DNA quality requirements for SingleMolecule sequencing Olga Vinnere












- Slides: 32

DNA quality requirements for Single-Molecule sequencing Olga Vinnere Pettersson, Ph. D Project coordinator NGI-Sweden / Sci. Life. Lab (UU)

National Genomics Infrastructure Sweden NGI Stockholm NGI Uppsala NGI staff: 60 -70 FTE, including head of facility, lab research engineers, bioinformaticians, IT-experts, project coordinators. Operates since 1998 (separate nodes); from 2006 as an infrastructure. Hosted by

NGI-Sci. Life. Lab is one of the most well-equipped NGS sites in Europe 10 17 3 1 2 6 2 2 1 2 Illumina Hi. Seq Xten Illumina Hi. Seq 2000/2500 Illumina Mi. Seq Illumina Next. Seq Life Technologies Ion Torrent Life Technologies Ion Proton Pacific Biosciences RSII Sanger ABI 3730 Argus Whole Genome Map. Syst. Oxford Nanopore Min. Ion

Outline • • • SMRT sequencing - the challenge Basics of DNA extraction Organism – specific extraction protocols Post-extraction clean-up Small adjustments that matter - examples • Conclusions

Pac. Bio told us not to, but we decided to try A WGS project of a mould Nothing…

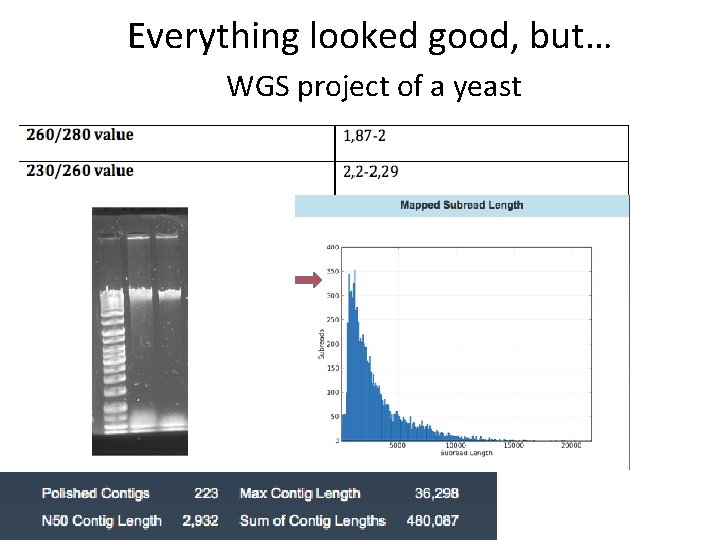
Everything looked good, but… WGS project of a yeast

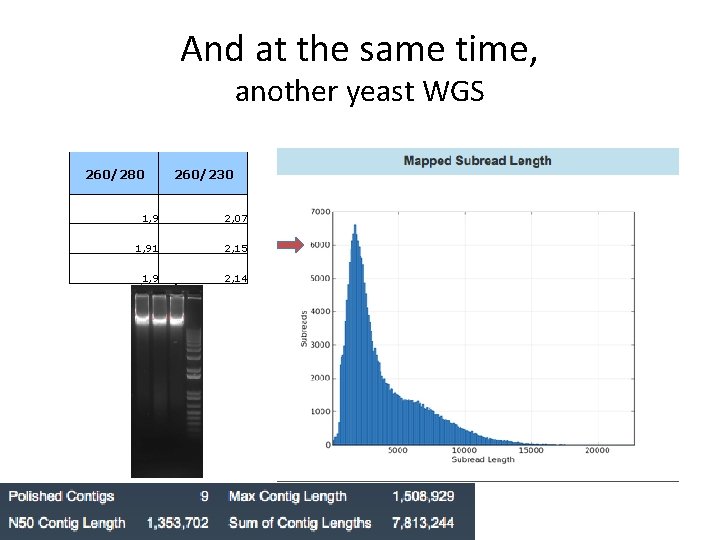
And at the same time, another yeast WGS 260/280 260/230 1, 9 2, 07 1, 91 2, 15 1, 9 2, 14

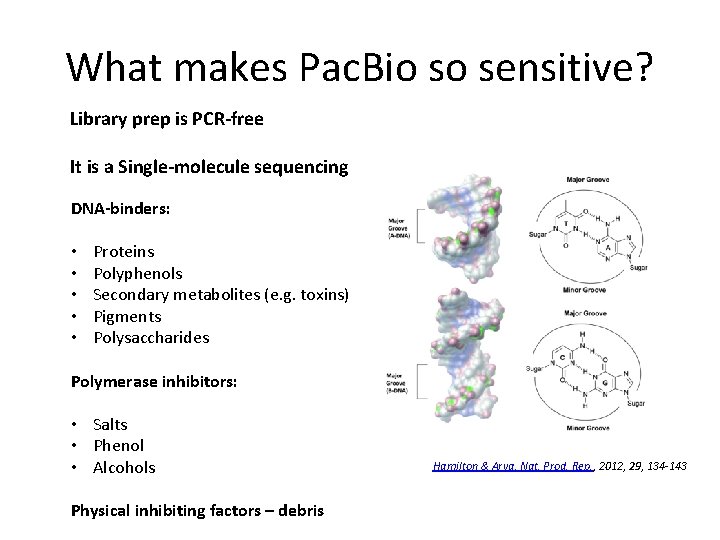
What makes Pac. Bio so sensitive? Library prep is PCR-free It is a Single-molecule sequencing DNA-binders: • • • Proteins Polyphenols Secondary metabolites (e. g. toxins) Pigments Polysaccharides Polymerase inhibitors: • Salts • Phenol • Alcohols Physical inhibiting factors – debris Hamilton & Arya, Nat. Prod. Rep. , 2012, 29, 134 -143

The DNA extraction process 1 2

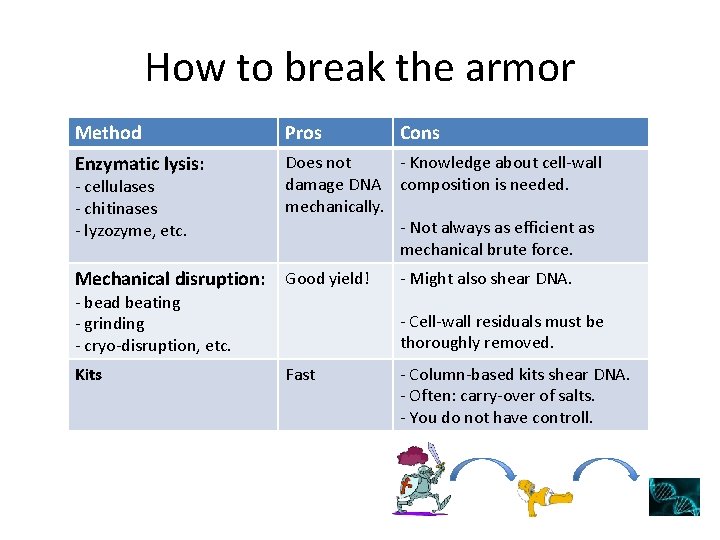
How to break the armor Method Pros Enzymatic lysis: Does not - Knowledge about cell-wall damage DNA composition is needed. mechanically. - Not always as efficient as mechanical brute force. - cellulases - chitinases - lyzozyme, etc. Mechanical disruption: Good yield! - bead beating - grinding - cryo-disruption, etc. Kits Cons - Might also shear DNA. - Cell-wall residuals must be thoroughly removed. Fast - Column-based kits shear DNA. - Often: carry-over of salts. - You do not have controll.

What are the main contaminants? Polysaccharides Lypopolysaccharides Growth media residuals Chitin Protein Secondary metabolites Pigments Growth media residuals Chitin Fats Proteins Pigments Polyphenols Polysaccharides Secondary metabolites Pigments

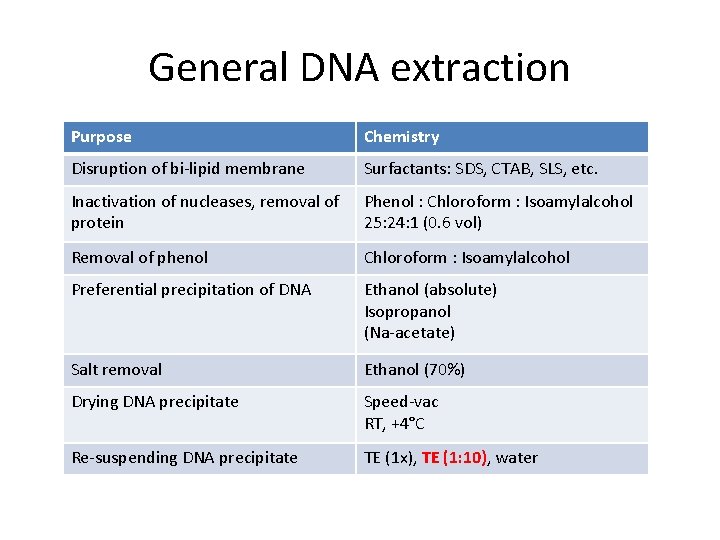
General DNA extraction Purpose Chemistry Disruption of bi-lipid membrane Surfactants: SDS, CTAB, SLS, etc. Inactivation of nucleases, removal of protein Phenol : Chloroform : Isoamylalcohol 25: 24: 1 (0. 6 vol) Removal of phenol Chloroform : Isoamylalcohol Preferential precipitation of DNA Ethanol (absolute) Isopropanol (Na-acetate) Salt removal Ethanol (70%) Drying DNA precipitate Speed-vac RT, +4°C Re-suspending DNA precipitate TE (1 x), TE (1: 10), water

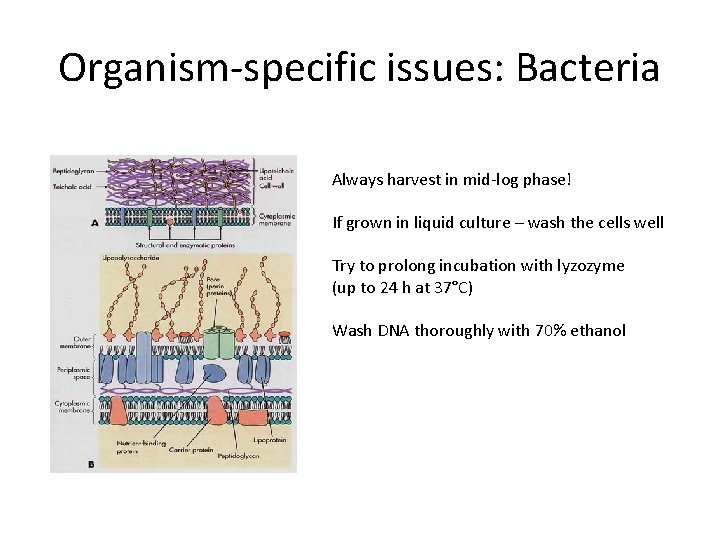
Organism-specific issues: Bacteria Always harvest in mid-log phase! If grown in liquid culture – wash the cells well Try to prolong incubation with lyzozyme (up to 24 h at 37°C) Wash DNA thoroughly with 70% ethanol

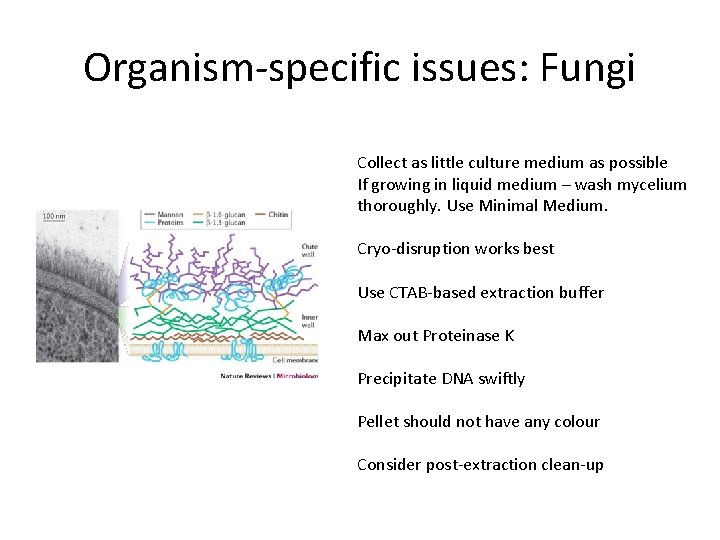
Organism-specific issues: Fungi Collect as little culture medium as possible If growing in liquid medium – wash mycelium thoroughly. Use Minimal Medium. Cryo-disruption works best Use CTAB-based extraction buffer Max out Proteinase K Precipitate DNA swiftly Pellet should not have any colour Consider post-extraction clean-up

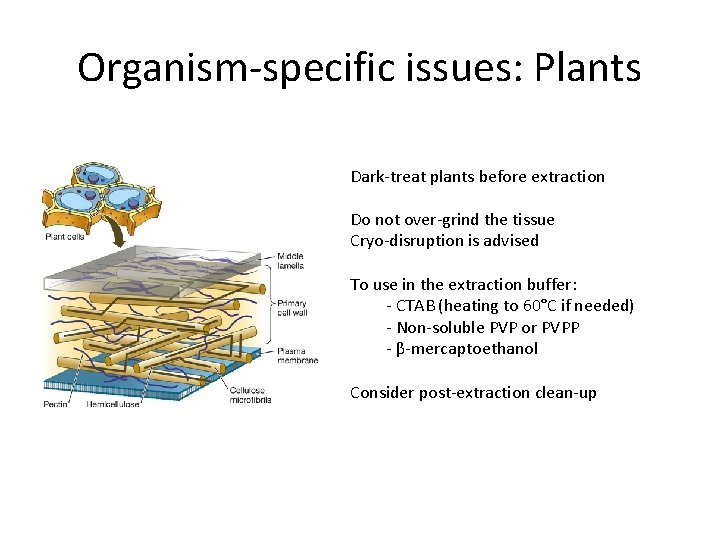
Organism-specific issues: Plants Dark-treat plants before extraction Do not over-grind the tissue Cryo-disruption is advised To use in the extraction buffer: - CTAB (heating to 60°C if needed) - Non-soluble PVP or PVPP - β-mercaptoethanol Consider post-extraction clean-up

Organism-specific issues: Insects Try to remove the shell completely Try cryo-disruption Try to boost your SDS & Prot. K concentrations (apply heating if necessary) Consider adding β-mercaptoethanol to the extraction buffer Precipitate DNA swiftly Consider post-extraction clean-up

Organism-specific issues: animal tissue Avoid high-fat tissues: brain, liver Use organs with no, or little connective tissue If using kits: check 260/230 value!

Notes on common, non-kit extraction protocols • Aim for high-yield • If working with Phenol: Chloroform: IAA protocol: make sure you remove phenol • Always use RNase • Proteinase K is tougher than you think • Isopropanol precipitation – RT and short time • Try to collect precipitate with glass-rod • Wash with 70% ethanol several times • Use 1: 10 TE buffer as DNA solvent

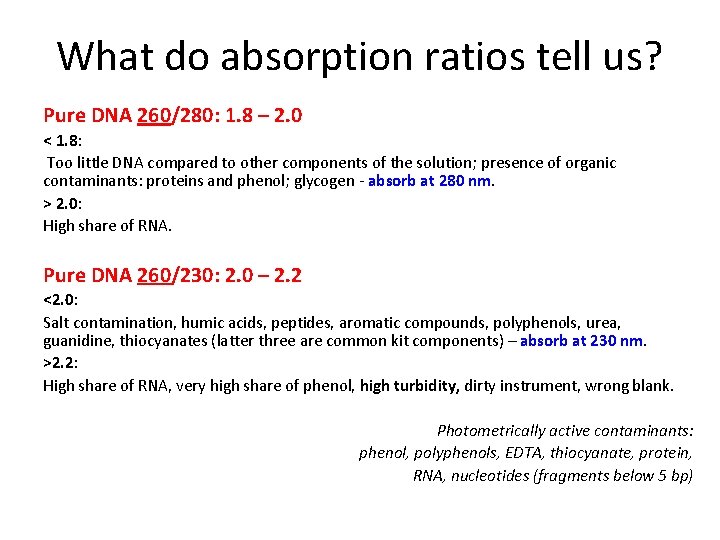
What do absorption ratios tell us? Pure DNA 260/280: 1. 8 – 2. 0 < 1. 8: Too little DNA compared to other components of the solution; presence of organic contaminants: proteins and phenol; glycogen - absorb at 280 nm. > 2. 0: High share of RNA. Pure DNA 260/230: 2. 0 – 2. 2 <2. 0: Salt contamination, humic acids, peptides, aromatic compounds, polyphenols, urea, guanidine, thiocyanates (latter three are common kit components) – absorb at 230 nm. >2. 2: High share of RNA, very high share of phenol, high turbidity, dirty instrument, wrong blank. Photometrically active contaminants: phenol, polyphenols, EDTA, thiocyanate, protein, RNA, nucleotides (fragments below 5 bp)

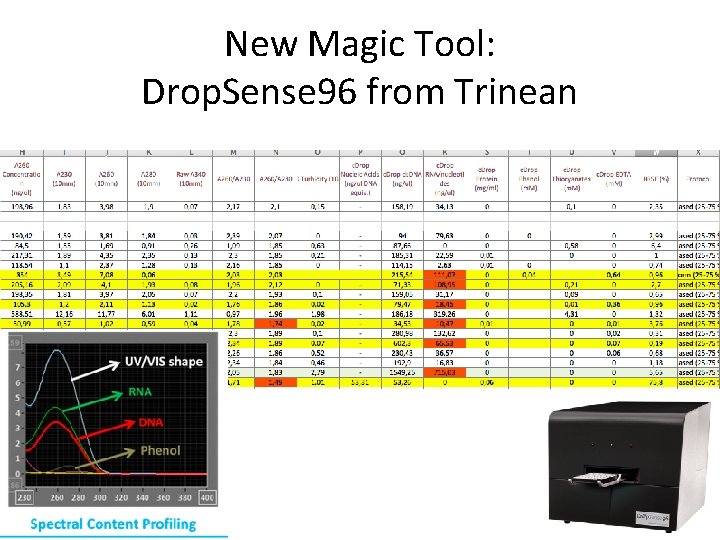
New Magic Tool: Drop. Sense 96 from Trinean

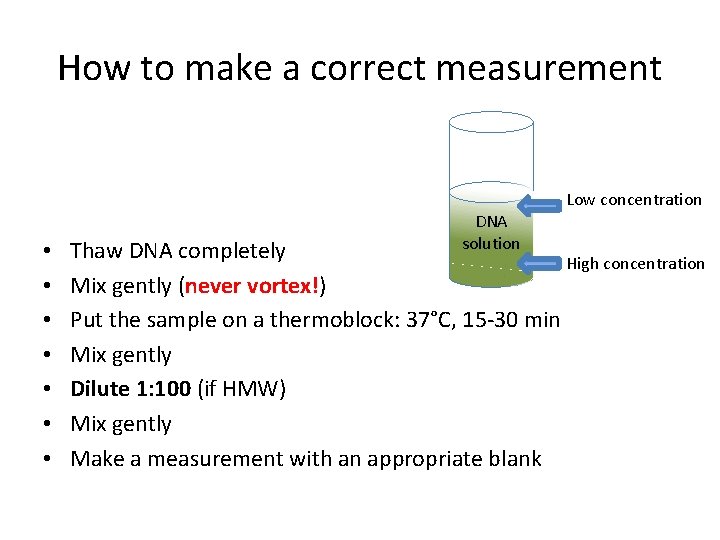
How to make a correct measurement Low concentration • • DNA solution Thaw DNA completely High concentration Mix gently (never vortex!) Put the sample on a thermoblock: 37°C, 15 -30 min Mix gently Dilute 1: 100 (if HMW) Mix gently Make a measurement with an appropriate blank

Recommended kit: Genomic Tip 500 Gravity-flow, anion-exchange Average size up to 50 -100 kb Other kits: Mag. Attract HMW DNA kit Qia. Gen Disclamer: Suggestions and advice are purely based on empirical evidence collected at Uppsala and that there is no conflict of interest.

Post-extraction cleanup Protein contamination - Apply phenol-chloroform Phenol carry-over or overloaded sample? RNA contamination - Apply RNase, followed by phenol-chloroform extraction If unsure, make dilution series. If problem persists – try Mo. Bio clean-up kit, or re-extract DNA

Post-extraction clean-up kits: Use for samples contaminated with humic acids, phenol, lipids, secondary metabolites, etc. Recover high-molecular weight DNA. Power. Clean®DNA clean-up kit Mo. Bio Genomic DNA Clean & Concentrator™ kit Zymo Research Be prepared to lose up to 80% of the sample…

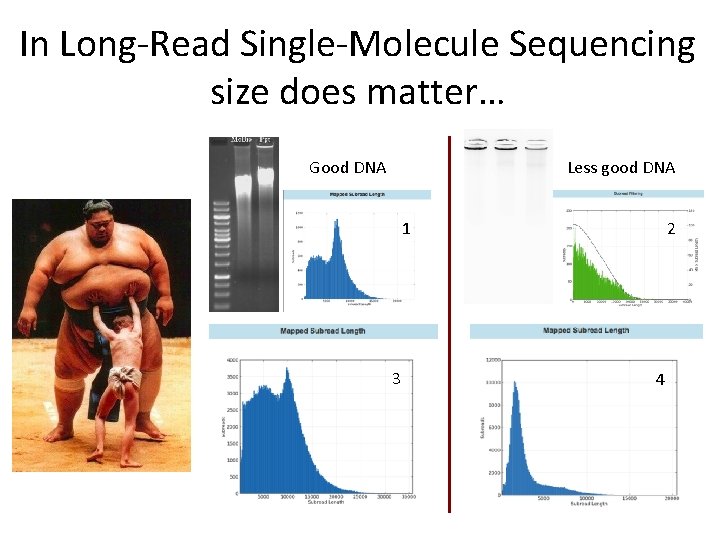
In Long-Read Single-Molecule Sequencing size does matter… Good DNA Less good DNA 1 3 2 4

General recommendations Treat DNA as a crystal vase: it is fragile when in solution As soon as DNA is released from the cells – use wide-bore tips Limit pipetting to minimum Never vortex! Do not heat above 65°C Reduce amount of freeze-thaw cycles to minimum Pac. Bio-grade DNA is quite different from PCR-grade DNA

To download a file with all these recommendations go to: https: //portal. scilifelab. se/genomics/node/1112

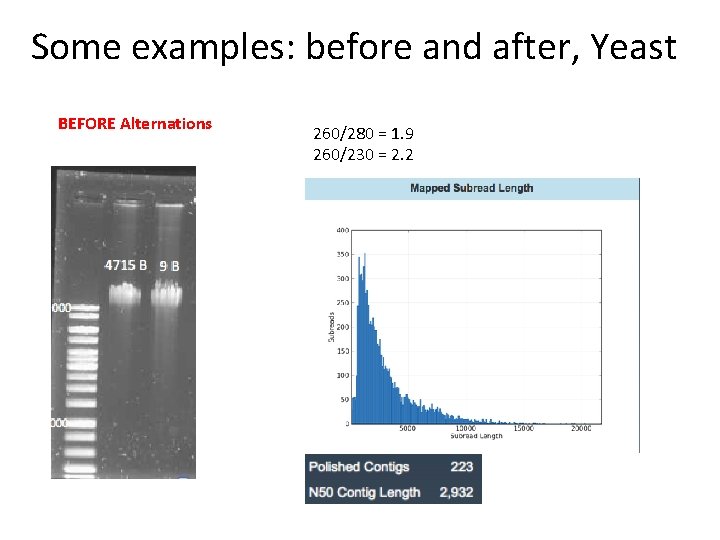
Some examples: before and after, Yeast BEFORE Alternations 260/280 = 1. 9 260/230 = 2. 2

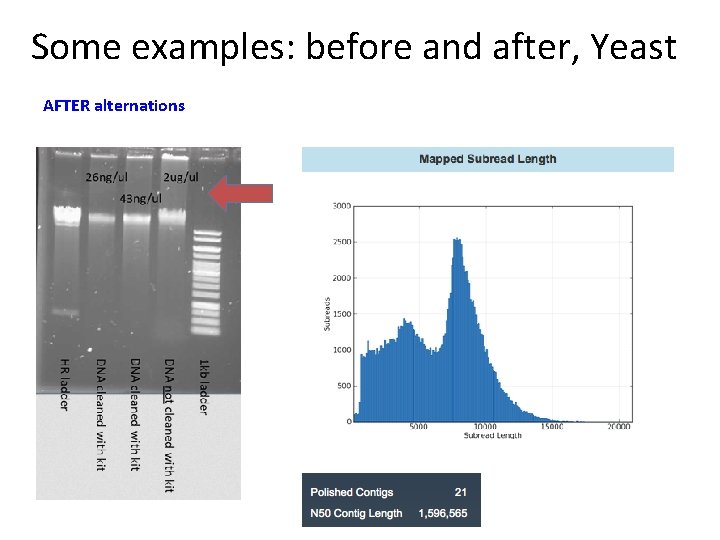
Some examples: before and after, Yeast AFTER alternations

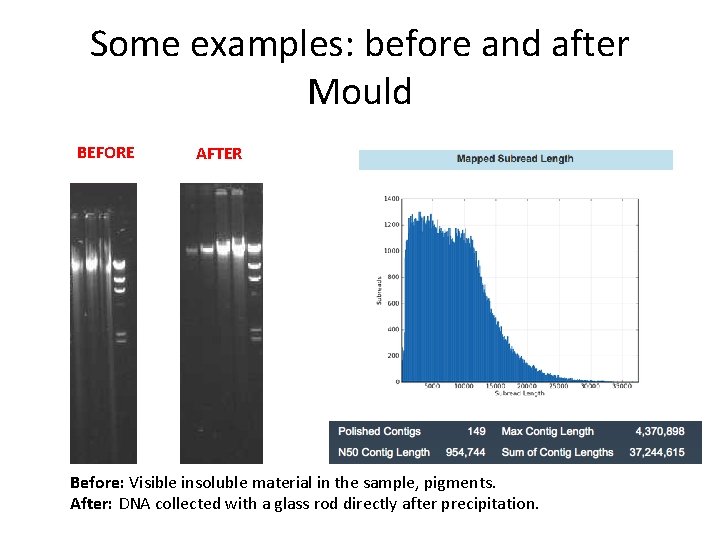
Some examples: before and after Mould BEFORE AFTER Before: Visible insoluble material in the sample, pigments. After: DNA collected with a glass rod directly after precipitation.

Acknowledgements • Swati Ranade, Pacific Biosciences • Jenny Ekholm, Pacific Biosciences • • Christian Tellgren-Roth, NGI Susana Häggqvist, NGI Ida Höijer, NGI Inger Jonasson, NGI

Questions, please!
 Olga v. pettersson
Olga v. pettersson Sanger
Sanger 3rd generation dna sequencing
3rd generation dna sequencing Dna sequencing methods
Dna sequencing methods Contigs
Contigs Dna sequencing
Dna sequencing Dna sequencing applications
Dna sequencing applications Function of dna polymerase 3
Function of dna polymerase 3 Bioflix activity dna replication dna replication diagram
Bioflix activity dna replication dna replication diagram Coding dna and non coding dna
Coding dna and non coding dna What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication What is conformance to requirements
What is conformance to requirements Epa requirements for quality assurance project plans
Epa requirements for quality assurance project plans Quality control and quality assurance
Quality control and quality assurance Quality management pmp
Quality management pmp Pmbok quality management
Pmbok quality management Ana quality assurance model
Ana quality assurance model Compliance vs quality
Compliance vs quality Concept of quality assurance
Concept of quality assurance Quality definition by quality gurus
Quality definition by quality gurus Quality is free: the art of making quality certain
Quality is free: the art of making quality certain What is tqm
What is tqm Fspos
Fspos Novell typiska drag
Novell typiska drag Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Vad står k.r.å.k.a.n för
Vad står k.r.å.k.a.n för Varför kallas perioden 1918-1939 för mellankrigstiden?
Varför kallas perioden 1918-1939 för mellankrigstiden? En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Adressändring ideell förening
Adressändring ideell förening Personlig tidbok
Personlig tidbok A gastrica
A gastrica Vad är densitet
Vad är densitet