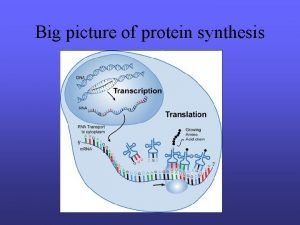
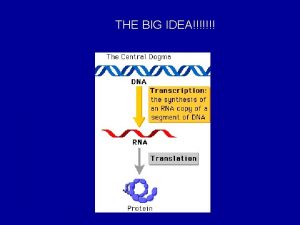
D Cell Specialization Regulation of Transcription Cell specialization



- Slides: 45

D. Cell Specialization: Regulation of Transcription Cell specialization in multicellular organisms results from differential gene expression

D. CELL SPECIALIZATION: Regulation of Transcription 1. Chromosome, Gene and RNA Architecture 2. Cell-Specific Regulation of Chromosome Structure 3. Cell-Specific Regulation of Transcription Activation

1. Review of Chromosome, Gene and RNA Architecture a. Review of Chromatin Structure b. Chromosomal Gene Arrangement c. Single Gene Components d. Nuclear RNA, m. RNA and Protein e. Other RNA Molecules f. Fast review of Transcription

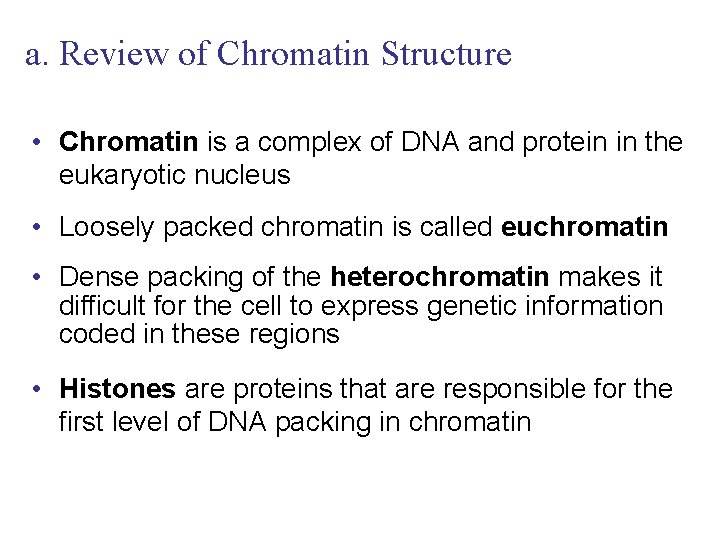
a. Review of Chromatin Structure • Chromatin is a complex of DNA and protein in the eukaryotic nucleus • Loosely packed chromatin is called euchromatin • Dense packing of the heterochromatin makes it difficult for the cell to express genetic information coded in these regions • Histones are proteins that are responsible for the first level of DNA packing in chromatin

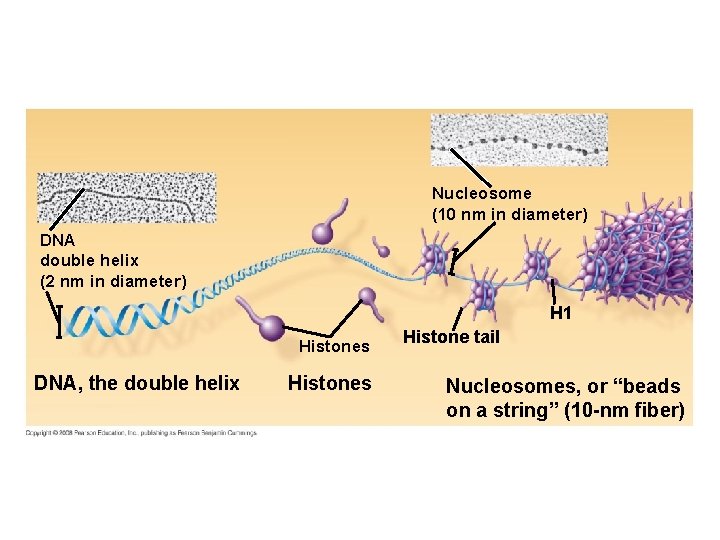
Nucleosome (10 nm in diameter) DNA double helix (2 nm in diameter) H 1 Histones DNA, the double helix Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)

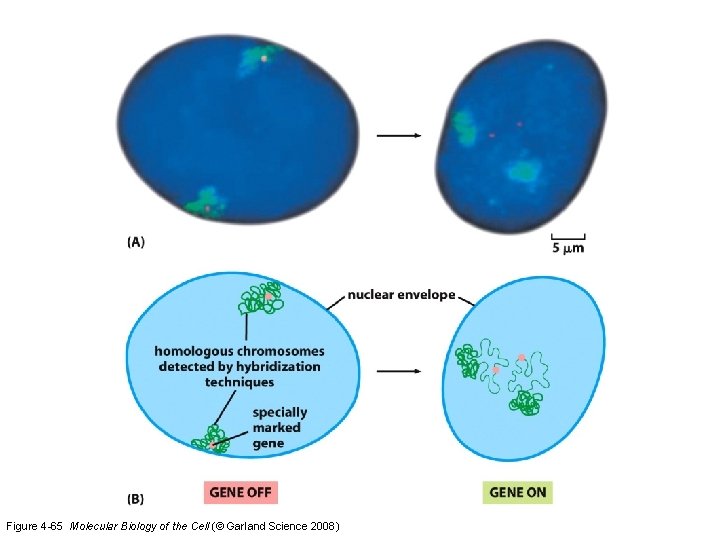
Figure 4 -65 Molecular Biology of the Cell (© Garland Science 2008)

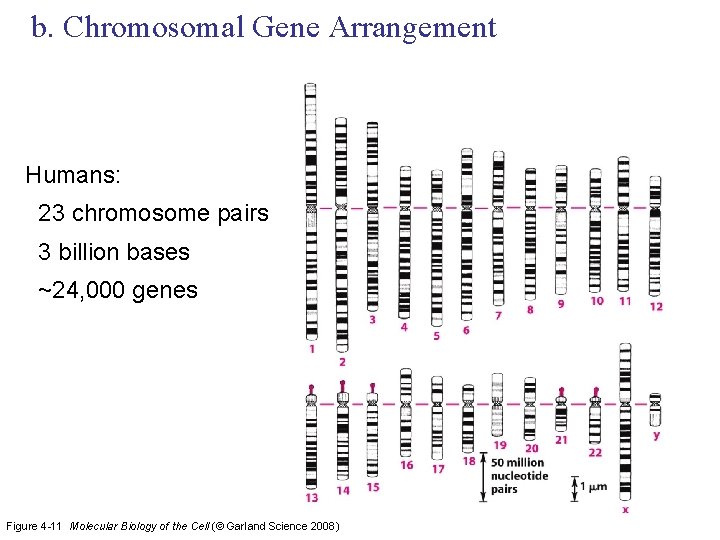
b. Chromosomal Gene Arrangement Humans: 23 chromosome pairs 3 billion bases ~24, 000 genes Figure 4 -11 Molecular Biology of the Cell (© Garland Science 2008)

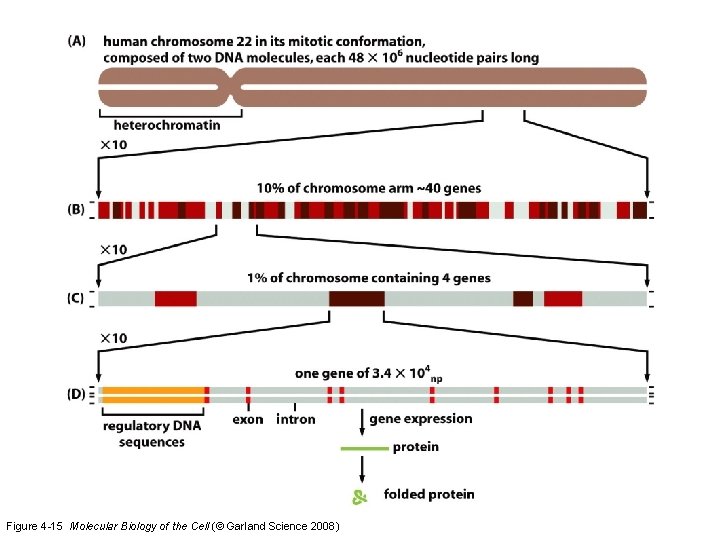
Figure 4 -15 Molecular Biology of the Cell (© Garland Science 2008)

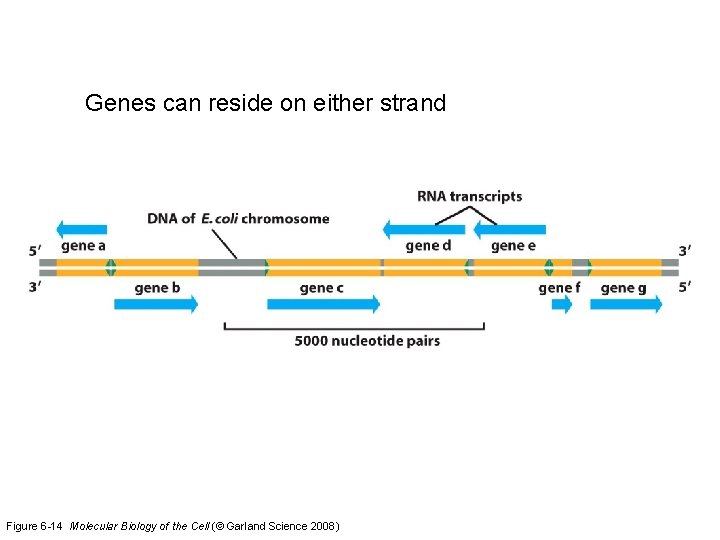
Genes can reside on either strand Figure 6 -14 Molecular Biology of the Cell (© Garland Science 2008)

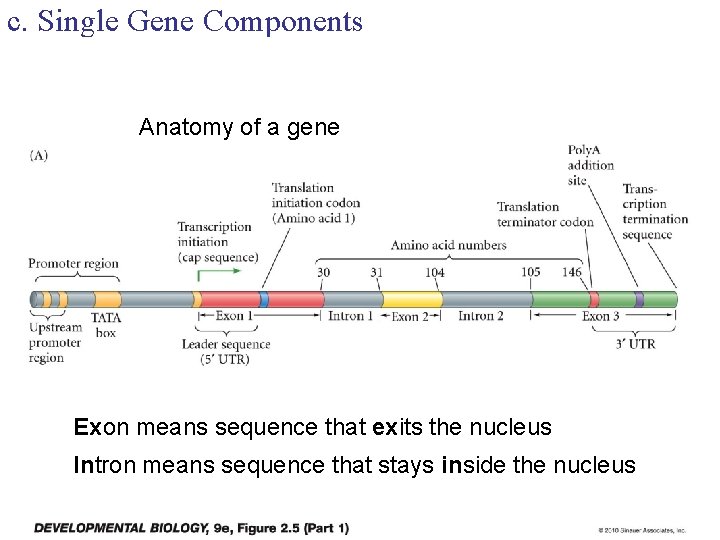
c. Single Gene Components Anatomy of a gene Exon means sequence that exits the nucleus Intron means sequence that stays inside the nucleus

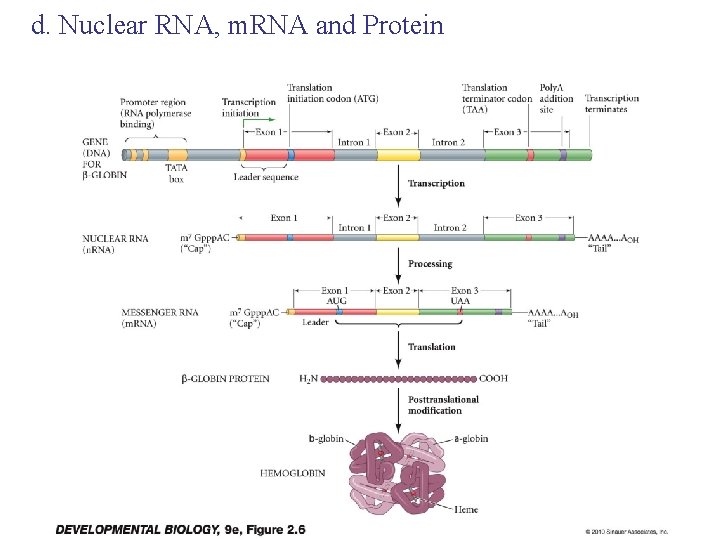
d. Nuclear RNA, m. RNA and Protein

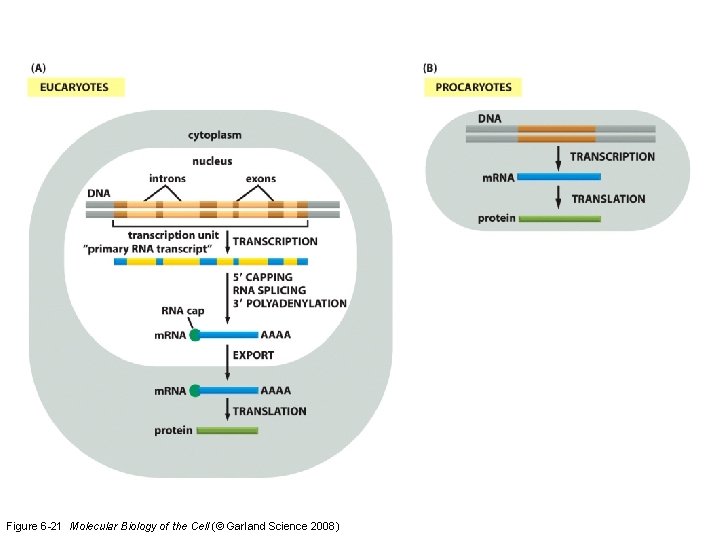
Figure 6 -21 Molecular Biology of the Cell (© Garland Science 2008)

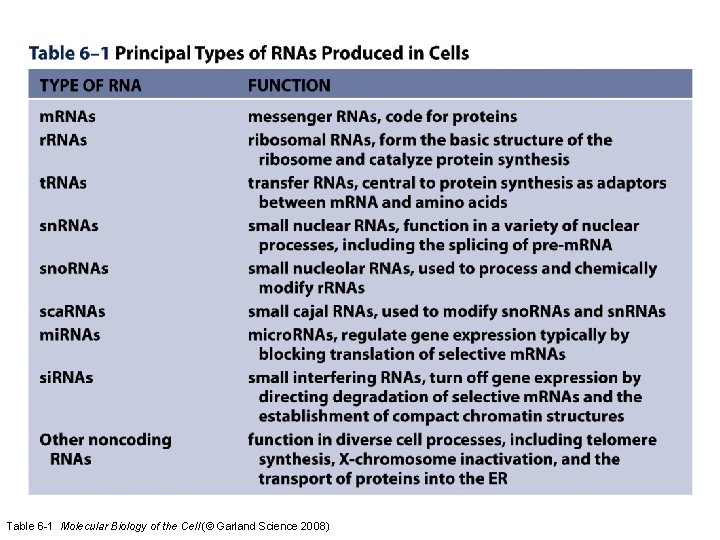
e. Other RNA Molecules 1. The Translational Apparatus 2. Nuclear Effectors 3. Cytosolic Effectors

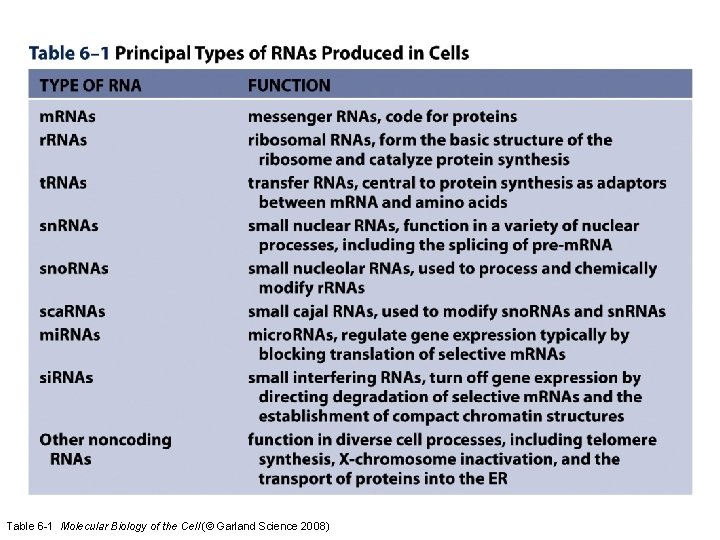
Table 6 -1 Molecular Biology of the Cell (© Garland Science 2008)

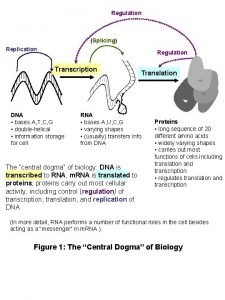
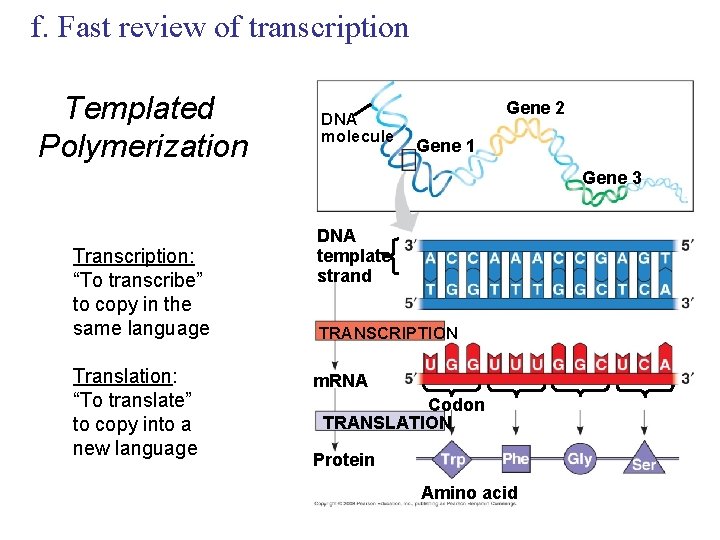
f. Fast review of transcription Templated Polymerization DNA molecule Gene 2 Gene 1 Gene 3 Transcription: “To transcribe” to copy in the same language Translation: “To translate” to copy into a new language DNA template strand TRANSCRIPTION m. RNA Codon TRANSLATION Protein Amino acid

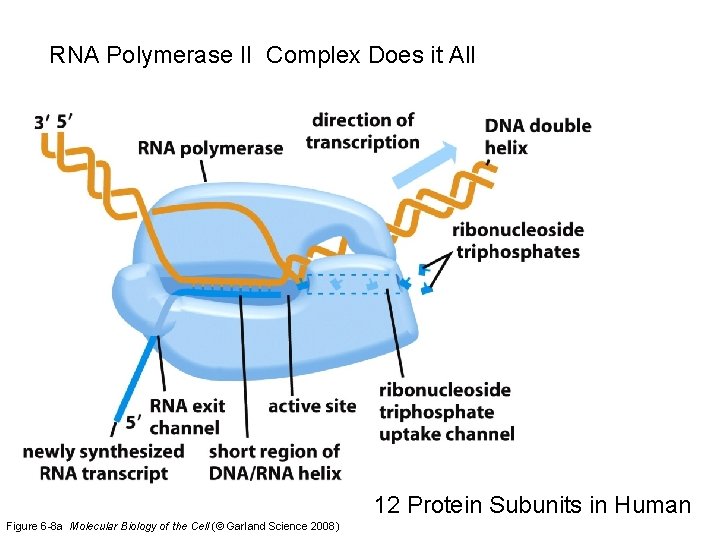
RNA Polymerase II Complex Does it All 12 Protein Subunits in Human Figure 6 -8 a Molecular Biology of the Cell (© Garland Science 2008)

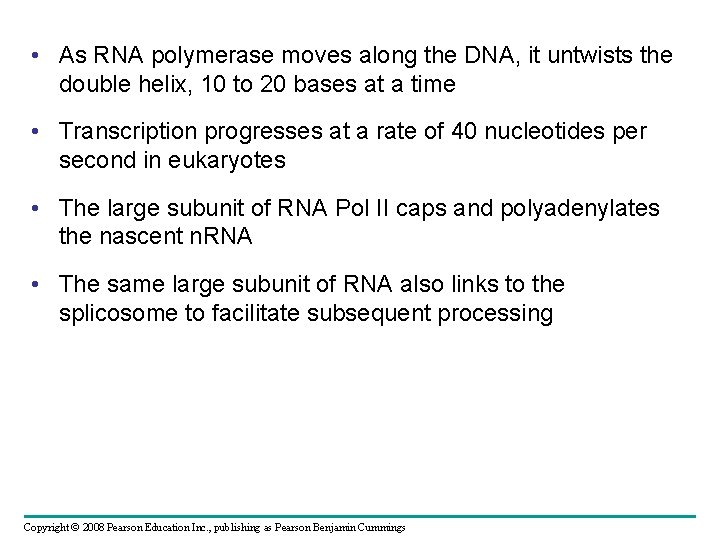
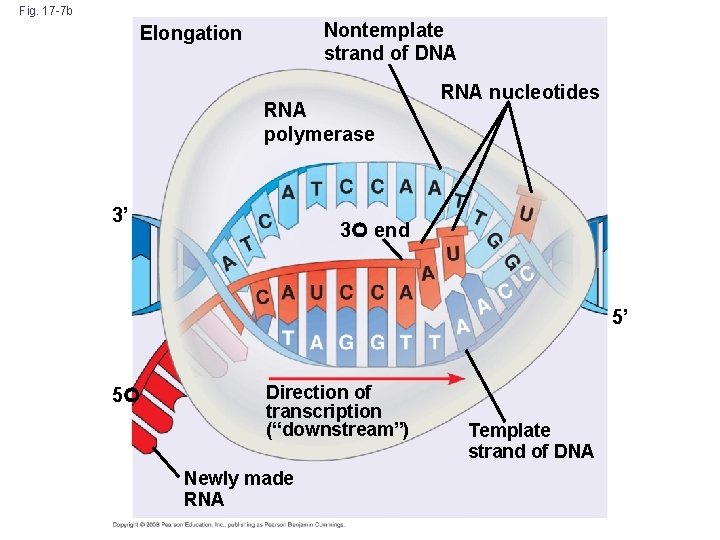
• As RNA polymerase moves along the DNA, it untwists the double helix, 10 to 20 bases at a time • Transcription progresses at a rate of 40 nucleotides per second in eukaryotes • The large subunit of RNA Pol II caps and polyadenylates the nascent n. RNA • The same large subunit of RNA also links to the splicosome to facilitate subsequent processing Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

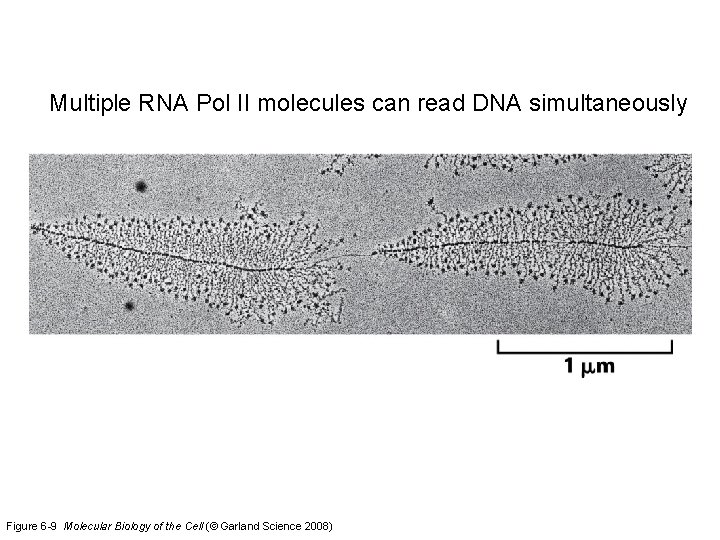
Multiple RNA Pol II molecules can read DNA simultaneously Figure 6 -9 Molecular Biology of the Cell (© Garland Science 2008)

• So, how do individual cells regulate which of the genes in their genome they will express? • Remember from Intro Bio that prokaryotes regulate expression through repressors/activators • Eukaryotes have more complex regulatory mechanisms • Histone modification regulates chromatin structure • DNA modification regulates promoter accessibility • Epigenetic modification can be copied and inherited • Transcription factors regulate promoter activation • Specialized transcriptional activities

2. Nucleosome and Histone Modification Regulates of Chromatin Structure • Chromatin is a complex of DNA and protein in the eukaryotic nucleus • Loosely packed chromatin is called euchromatin • Dense packing of the heterochromatin makes it difficult for the cell to express genetic information coded in these regions • Histones are proteins that are responsible for the first level of DNA packing in chromatin

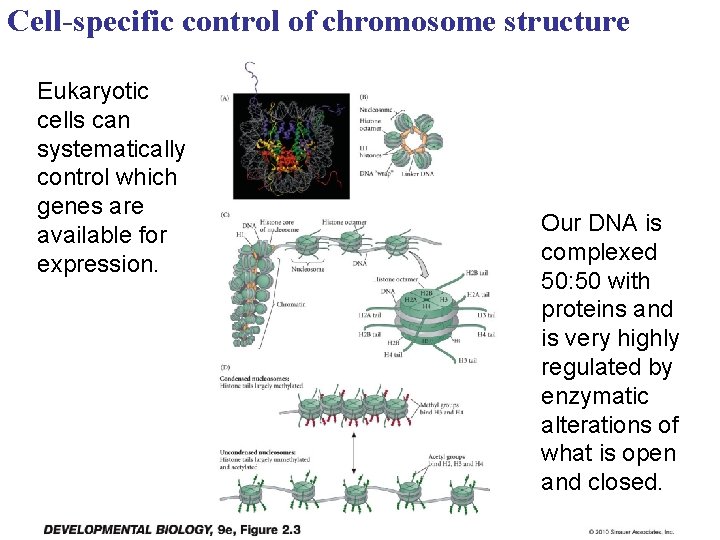
Cell-specific control of chromosome structure Eukaryotic cells can systematically control which genes are available for expression. Our DNA is complexed 50: 50 with proteins and is very highly regulated by enzymatic alterations of what is open and closed.

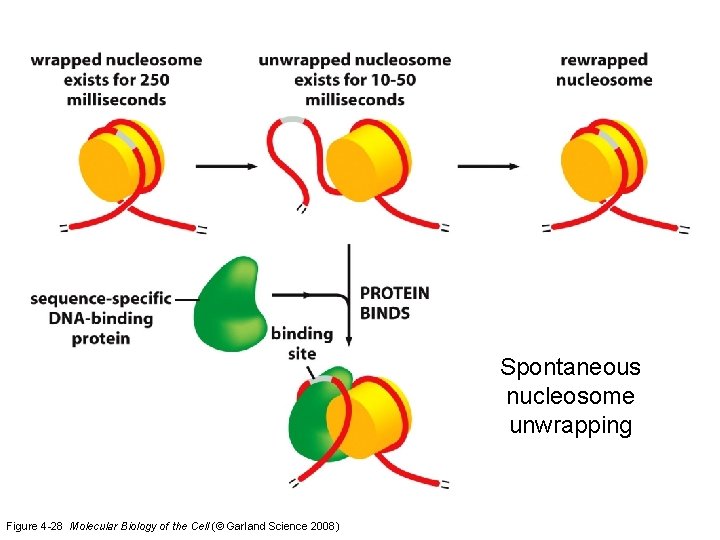
Spontaneous nucleosome unwrapping Figure 4 -28 Molecular Biology of the Cell (© Garland Science 2008)

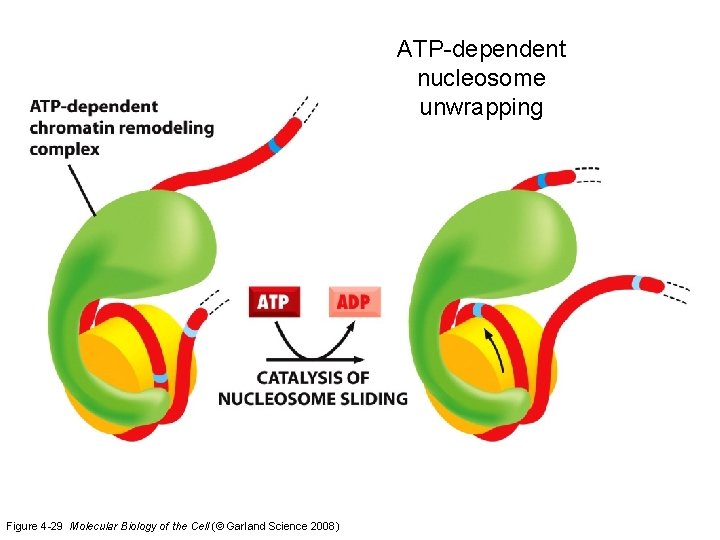
ATP-dependent nucleosome unwrapping Figure 4 -29 Molecular Biology of the Cell (© Garland Science 2008)

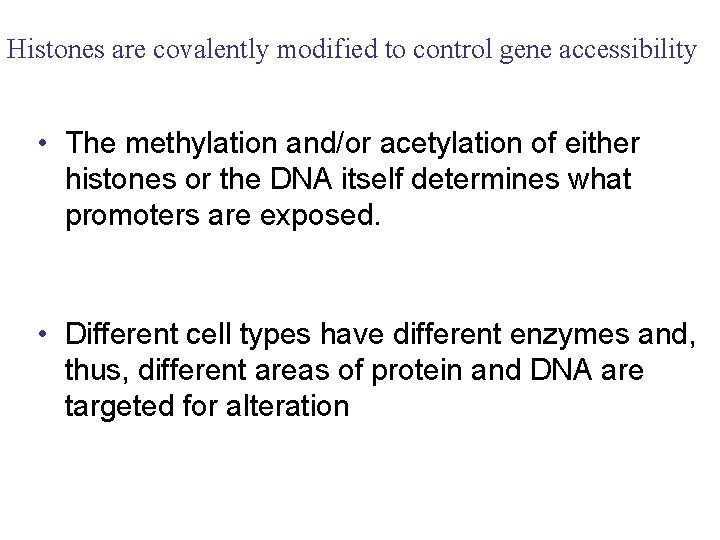
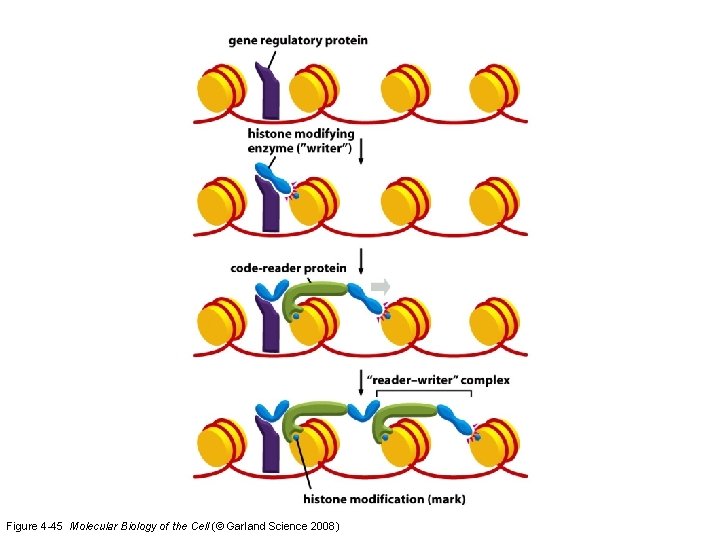
Histones are covalently modified to control gene accessibility • The methylation and/or acetylation of either histones or the DNA itself determines what promoters are exposed. • Different cell types have different enzymes and, thus, different areas of protein and DNA are targeted for alteration

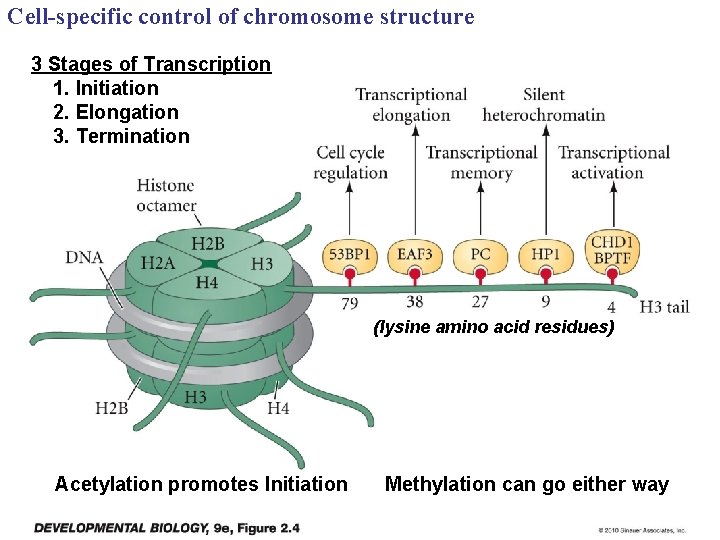
Cell-specific control of chromosome structure 3 Stages of Transcription 1. Initiation 2. Elongation 3. Termination (lysine amino acid residues) Acetylation promotes Initiation Methylation can go either way

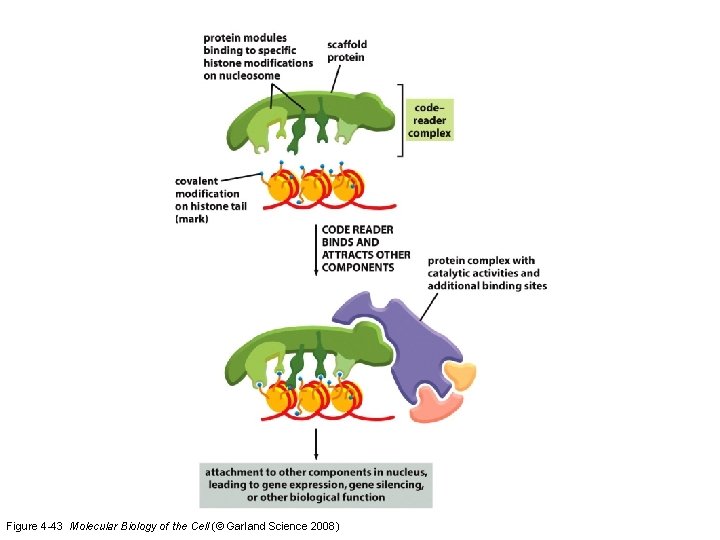
3. The “Histone Code” Hypothesis • Combinations of covalent modifications have specific information for the cell – This DNA is newly replicated – This DNA is damaged and needs repair – Express this DNA – Put this DNA into heterochromatin storage

Figure 4 -43 Molecular Biology of the Cell (© Garland Science 2008)

Figure 4 -45 Molecular Biology of the Cell (© Garland Science 2008)

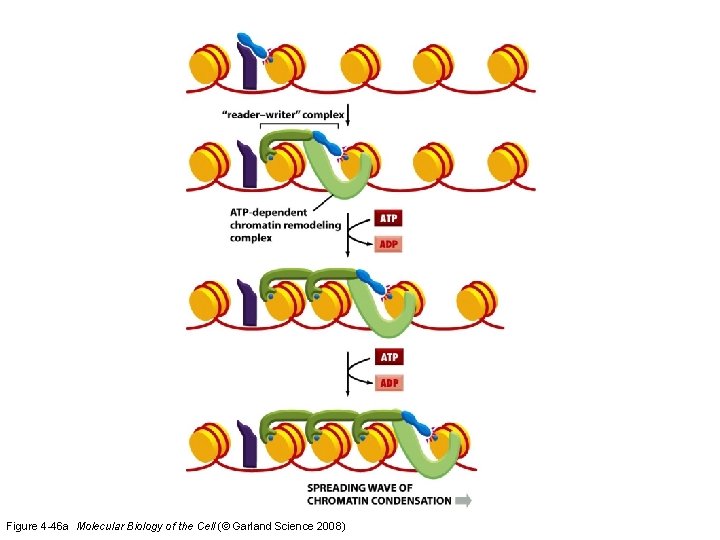
Figure 4 -46 a Molecular Biology of the Cell (© Garland Science 2008)

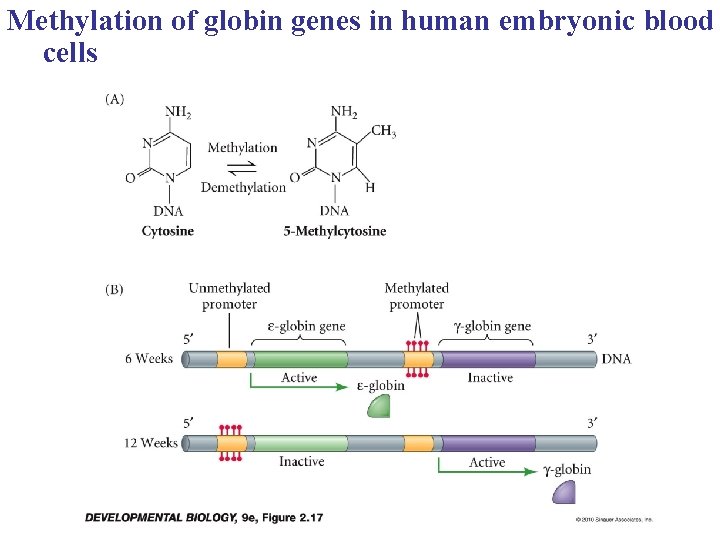
b. Direct covalent modifications of DNA can also control expression from genes in the euchromatin

Methylation of globin genes in human embryonic blood cells

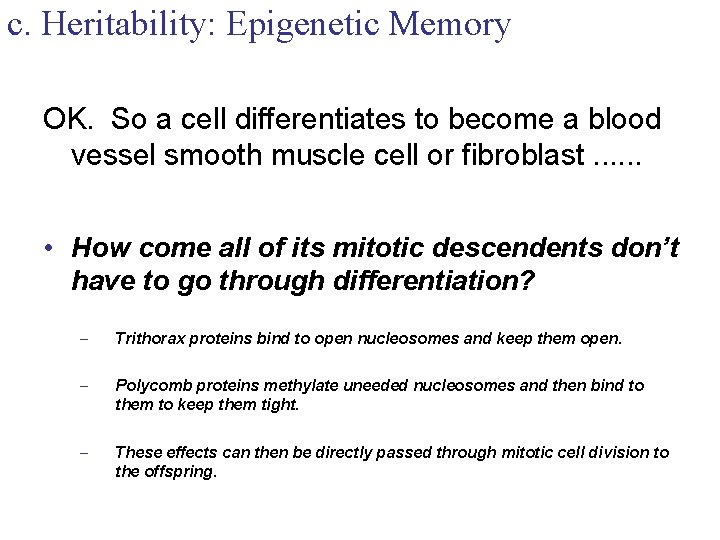
c. Heritability: Epigenetic Memory OK. So a cell differentiates to become a blood vessel smooth muscle cell or fibroblast. . . • How come all of its mitotic descendents don’t have to go through differentiation? – Trithorax proteins bind to open nucleosomes and keep them open. – Polycomb proteins methylate uneeded nucleosomes and then bind to them to keep them tight. – These effects can then be directly passed through mitotic cell division to the offspring.

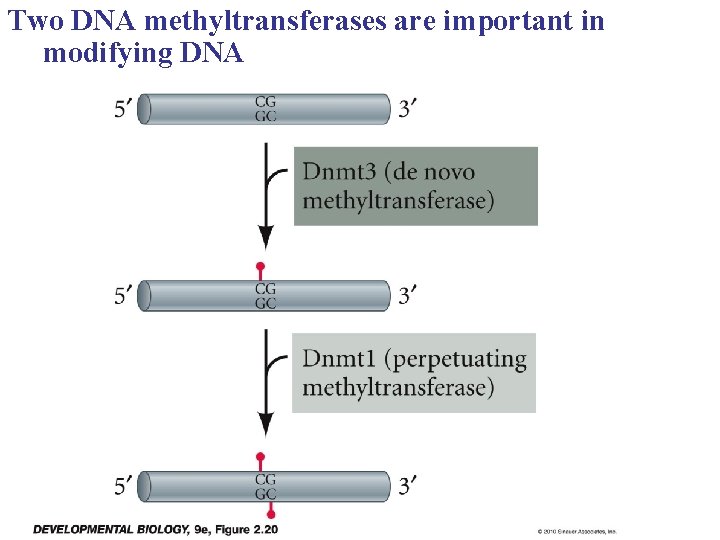
Two DNA methyltransferases are important in modifying DNA

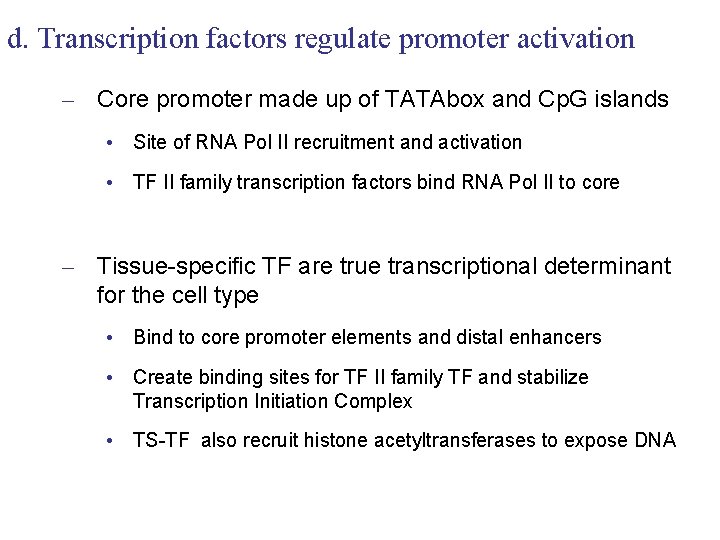
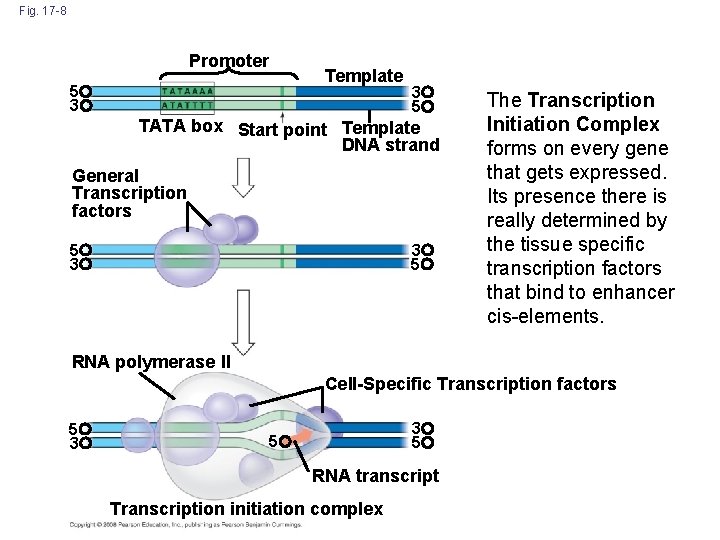
d. Transcription factors regulate promoter activation – Core promoter made up of TATAbox and Cp. G islands • Site of RNA Pol II recruitment and activation • TF II family transcription factors bind RNA Pol II to core – Tissue-specific TF are true transcriptional determinant for the cell type • Bind to core promoter elements and distal enhancers • Create binding sites for TF II family TF and stabilize Transcription Initiation Complex • TS-TF also recruit histone acetyltransferases to expose DNA

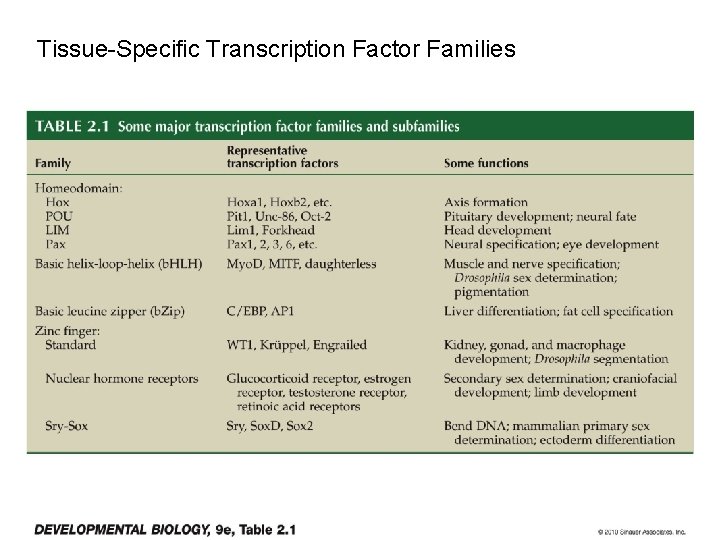
Tissue-Specific Transcription Factor Families

Fig. 17 -8 Promoter Template 5 3 3 5 TATA box Start point Template DNA strand General Transcription factors 5 3 3 5 The Transcription Initiation Complex forms on every gene that gets expressed. Its presence there is really determined by the tissue specific transcription factors that bind to enhancer cis-elements. RNA polymerase II Cell-Specific Transcription factors 5 3 3 5 5 RNA transcript Transcription initiation complex

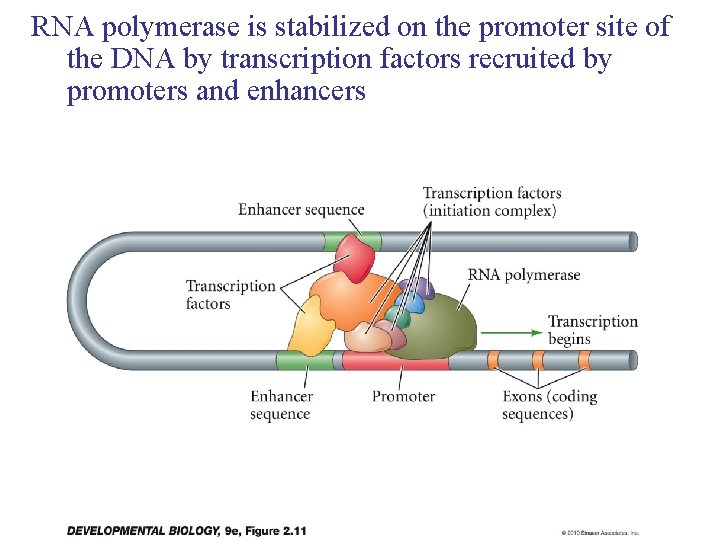
RNA polymerase is stabilized on the promoter site of the DNA by transcription factors recruited by promoters and enhancers

Fig. 17 -7 b Nontemplate strand of DNA Elongation RNA polymerase 3’ RNA nucleotides 3 end 5’ 5 Direction of transcription (“downstream”) Newly made RNA Template strand of DNA

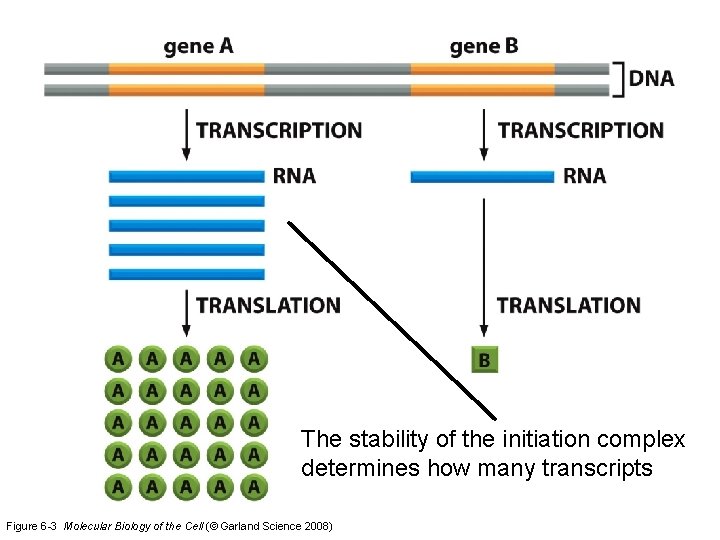
The stability of the initiation complex determines how many transcripts Figure 6 -3 Molecular Biology of the Cell (© Garland Science 2008)

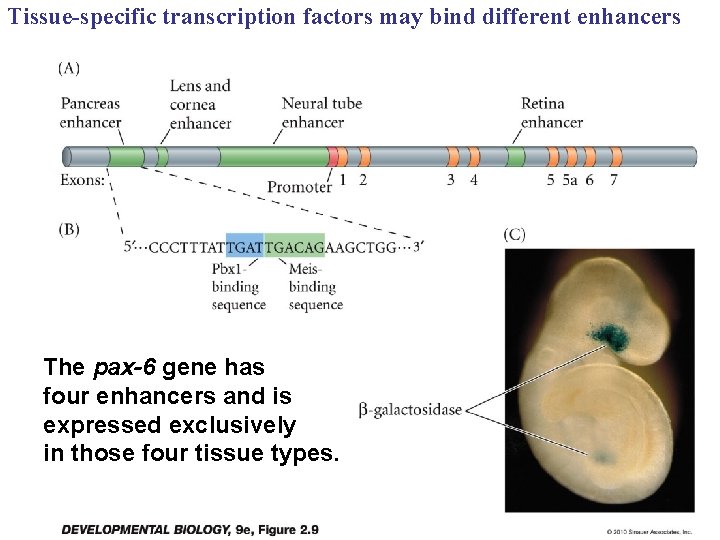
Tissue-specific transcription factors may bind different enhancers The pax-6 gene has four enhancers and is expressed exclusively in those four tissue types.

TS TFs can even control differentiation stability A really important idea in cell differentiation is that there must be a molecular mechanism that keeps a cell differentiated. – The pax-6 gene has a Pax-6 site in its enhancer – When it is present the transcription rate is maximal – This mechanism is repeated in several cell types – A rare positive feedback loop

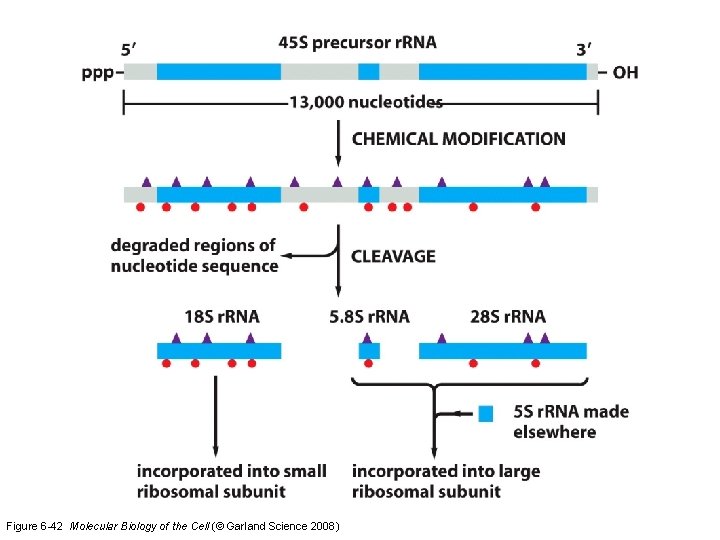
e. Specialized transcriptional activities • Only about 3 -5% of RNA in a cell is m. RNA • Up to 80% of RNA is ribosomal RNA – As many as 10 million ribosomes per cell – humans have 400 r. RNA gene copies on 5 chromosome pairs (frogs have 1200) – 4 eukaryotic subunits: 18 S, 5. 8 S, 28 S, 5 S – First 3 from one gene with RNA Pol I – 5 S is from a separate gene with RNA Pol III

Figure 6 -42 Molecular Biology of the Cell (© Garland Science 2008)

Table 6 -1 Molecular Biology of the Cell (© Garland Science 2008)

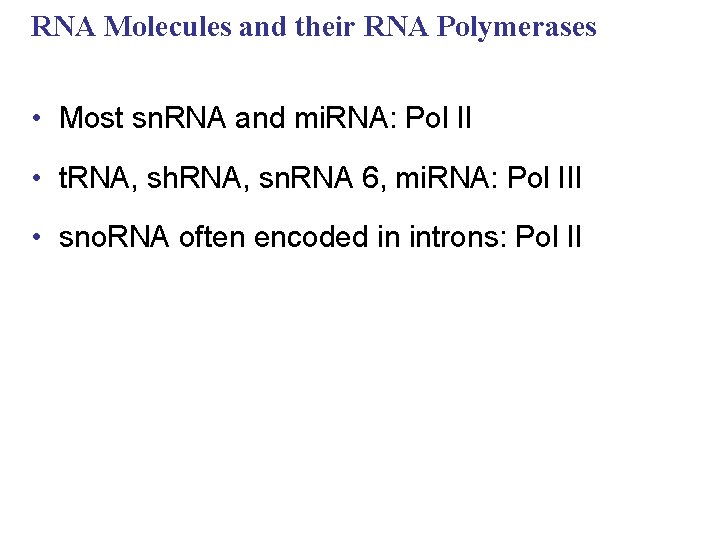
RNA Molecules and their RNA Polymerases • Most sn. RNA and mi. RNA: Pol II • t. RNA, sh. RNA, sn. RNA 6, mi. RNA: Pol III • sno. RNA often encoded in introns: Pol II
 Cell specialization definition
Cell specialization definition Muscle cell specialization
Muscle cell specialization Example of cell specialization
Example of cell specialization Transcription
Transcription Types of dna polymerase in eukaryotes
Types of dna polymerase in eukaryotes Cot phonetic transcription
Cot phonetic transcription Virusmax
Virusmax Dna construct
Dna construct Dna transcription
Dna transcription Transcription or translation
Transcription or translation Rna matching
Rna matching Transcription
Transcription Eukaryotic vs prokaryotic cells
Eukaryotic vs prokaryotic cells Translation or transcription
Translation or transcription Picture of protein synthesis
Picture of protein synthesis Dna transcription
Dna transcription Odd word out
Odd word out Transcription in prokaryotes
Transcription in prokaryotes Dna code
Dna code Dna replication transcription and translation
Dna replication transcription and translation Language
Language Phonetic transcription
Phonetic transcription Transcription of jeopardy
Transcription of jeopardy Transcription
Transcription Basal transcription apparatus
Basal transcription apparatus Transcription translation replication
Transcription translation replication Termination of transcription in prokaryotes
Termination of transcription in prokaryotes Activation domain of transcription factors
Activation domain of transcription factors Translation transcription
Translation transcription What kind of mutation
What kind of mutation Eukaryotic transcription
Eukaryotic transcription Dna transcription and translation
Dna transcription and translation Dna to rna transcription
Dna to rna transcription Transcription
Transcription Rna transcription
Rna transcription Ocr transcription
Ocr transcription Transcription error
Transcription error Transcription
Transcription Transcription
Transcription Translation transcription
Translation transcription Hiv reverse transcription
Hiv reverse transcription Read the transcription and write the words
Read the transcription and write the words Prokaryotic
Prokaryotic Idea transcription
Idea transcription Semiconservative replication
Semiconservative replication Phonetic transcription of tomb
Phonetic transcription of tomb