surprising. MPBE on Bob’s dataset suggested

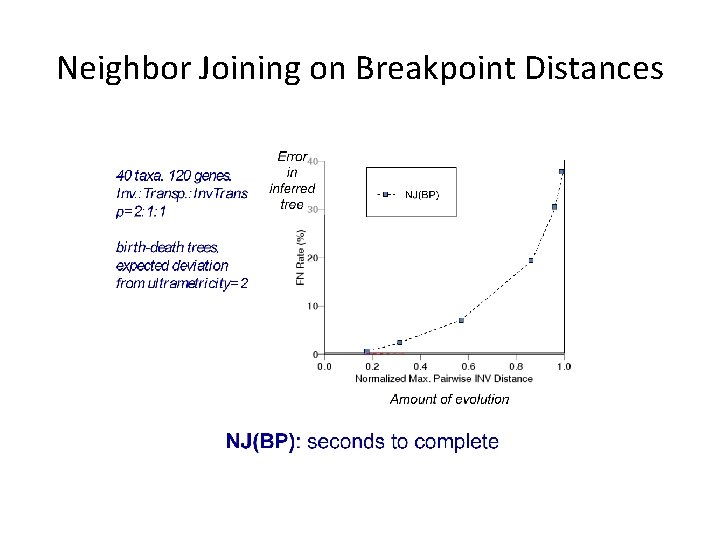
Neighbor Joining on Breakpoint Distances
![Phylogeny reconstruction in 1999 Distancebased Breakpoint BP distances Blanchette Kunisawa Sankoff 1999 Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999]](https://slidetodoc.com/presentation_image_h2/a68ccc29b906da2402ef93f46bd45e1e/image-15.jpg)
Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999] • Breakpoint tree (NP-hard, even for three taxa) – BPAnalysis: [Sankoff & Blanchette 1998]: exhaustive search through treespace to find the minimum breakpoint length (the number of breakpoints on the tree) • MPBE [Cosner 1993]: maximum parsimony on binary encoding
![Phylogeny reconstruction in 1999 Distancebased Breakpoint BP distances Blanchette Kunisawa Sankoff 1999 Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999]](https://slidetodoc.com/presentation_image_h2/a68ccc29b906da2402ef93f46bd45e1e/image-16.jpg)
Phylogeny reconstruction in 1999 • Distance-based – Breakpoint (BP) distances [Blanchette, Kunisawa, Sankoff 1999] – fast but high error • Breakpoint tree (NP-hard, even for three taxa) – BPAnalysis: [Sankoff & Blanchette 1998]: exhaustive search through treespace to find the minimum breakpoint length (the number of breakpoints on the tree) – too slow • MPBE [Cosner 1993]: maximum parsimony on binary encoding: can find infeasible ancestors

The challenges! 1. Find all the best genome trees for Bob’s dataset, and determine if inversions suffice (or if we really do need transpositions). 2. Design statistically rigorous methods for genome rearrangement phylogeny. 3. Design efficient techniques to enable genome-scale phylogeny for large datasets that are difficult to analyze.

Genomes Evolve by Rearrangements 1 2 3 4 5 6 7 9 10 1 2 3 – 8 9 – 7 -8 4 – 6 – 7 5 – 6 6 -4 – 5 7 – 4 8 9 10 • Inversion (Reversal) • Transposition • Inverted Transposition 8

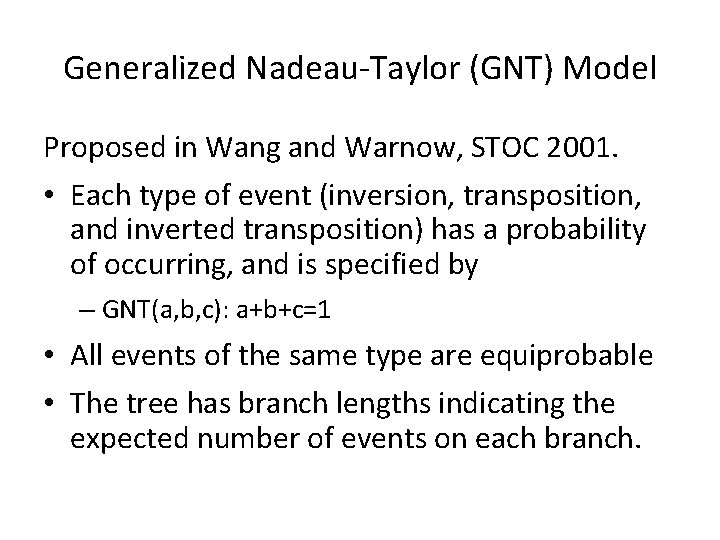
Generalized Nadeau-Taylor (GNT) Model Proposed in Wang and Warnow, STOC 2001. • Each type of event (inversion, transposition, and inverted transposition) has a probability of occurring, and is specified by – GNT(a, b, c): a+b+c=1 • All events of the same type are equiprobable • The tree has branch lengths indicating the expected number of events on each branch.

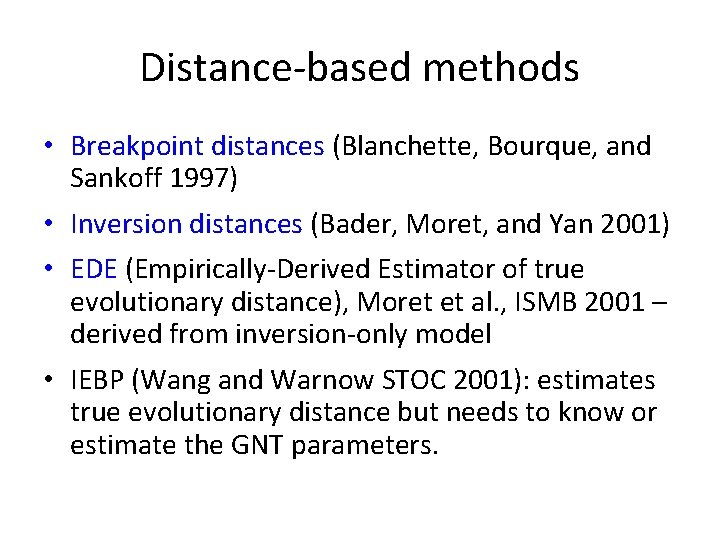
Distance-based methods • Breakpoint distances (Blanchette, Bourque, and Sankoff 1997) • Inversion distances (Bader, Moret, and Yan 2001) • EDE (Empirically-Derived Estimator of true evolutionary distance), Moret et al. , ISMB 2001 – derived from inversion-only model • IEBP (Wang and Warnow STOC 2001): estimates true evolutionary distance but needs to know or estimate the GNT parameters.

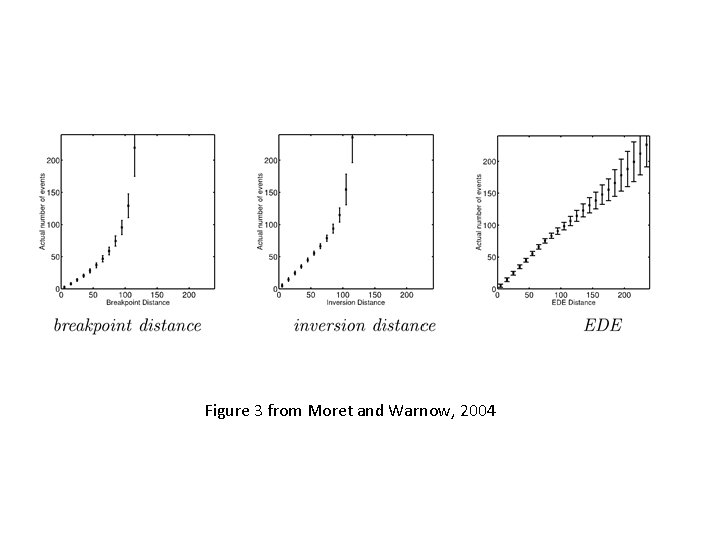
Figure 3 from Moret and Warnow, 2004

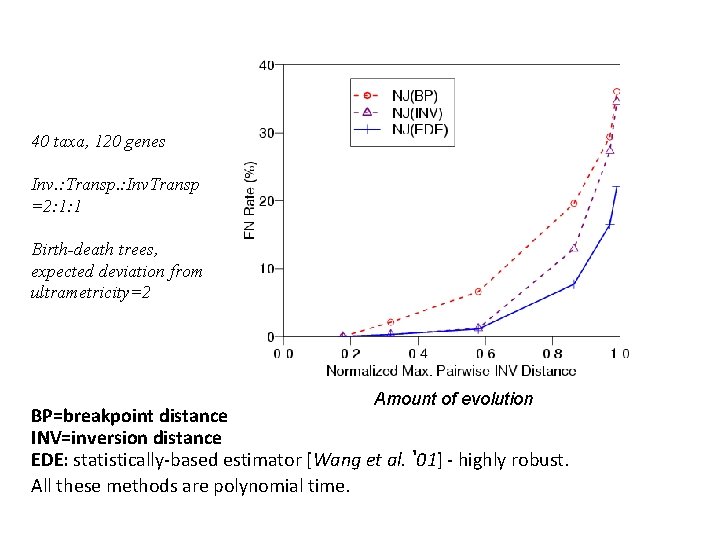
40 taxa, 120 genes Inv. : Transp. : Inv. Transp =2: 1: 1 Birth-death trees, expected deviation from ultrametricity=2 Amount of evolution BP=breakpoint distance INV=inversion distance EDE: statistically-based estimator [Wang et al. ‘ 01] - highly robust. All these methods are polynomial time.

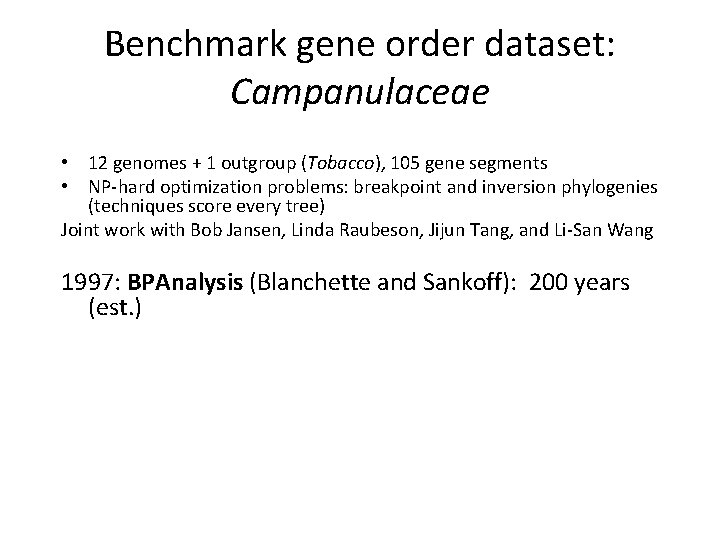
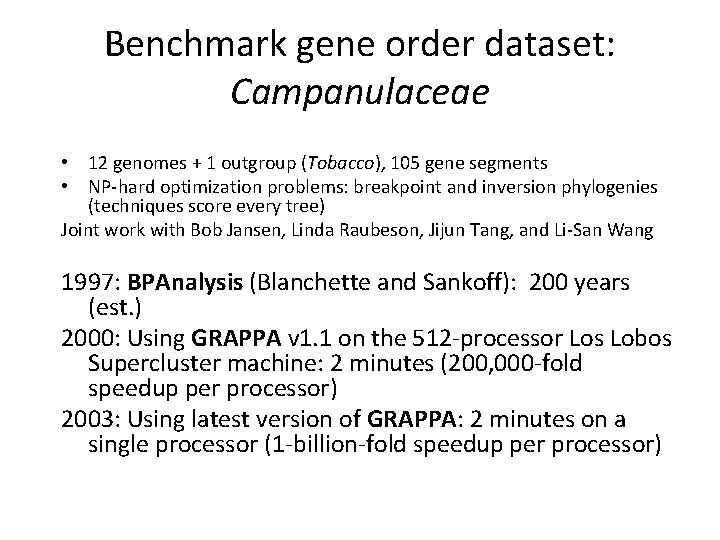
Benchmark gene order dataset: Campanulaceae • 12 genomes + 1 outgroup (Tobacco), 105 gene segments • NP-hard optimization problems: breakpoint and inversion phylogenies (techniques score every tree) Joint work with Bob Jansen, Linda Raubeson, Jijun Tang, and Li-San Wang 1997: BPAnalysis (Blanchette and Sankoff): 200 years (est. ) 2000: Using GRAPPA v 1. 1 on the 512 -processor Los Lobos Supercluster machine: 2 minutes (200, 000 -fold speedup per processor) 2003: Using latest version of GRAPPA: 2 minutes on a single processor (1 -billion-fold speedup per processor)

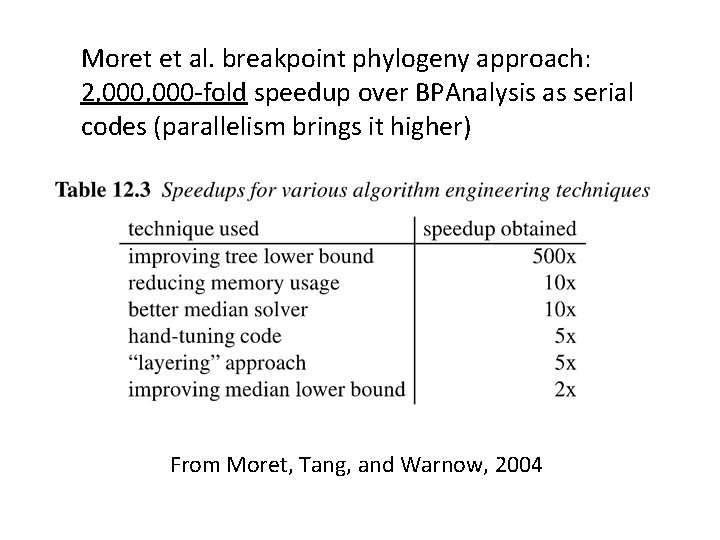
Moret et al. breakpoint phylogeny approach: 2, 000 -fold speedup over BPAnalysis as serial codes (parallelism brings it higher) From Moret, Tang, and Warnow, 2004

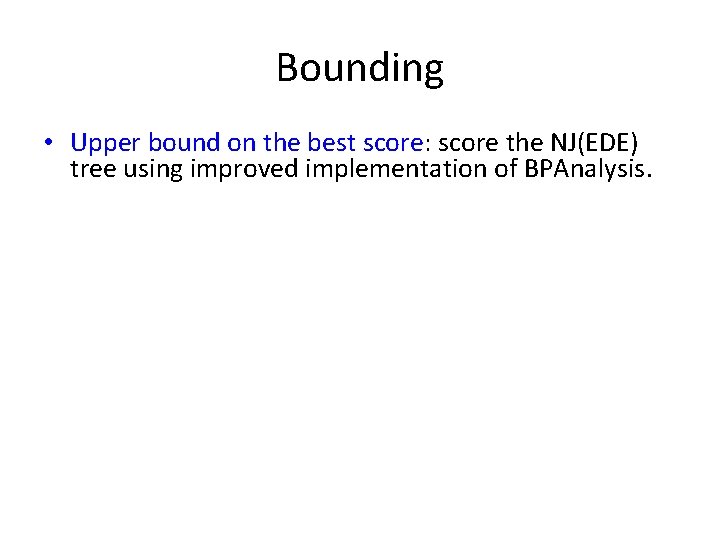
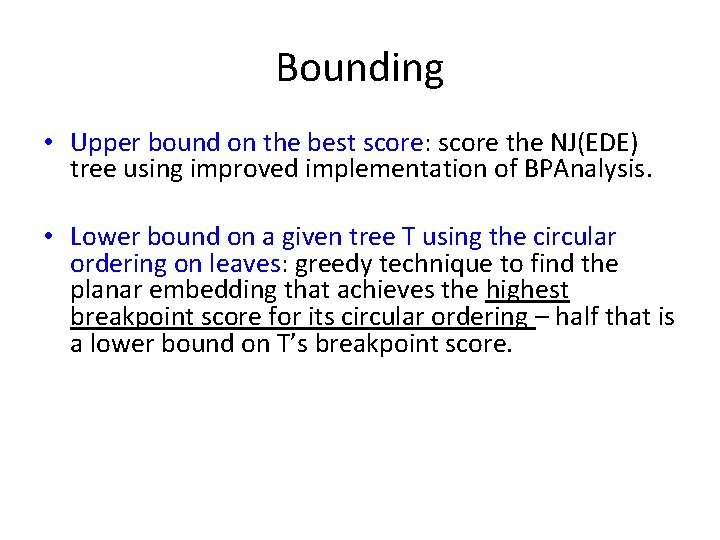
Bounding • Upper bound on the best score: score the NJ(EDE) tree using improved implementation of BPAnalysis. • Lower bound on a given tree T using the circular ordering on leaves: greedy technique to find the planar embedding that achieves the highest breakpoint score for its circular odering – half that is a lower bound on T’s breakpoint score. By the way, Bernard and I had a big argument about using this lower bound… he didn’t think it would work, but it did!

Bounding • Upper bound on the best score: score the NJ(EDE) tree using improved implementation of BPAnalysis. • Lower bound on a given tree T using the circular ordering on leaves: greedy technique to find the planar embedding that achieves the highest breakpoint score for its circular ordering – half that is a lower bound on T’s breakpoint score. By the way, Bernard and I had a big argument about using this lower bound… he didn’t think it would work, but it did!

GRAPPA http: //www. cs. unm. edu/~moret/GRAPPA/ • Genome Rearrangement Analysis under Parsimony and other Phylogenetic Algorithms • Heuristics for NP-hard optimization problems • Uses high-level algorithmic ideas with low-level algorithms engineering to dramatically speed-up the searches for the breakpoint and inversion phylogenies. • 2000 -2004 • Project leader: Bernard Moret

Benchmark gene order dataset: Campanulaceae • 12 genomes + 1 outgroup (Tobacco), 105 gene segments • NP-hard optimization problems: breakpoint and inversion phylogenies (techniques score every tree) Joint work with Bob Jansen, Linda Raubeson, Jijun Tang, and Li-San Wang 1997: BPAnalysis (Blanchette and Sankoff): 200 years (est. ) 2000: Using GRAPPA v 1. 1 on the 512 -processor Los Lobos Supercluster machine: 2 minutes (200, 000 -fold speedup per processor) 2003: Using latest version of GRAPPA: 2 minutes on a single processor (1 -billion-fold speedup per processor)

In 1999, Bob needed help with 13 Campanulaceae genomes

The challenges! 1. Find all the best genome trees for Bob’s dataset, and determine if inversions suffice (or if we really do need transpositions). 2. Design statistically rigorous methods for genome rearrangement phylogeny. 3. Design efficient techniques to enable genome-scale phylogeny for large datasets that are difficult to analyze.

What we found • Optimal solutions for the breakpoint phylogeny, for the inversion-only phylogeny, and for a weighted sum of inversions and transpositions. • 67 inversions suffices for this dataset, and no transpositions are needed!

This was just the beginning… Other events, such as • Duplications, Insertions, and Deletions • Fissions and Fusions Other models • HP (Hannenhali and Pevzner) • Double Cut-and-Join (Yancopoulos et al. ) Other techniques • DCM-boosting to scale GRAPPA to large datasets (1000 species) • Estimating true evolutionary distances under these complex models • Inferring ancestral genomes under these complex models

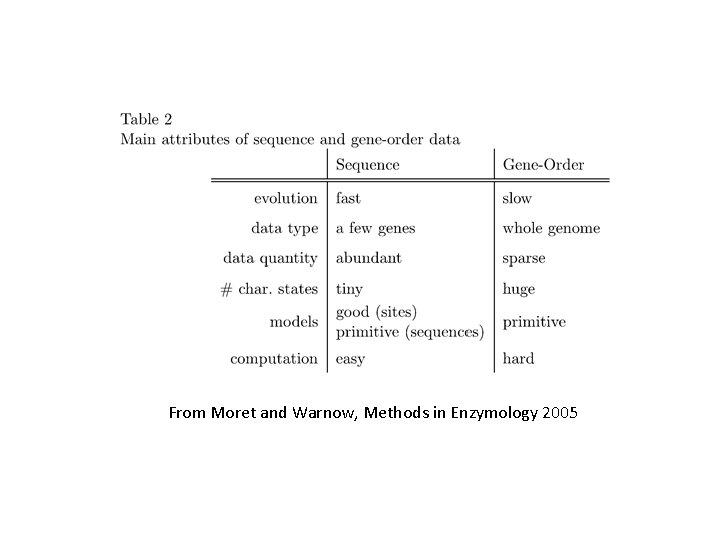
From Moret and Warnow, Methods in Enzymology 2005

March 29 to April 5 • March 29: Jinfeng, Wei, and Srilakshmi present papers. • April 3: Rishika and Aniket present papers. • April 5: Omri, Jeffrey, and Dikshant present papers. If you are presenting: send me the PPTX or PDF of your presentation by 10 PM the night before. I will post the presentations in the class webpage. All: read the papers *before* coming to class, and be prepared to ask questions. Please remember that class participation is 10% of the grade. Your class presentations are half of this class participation grade.
 Fineboosting
Fineboosting Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Tandy warnow
Tandy warnow Genome-to-genome distance calculator
Genome-to-genome distance calculator Tandy trower
Tandy trower Tandy trower
Tandy trower Mclafferty rearrangement
Mclafferty rearrangement Demjanov rearrangement
Demjanov rearrangement Once a lad went to a famous teacher rearrange
Once a lad went to a famous teacher rearrange Imine hydrolysis
Imine hydrolysis Carbonium ion rearrangement
Carbonium ion rearrangement Semipinacol rearrangement
Semipinacol rearrangement Catabolism stages
Catabolism stages Openreach appointment tracker
Openreach appointment tracker Rearrangement of atoms in a chemical reaction
Rearrangement of atoms in a chemical reaction Predict the product for the following cope rearrangement.
Predict the product for the following cope rearrangement. Sine rule
Sine rule Migratory aptitude is greater for
Migratory aptitude is greater for Deer family tree
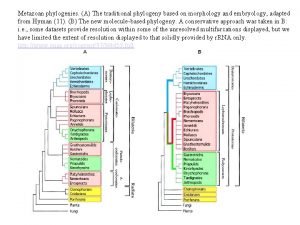
Deer family tree Phylogeny and the tree of life chapter 26
Phylogeny and the tree of life chapter 26 Bryozoa
Bryozoa Phylogenetic bracketing
Phylogenetic bracketing Crab cladogram
Crab cladogram Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Enantiornithes
Enantiornithes Fur
Fur Outgroup biology definition
Outgroup biology definition Fish phylogeny
Fish phylogeny Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Cladogram animal kingdom
Cladogram animal kingdom