Transcription Regulation Transcription Factor Motif Finding Xiaole Shirley















- Slides: 38

Transcription Regulation Transcription Factor Motif Finding Xiaole Shirley Liu STAT 115/215, BIO/BIST 282

Biology of Transcription Regulation Hemoglobin Beta . . . acatttgcttctgacacaactgtgttcactagcaacctca. . . aacagacacc. ATGGTGCACCTGACTCCTGAGGAGAAGTCT. . . atttgctt ttcact gcaacct Hemoglobin Zeta aactccagt. . . agcaggcccaactccagtgcagctgcaacctgcccactcc. . . ggcagcgcac. ATGTCTCTGACCAAGACTGAGAGTGCCGTC. . . gcaacct Hemoglobin Alpha actca. . . cgctcgcgggccggcactcttctggtccccacagactcag. . . gatacccaccg. ATGGTGCTGTCTCCTGCCGACAAGACCAA. . . gcaacct Hemoglobin Gamma . . . gccccgccagcgccgctaccgccctgcccccgggcgagcg. . . gatgcgcgagt. ATGGTGCTGTCTCCTGCCGACAAGACCAA. . . gcaacct ccagcgccg Transcription Factor (TF) gcaacct TF Binding Motif can only be computational discovered when there are enough cases for machine learning 2

Computational Motif Finding • Input data: – Upstream sequences of gene expression profile cluster – 20 -800 sequences, each 300 -5000 bps long • Output: enriched sequence patterns (motifs) • Ultimate goals: – Which TFs are involved and their binding motifs and effects (enhance / repress gene expression)? – Which genes are regulated by this TF, why is there disease when a TF goes wrong? – Are there binding partner / competitor for a TF? 3

TF Motif Representation • Regular expression: Consensus binary decision CACAAAA Degenerate CRCAAAW IUPAC A/G A/T 4

TF Motif Representation • Regular expression: Consensus binary decision CACAAAA Degenerate CRCAAAW IUPAC A/G A/T • Position weight matrix (PWM): need score cutoff Sites Pos 12345678 ATGGCATG AGGGTGCG ATCGCATG TTGCCACG ATGGTATT ATTGCACG AGGGCGTT ATGACATG ATGGCATG ACTGGATG Motif Matrix Segment ATGCAGCT score = p(generate ATGCAGCT from motif matrix) p(generate ATGCAGCT from background) p 0 A p 0 T p 0 G p 0 C p 0 A p 0 G p 0 C p 0 T 5

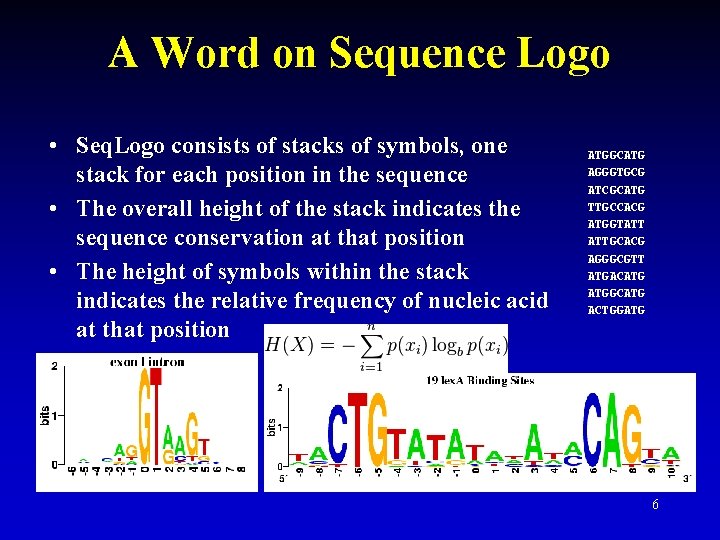
A Word on Sequence Logo • Seq. Logo consists of stacks of symbols, one stack for each position in the sequence • The overall height of the stack indicates the sequence conservation at that position • The height of symbols within the stack indicates the relative frequency of nucleic acid at that position ATGGCATG AGGGTGCG ATCGCATG TTGCCACG ATGGTATT ATTGCACG AGGGCGTT ATGACATG ATGGCATG ACTGGATG 6

TF Motif Database • JASPAR: literature curation • Uni. Probe: Protein Binding Microarrays – Badis et al, Science 2009 • Factor Book and HOMER: – Ch. IP-seq (later) 7

De novo Sequence Motif Finding • Goal: look for common sequence patterns enriched in the input data (compared to the genome background) • Regular expression enumeration – Pattern driven approach – Enumerate k-mers, check significance in dataset • Position weight matrix update – Data driven approach, use data to refine motifs – EM & Gibbs sampling – Motif score and Markov background 8

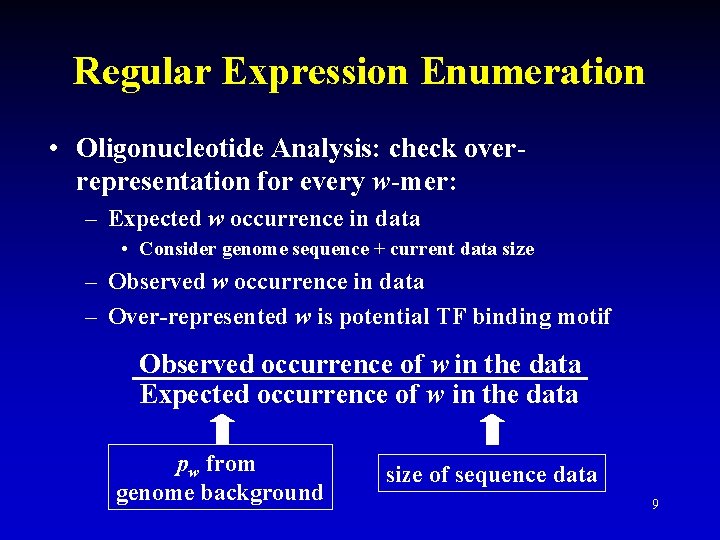
Regular Expression Enumeration • Oligonucleotide Analysis: check overrepresentation for every w-mer: – Expected w occurrence in data • Consider genome sequence + current data size – Observed w occurrence in data – Over-represented w is potential TF binding motif Observed occurrence of w in the data Expected occurrence of w in the data pw from genome background size of sequence data 9

Regular Expression Enumeration • Exhaustive, guaranteed to find global optimum, and can find multiple motifs • Not as flexible with base substitutions, long list of similar good motifs, and limited with motif width • Popular method: WEEDER – Suffix tree implementation of RE motif hits, very fast 10

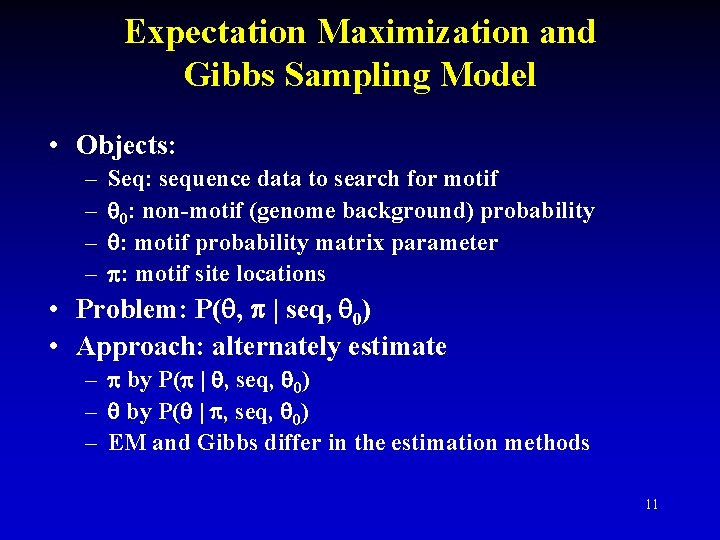
Expectation Maximization and Gibbs Sampling Model • Objects: – – Seq: sequence data to search for motif 0: non-motif (genome background) probability : motif probability matrix parameter : motif site locations • Problem: P( , | seq, 0) • Approach: alternately estimate – by P( | , seq, 0) – EM and Gibbs differ in the estimation methods 11

Expectation Maximization • E step: | , seq, 0 TTGACGACTGCACGT TTGAC p 1 TGACG p 2 GACGA p 3 ACGAC p 4 CGACT p 5 GACTG p 6 ACTGC p 7 CTGCA p 8. . . P 1 = likelihood ratio = P(TTGAC| ) P(TTGAC| 0) p 0 T p 0 G p 0 A p 0 C = 0. 3 0. 2 12

Expectation Maximization • E step: | , seq, 0 TTGACGACTGCACGT TTGAC p 1 TGACG p 2 GACGA p 3 ACGAC p 4 CGACT p 5 GACTG p 6 ACTGC p 7 CTGCA p 8. . . • M step: | , seq, 0 p 1 TTGAC p 2 TGACG p 3 GACGA p 4 ACGAC. . . • Scale ACGT at each position, reflects weighted average of 13

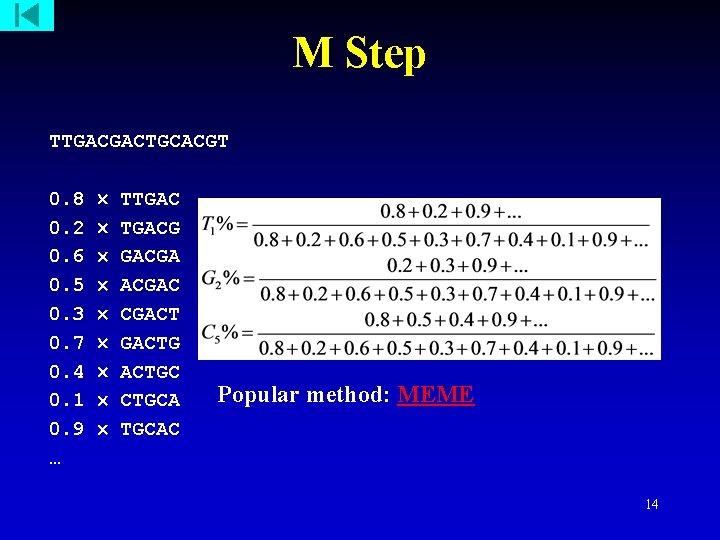
M Step TTGACGACTGCACGT 0. 8 0. 2 0. 6 0. 5 0. 3 0. 7 0. 4 0. 1 0. 9 … TTGACG GACGAC CGACTG ACTGCA TGCAC Popular method: MEME 14

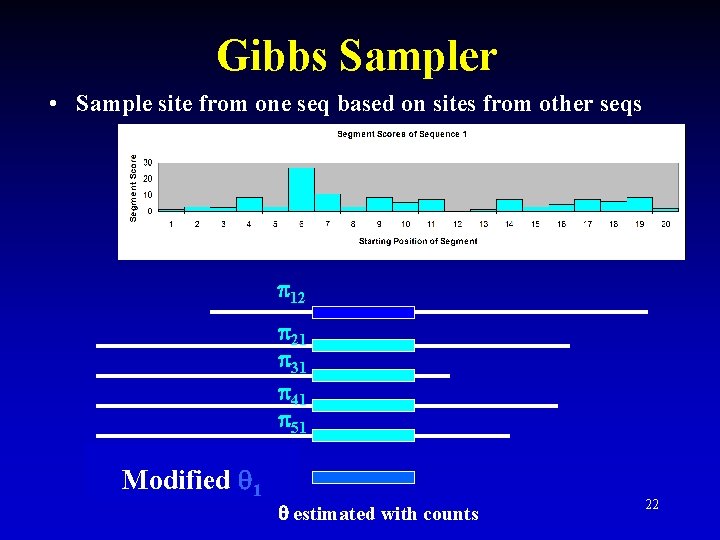
Gibbs Sampling • Stochastic process, although still may need multiple initializations – Sample from P( | , seq, 0) • Collapsed form: – estimated with counts, not sampling from Dirichlet – Sample site from one seq based on sites from other seqs • Converged motif matrix and converged motif sites represent stationary distribution of a Markov Chain 15

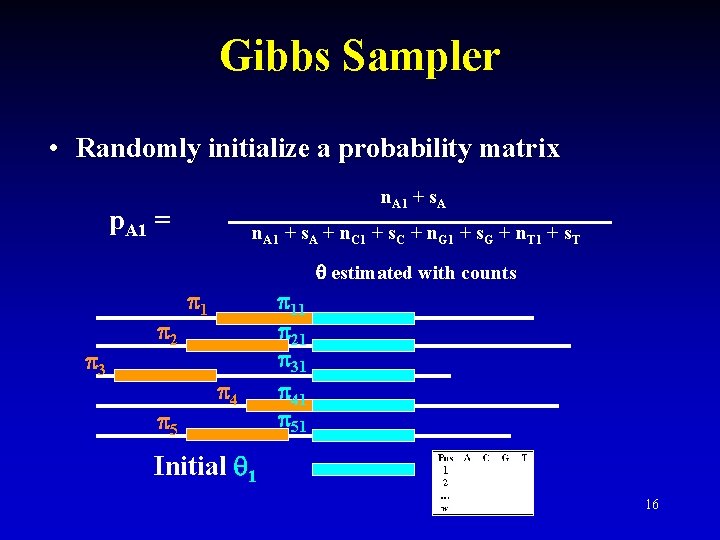
Gibbs Sampler • Randomly initialize a probability matrix n. A 1 + s. A p. A 1 = n. A 1 + s. A + n. C 1 + s. C + n. G 1 + s. G + n. T 1 + s. T estimated with counts 3 2 5 1 4 11 21 31 41 51 Initial 1 16

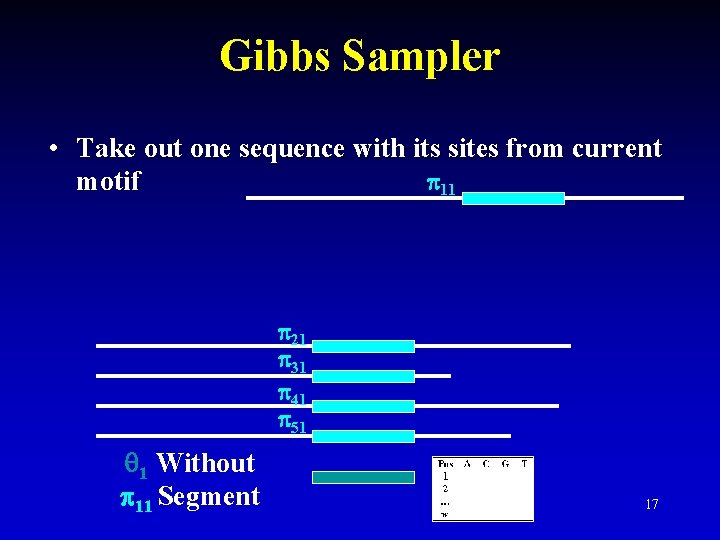
Gibbs Sampler • Take out one sequence with its sites from current 11 motif 21 31 41 51 1 Without 11 Segment 17

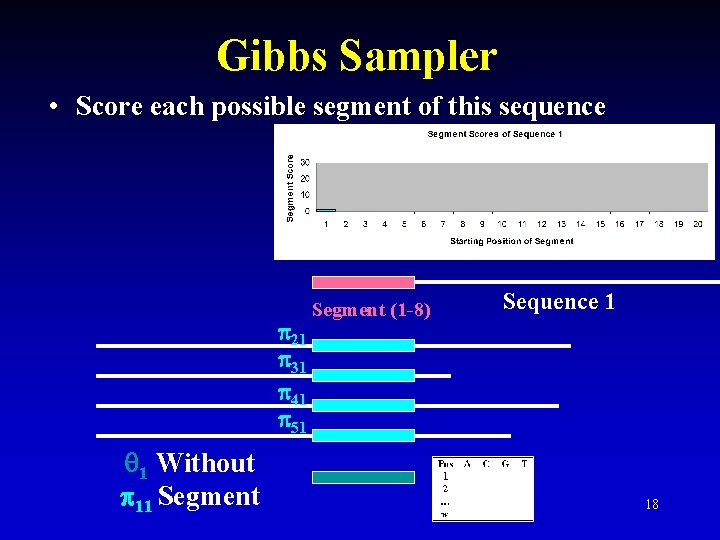
Gibbs Sampler • Score each possible segment of this sequence 21 31 41 51 1 Without 11 Segment (1 -8) Sequence 1 18

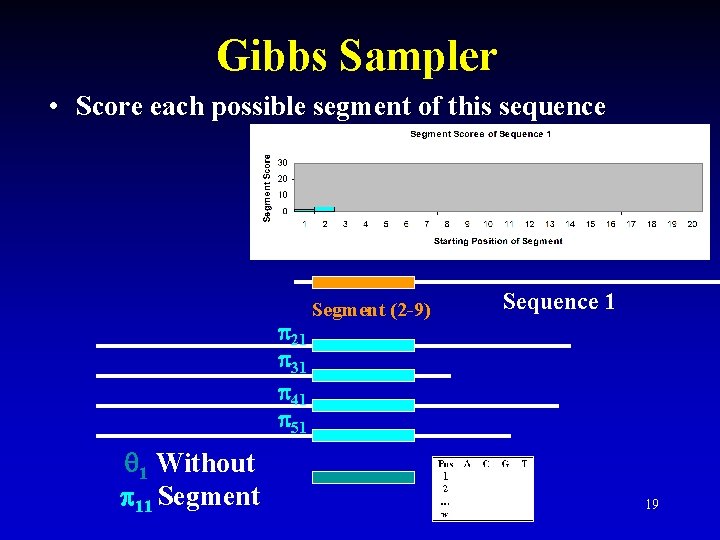
Gibbs Sampler • Score each possible segment of this sequence 21 31 41 51 1 Without 11 Segment (2 -9) Sequence 1 19

Segment Score • Use current motif matrix to score a segment Sites Pos 12345678 ATGGCATG AGGGTGCG ATCGCATG TTGCCACG ATGGTATT ATTGCACG AGGGCGTT ATGACATG ATGGCATG ACTGGATG Motif Matrix Segment ATGCAGCT score = p(generate ATGCAGCT from motif matrix) p(generate ATGCAGCT from background) p 0 A p 0 T p 0 G p 0 C p 0 A p 0 G p 0 C p 0 T 20

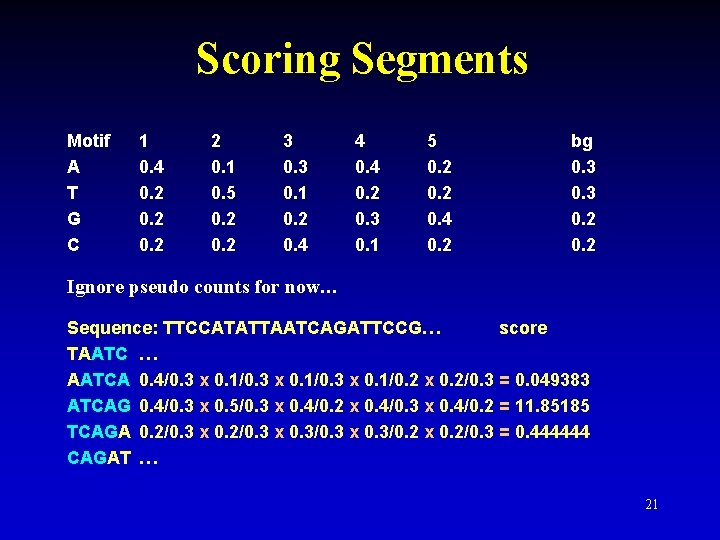
Scoring Segments Motif A T G C 1 0. 4 0. 2 2 0. 1 0. 5 0. 2 3 0. 1 0. 2 0. 4 4 0. 2 0. 3 0. 1 5 0. 2 0. 4 0. 2 bg 0. 3 0. 2 Ignore pseudo counts for now… Sequence: TTCCATATTAATCAGATTCCG… score TAATC … AATCA 0. 4/0. 3 x 0. 1/0. 2 x 0. 2/0. 3 = 0. 049383 ATCAG 0. 4/0. 3 x 0. 5/0. 3 x 0. 4/0. 2 x 0. 4/0. 3 x 0. 4/0. 2 = 11. 85185 TCAGA 0. 2/0. 3 x 0. 3/0. 2 x 0. 2/0. 3 = 0. 444444 CAGAT … 21

Gibbs Sampler • Sample site from one seq based on sites from other seqs 12 21 31 41 51 Modified 1 estimated with counts 22

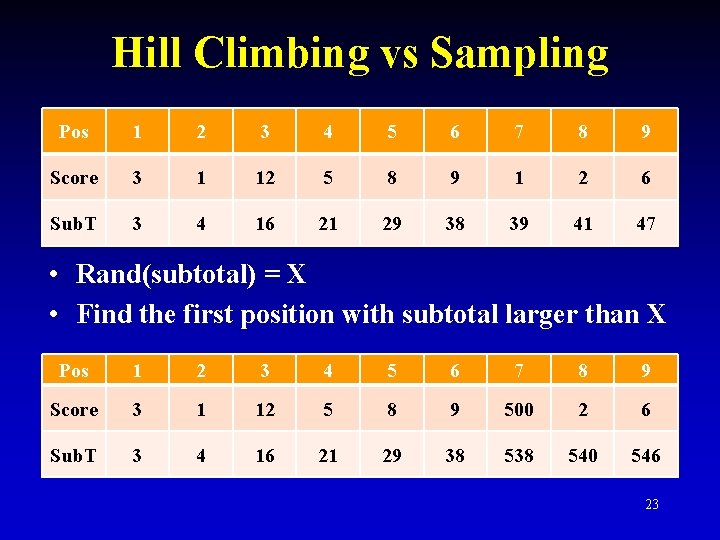
Hill Climbing vs Sampling Pos 1 2 3 4 5 6 7 8 9 Score 3 1 12 5 8 9 1 2 6 Sub. T 3 4 16 21 29 38 39 41 47 • Rand(subtotal) = X • Find the first position with subtotal larger than X Pos 1 2 3 4 5 6 7 8 9 Score 3 1 12 5 8 9 500 2 6 Sub. T 3 4 16 21 29 38 540 546 23

Gibbs Sampler • Repeat the process until motif converges 21 12 31 41 51 1 Without 21 Segment 24

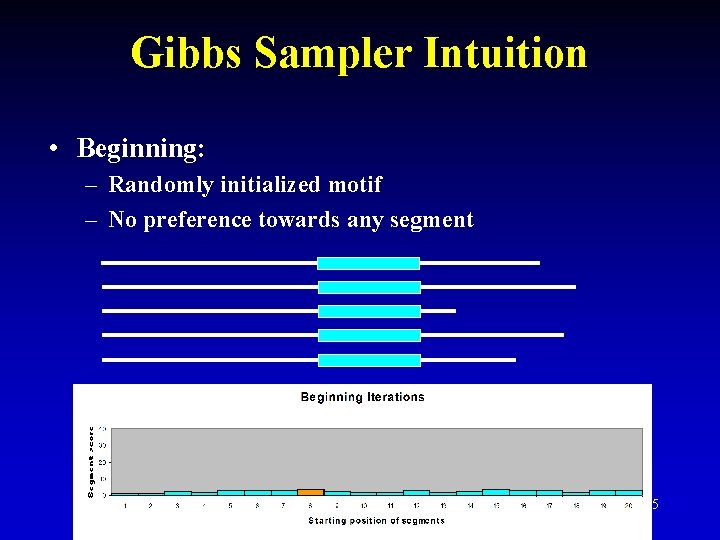
Gibbs Sampler Intuition • Beginning: – Randomly initialized motif – No preference towards any segment 25

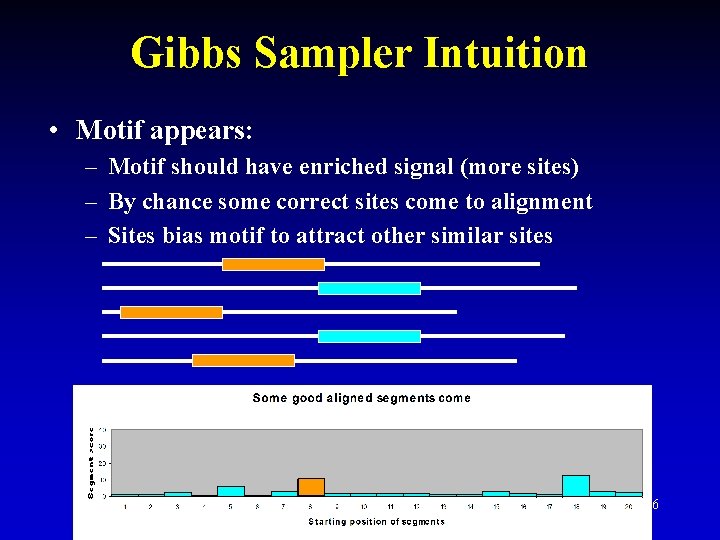
Gibbs Sampler Intuition • Motif appears: – Motif should have enriched signal (more sites) – By chance some correct sites come to alignment – Sites bias motif to attract other similar sites 26

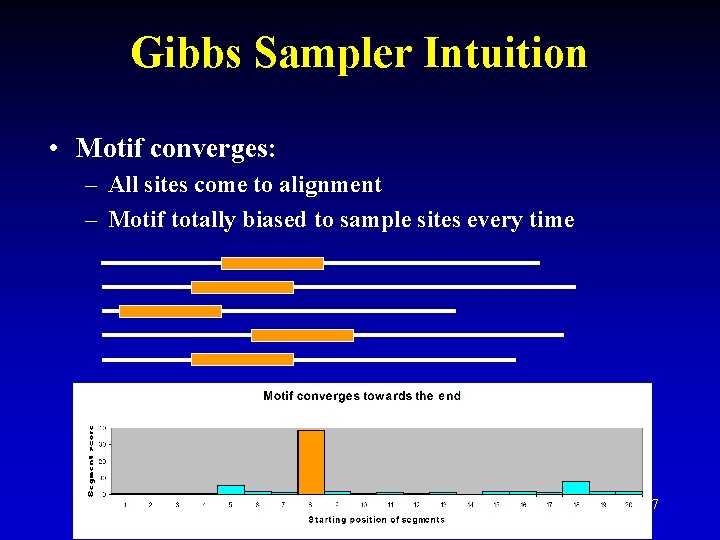
Gibbs Sampler Intuition • Motif converges: – All sites come to alignment – Motif totally biased to sample sites every time 27

Gibbs Sampler • Column shift 11 i 22 i 33 i 44 i 55 i • Metropolis algorithm: – Propose * as shifted 1 column to left or right – Calculate motif score u( ) and u( *) – Accept * with prob = min(1, u( *) / u( )) 28

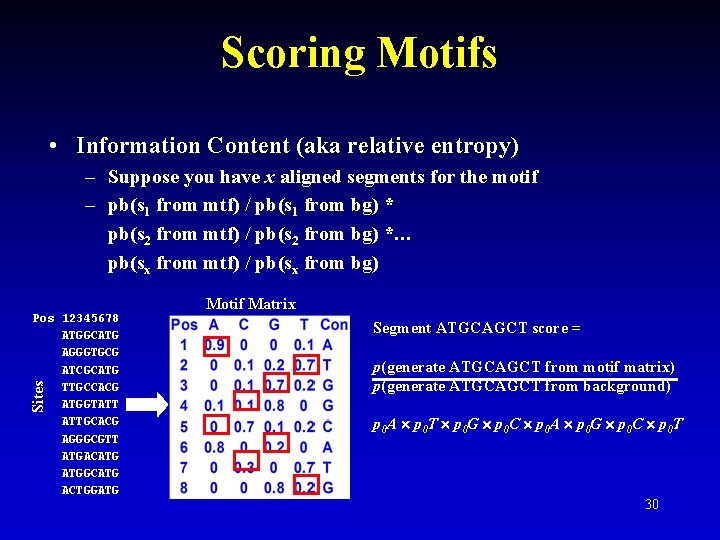
Scoring Motifs • Information Content (aka relative entropy) – Suppose you have x aligned segments for the motif – pb(s 1 from mtf) / pb(s 1 from bg) * pb(s 2 from mtf) / pb(s 2 from bg) *… pb(sx from mtf) / pb(sx from bg) Sites Pos 12345678 ATGGCATG AGGGTGCG ATCGCATG TTGCCACG ATGGTATT ATTGCACG AGGGCGTT ATGACATG ATGGCATG ACTGGATG Motif Matrix Segment ATGCAGCT score = p(generate ATGCAGCT from motif matrix) p(generate ATGCAGCT from background) p 0 A p 0 T p 0 G p 0 C p 0 A p 0 G p 0 C p 0 T 29

Scoring Motifs • Information Content (aka relative entropy) – Suppose you have x aligned segments for the motif – pb(s 1 from mtf) / pb(s 1 from bg) * pb(s 2 from mtf) / pb(s 2 from bg) *… pb(sx from mtf) / pb(sx from bg) Sites Pos 12345678 ATGGCATG AGGGTGCG ATCGCATG TTGCCACG ATGGTATT ATTGCACG AGGGCGTT ATGACATG ATGGCATG ACTGGATG Motif Matrix Segment ATGCAGCT score = p(generate ATGCAGCT from motif matrix) p(generate ATGCAGCT from background) p 0 A p 0 T p 0 G p 0 C p 0 A p 0 G p 0 C p 0 T 30

Scoring Motifs Pos 12345678 pb(s 1 from mtf) / pb(s 1 from bg) * ATGGCATG pb(s 2 from mtf) / pb(s 2 from bg) *… AGGGTGCG ATCGCATG pb(sx from mtf) / pb(sx from bg) TTGCCACG A 1 T 2 G 2 = (p. A 1/p. A 0) (p. T 1/p. T 0) (p. T 2/p. T 0) (p. G 2/p. G 0) ATGGTATT ATTGCACG (p. C 2/p. C 0)C 2… AGGGCGTT Take log of this: ATGACATG ATGGCATG = A 1 log (p. A 1/p. A 0) + T 1 log (p. T 1/p. T 0) + ACTGGATG T 2 log (p. T 2/p. T 0) + G 2 log (p. G 2/p. G 0) + … Divide by the number of segments (if all the motifs have same number of segments) = p. A 1 log (p. A 1/p. A 0) + p. T 1 log (p. T 1/p. T 0) + p. T 2 log (p. T 2/p. T 0)… 31

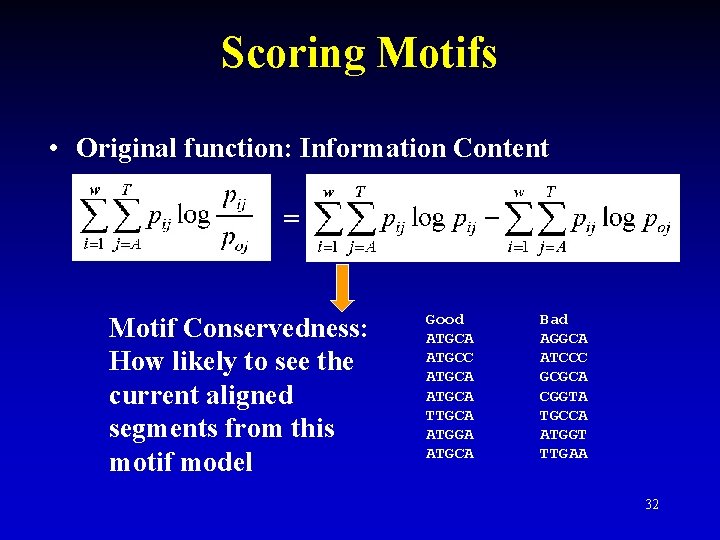
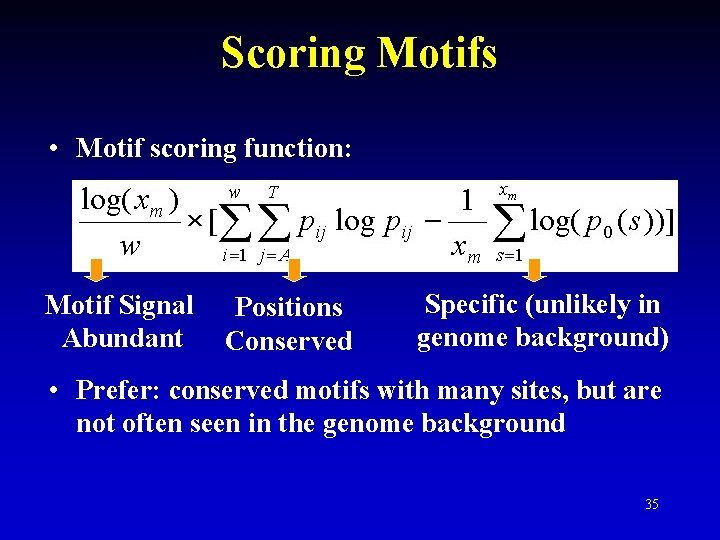
Scoring Motifs • Original function: Information Content = Motif Conservedness: How likely to see the current aligned segments from this motif model Good ATGCA ATGCC ATGCA TTGCA ATGGA ATGCA Bad AGGCA ATCCC GCGCA CGGTA TGCCA ATGGT TTGAA 32

Scoring Motifs • Original function: Information Content = Good AGTCC AGTCC Bad ATAAA ATAAA Motif Specificity: How likely to see the current aligned segments from background 33

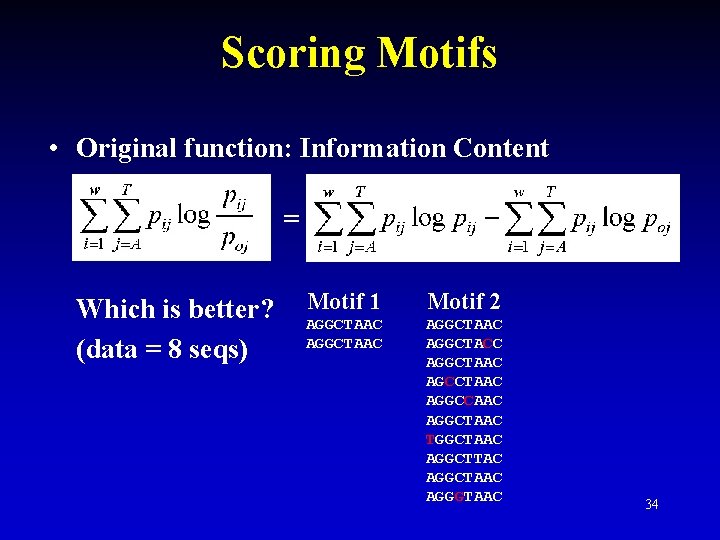
Scoring Motifs • Original function: Information Content = Which is better? (data = 8 seqs) Motif 1 Motif 2 AGGCTAAC AGGCTACC AGGCTAAC AGCCTAAC AGGCCAAC AGGCTAAC TGGCTAAC AGGCTTAC AGGCTAAC AGGGTAAC 34

Scoring Motifs • Motif scoring function: Motif Signal Abundant Positions Conserved Specific (unlikely in genome background) • Prefer: conserved motifs with many sites, but are not often seen in the genome background 35

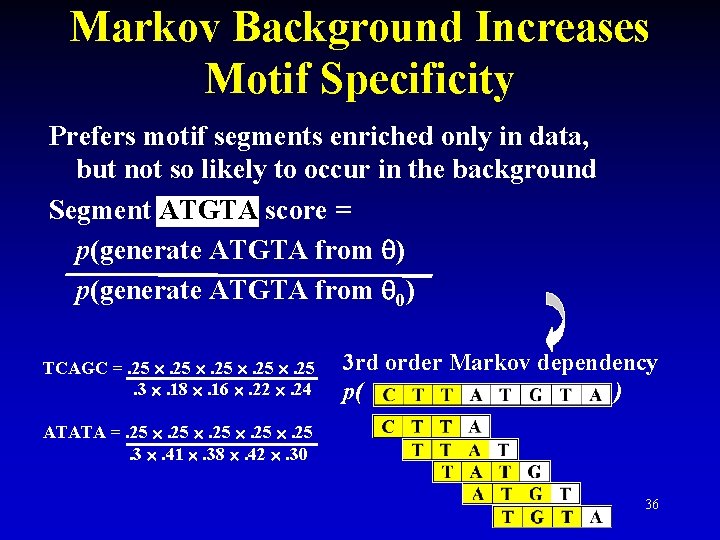
Markov Background Increases Motif Specificity Prefers motif segments enriched only in data, but not so likely to occur in the background Segment ATGTA score = p(generate ATGTA from ) p(generate ATGTA from 0) TCAGC =. 25 . 25. 3 . 18 . 16 . 22 . 24 3 rd order Markov dependency p( ) ATATA =. 25 . 25. 3 . 41 . 38 . 42 . 30 36

Position Weight Matrix Update • Advantage – Can look for motifs of any widths – Flexible with base substitutions • Disadvantage: – EM and Gibbs sampling: no guaranteed convergence time – No guaranteed global optimum 37

Summary • Biology and challenge of transcription regulation • Known TF motif databases • De novo method – Regular expression enumeration – Position weight matrix update • EM (iterate , ; ~ weighted average) • Gibbs Sampler (sample , ; Markov chain convergence) • Shifts, Markov background, motif score 38
 Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Motif berasal dari bahasa inggris yaitu motif yang berarti
Motif berasal dari bahasa inggris yaitu motif yang berarti Remaining factor
Remaining factor Constant phase difference
Constant phase difference What factor
What factor Factor the common factor out of each expression
Factor the common factor out of each expression What is factored form
What is factored form Factor out the greatest common factor
Factor out the greatest common factor Common factors of 18 and 27
Common factors of 18 and 27 Shirley ardell mason art
Shirley ardell mason art Shirley island
Shirley island Lida zheng md
Lida zheng md Cornelia m ruland
Cornelia m ruland The lottery by shirley jackson discussion questions
The lottery by shirley jackson discussion questions Shirley phan
Shirley phan The lottery shirley jackson theme
The lottery shirley jackson theme Dr shirley d'sa
Dr shirley d'sa Shirley hall farm
Shirley hall farm The lottery by shirley jackson plot diagram
The lottery by shirley jackson plot diagram The lottery shirley jackson questions
The lottery shirley jackson questions Dr shirley wray
Dr shirley wray What is the exposition of charles by shirley jackson
What is the exposition of charles by shirley jackson Sybil dorsett
Sybil dorsett Shirley zimmerman functions of the family
Shirley zimmerman functions of the family Sybils group inc
Sybils group inc Paraganglioma radiopaedia
Paraganglioma radiopaedia Shirley chilet cama
Shirley chilet cama The lottery shirley jackson literary devices
The lottery shirley jackson literary devices Symbols in the lottery
Symbols in the lottery Shirley hills primary
Shirley hills primary Allusion in the lottery
Allusion in the lottery Shirley bloom
Shirley bloom Shirley cvc
Shirley cvc Cullman middle school
Cullman middle school Shirley toulson
Shirley toulson Shirley jackson seven types of ambiguity
Shirley jackson seven types of ambiguity The cardboard shows me how it was poet name
The cardboard shows me how it was poet name