Directed Mutagenesis and Protein Engineering Improving Properties of





- Slides: 21

Directed Mutagenesis and Protein Engineering

Improving Properties of Proteins for Therapeutic and Industrial Applications n Using a set of techniques that specifically change amino acids encoded by a cloned gene, proteins with properties that are better suited than naturally-occurring counterparts can be created for therapeutic and industrial applications.

Improving Properties of Proteins for Therapeutic and Industrial Applications n n n Increasing catalytic activity Increasing stability at high temperature, extreme p. H, or in organic solvent Elimination of cofactor requirement Increasing substrate specificity Increasing resistance to cellular protease Diminishing feedback inhibition by altering the allosteric regulation

Directed Mutagenesis ~ a process for generating amino acid coding changes at the DNA level n Deciding the sites for mutagenesis ¨ ¨ n Site-directed mutagenesis using oligonucleotide ¨ ¨ n Based on structural information Computer-assisted design With M 13 DNA Using PCR Random mutagenesis ¨ ¨ ¨ With oligonucleotide analogues Error-prone PCR DNA shuffling

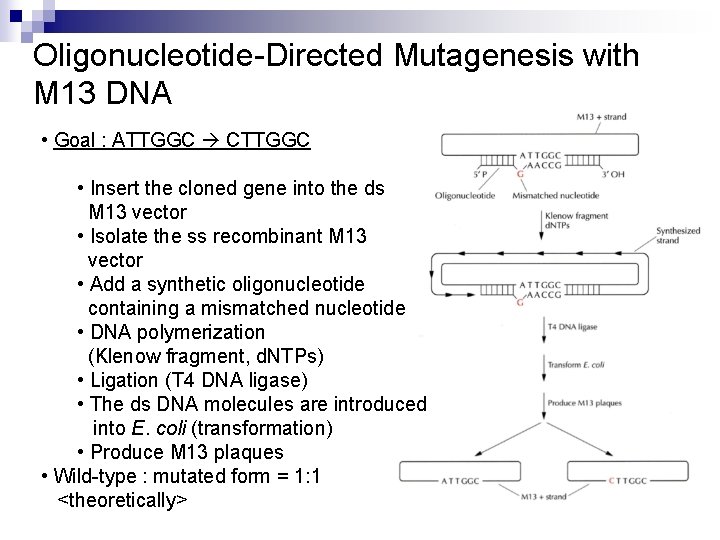
Oligonucleotide-Directed Mutagenesis with M 13 DNA • Goal : ATTGGC CTTGGC • Insert the cloned gene into the ds M 13 vector • Isolate the ss recombinant M 13 vector • Add a synthetic oligonucleotide containing a mismatched nucleotide • DNA polymerization (Klenow fragment, d. NTPs) • Ligation (T 4 DNA ligase) • The ds DNA molecules are introduced into E. coli (transformation) • Produce M 13 plaques • Wild-type : mutated form = 1: 1 <theoretically>

Mutagenesis with M 13 DNA n Recovery of the mutated DNA ¨ Expected: 50% ¨ Result: 1~5% (for technical reasons)

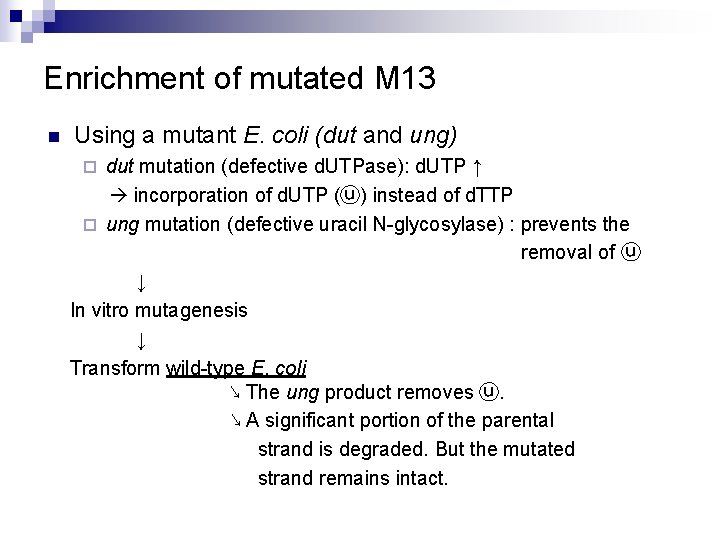
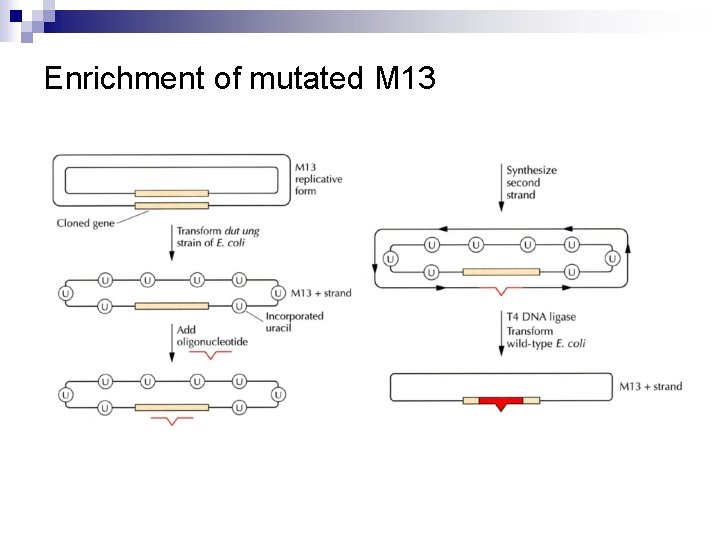
Enrichment of mutated M 13 n Using a mutant E. coli (dut and ung) dut mutation (defective d. UTPase): d. UTP ↑ incorporation of d. UTP (ⓤ) instead of d. TTP ¨ ung mutation (defective uracil N-glycosylase) : prevents the removal of ⓤ ↓ In vitro mutagenesis ↓ Transform wild-type E. coli ↘ The ung product removes ⓤ. ↘ A significant portion of the parental strand is degraded. But the mutated strand remains intact. ¨

Enrichment of mutated M 13

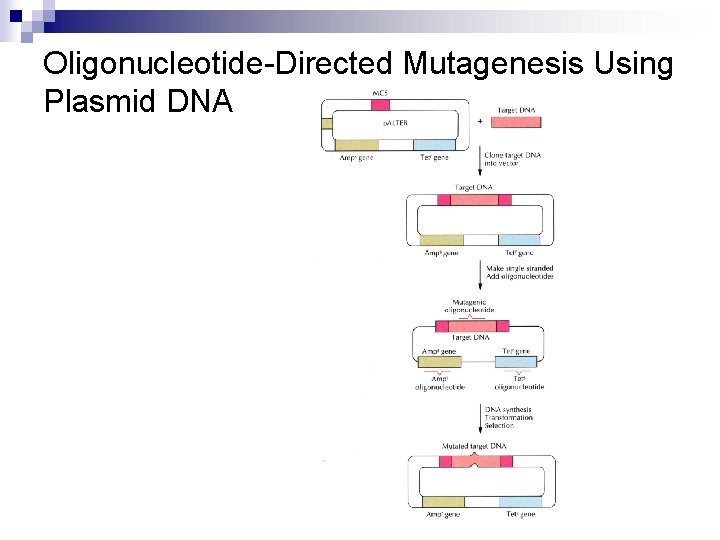
Oligonucleotide-Directed Mutagenesis Using Plasmid DNA

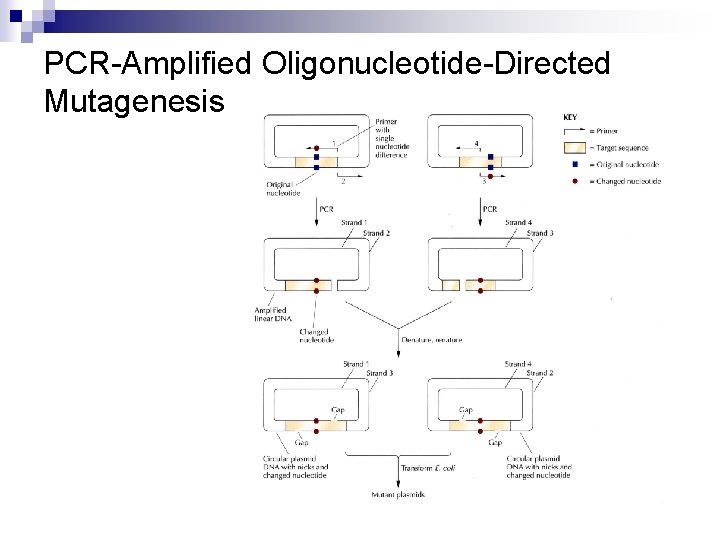
PCR-Amplified Oligonucleotide-Directed Mutagenesis

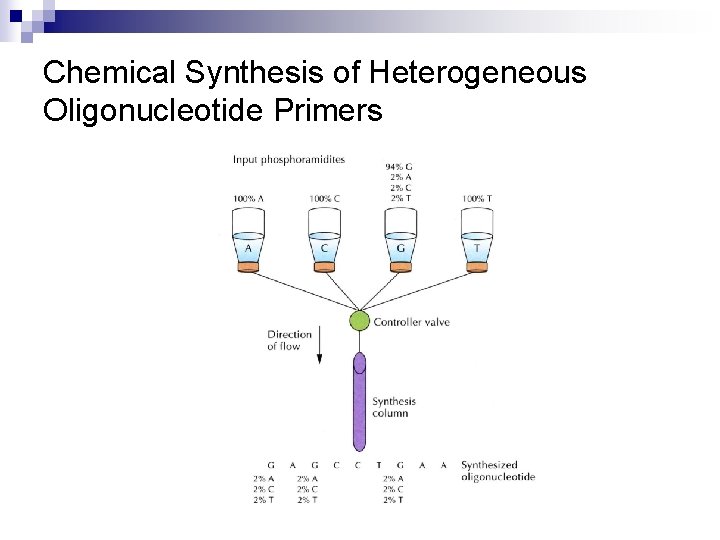
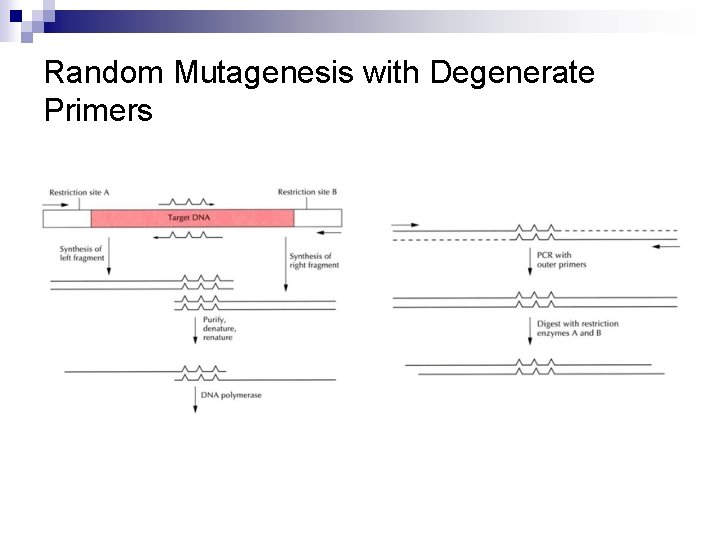
Random Mutagenesis with Degenerate Primers n n n To generate all the possible amino acid changes at one particular site Using heterogeneous oligonucleotide primers synthesized with any of the four nucleotides at defined positions (Figure 8. 5) Random mutagenesis with degenerate oligonucleotide primers (Figure 8. 6)

Chemical Synthesis of Heterogeneous Oligonucleotide Primers

Random Mutagenesis with Degenerate Primers

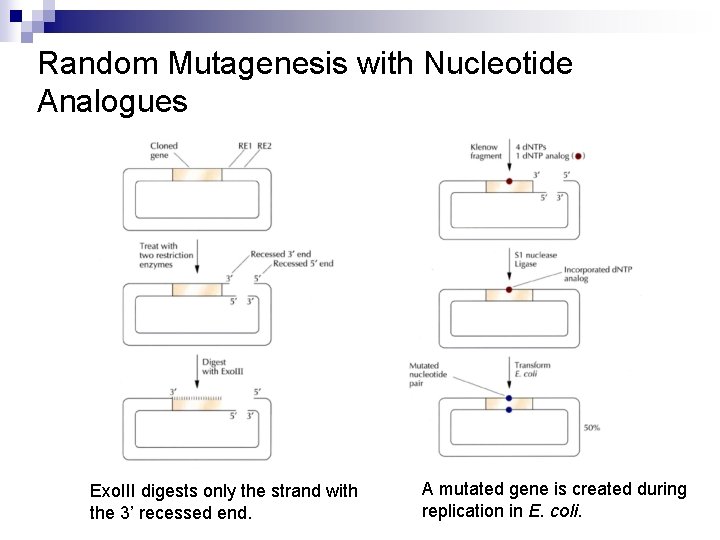
Random Mutagenesis with Nucleotide Analogues Exo. III digests only the strand with the 3’ recessed end. A mutated gene is created during replication in E. coli.

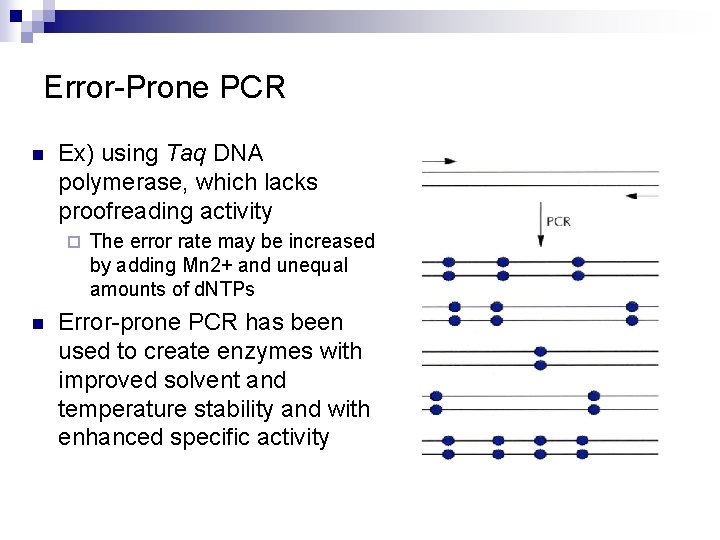
Error-Prone PCR n Ex) using Taq DNA polymerase, which lacks proofreading activity ¨ n The error rate may be increased by adding Mn 2+ and unequal amounts of d. NTPs Error-prone PCR has been used to create enzymes with improved solvent and temperature stability and with enhanced specific activity

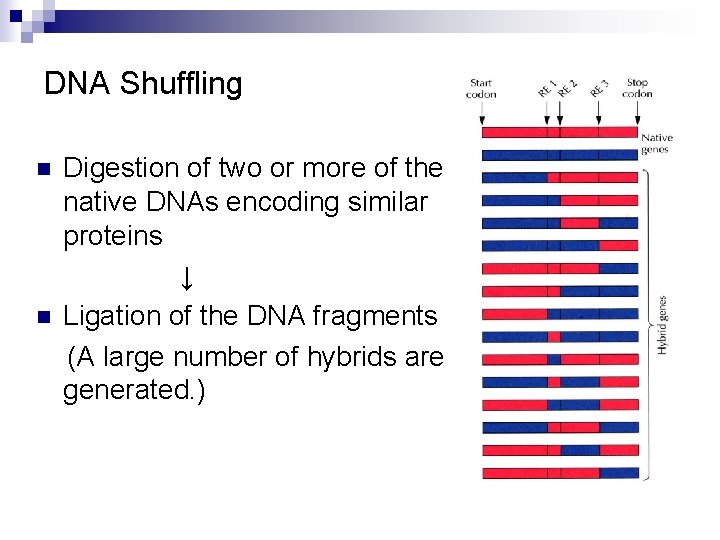
DNA Shuffling n n Digestion of two or more of the native DNAs encoding similar proteins ↓ Ligation of the DNA fragments (A large number of hybrids are generated. )

DNA Shuffling n n Fragmentation of DNA (several members of a gene family) with DNase I (endonuclease) ↓ Selection of smaller DNA fragments ↓ PCR without primers (DNA fragments cross-prime each other. ) ↓ PCR with ‘terminal primers’ to establish full-length hybrid DNAs.

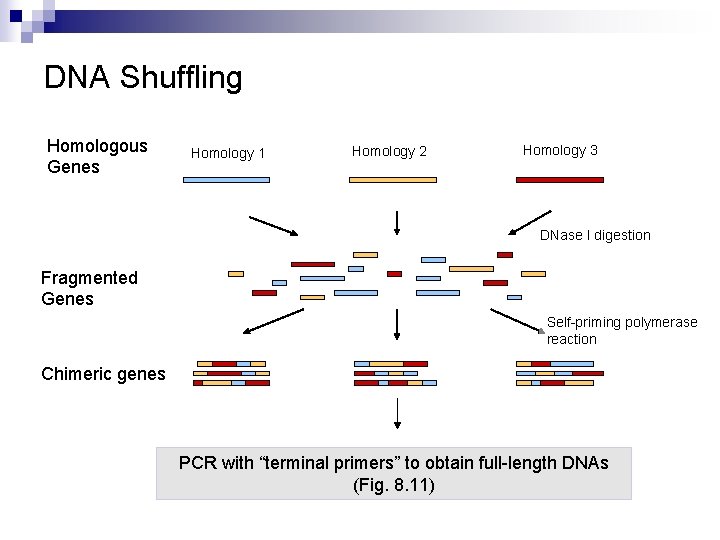
DNA Shuffling Homologous Genes Homology 1 Homology 2 Homology 3 DNase I digestion Fragmented Genes Self-priming polymerase reaction Chimeric genes PCR with “terminal primers” to obtain full-length DNAs (Fig. 8. 11)

DNA Shuffling

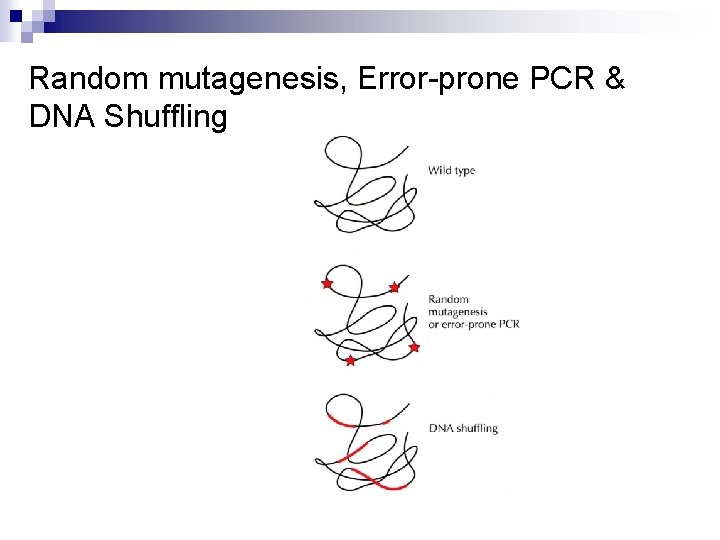
Random mutagenesis, Error-prone PCR & DNA Shuffling

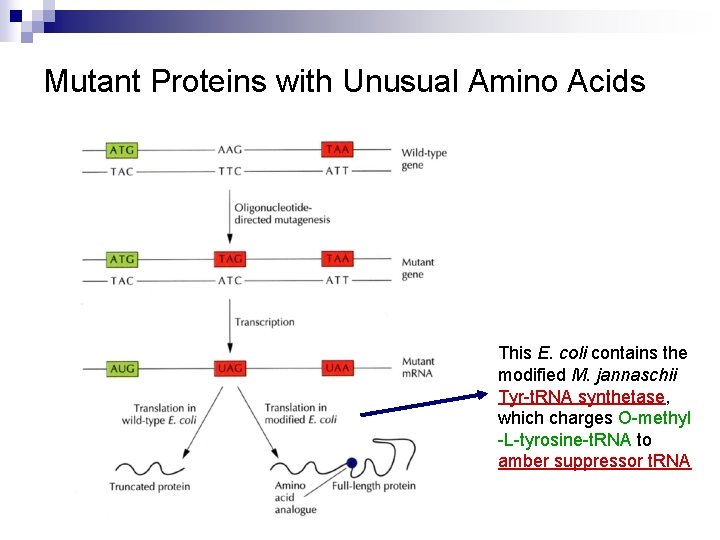
Mutant Proteins with Unusual Amino Acids This E. coli contains the modified M. jannaschii Tyr-t. RNA synthetase, which charges O-methyl -L-tyrosine-t. RNA to amber suppressor t. RNA