Eukaryotic Transcription Eukaryotic Transcriptional Transcription control is the




- Slides: 14

Eukaryotic Transcription

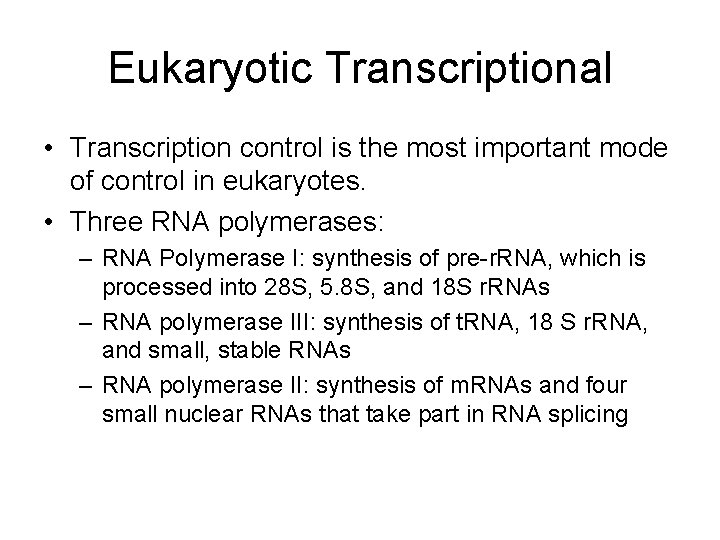
Eukaryotic Transcriptional • Transcription control is the most important mode of control in eukaryotes. • Three RNA polymerases: – RNA Polymerase I: synthesis of pre-r. RNA, which is processed into 28 S, 5. 8 S, and 18 S r. RNAs – RNA polymerase III: synthesis of t. RNA, 18 S r. RNA, and small, stable RNAs – RNA polymerase II: synthesis of m. RNAs and four small nuclear RNAs that take part in RNA splicing

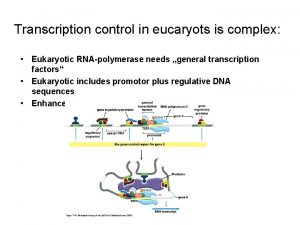
Eukaryotic Transcriptional • The main purpose is the execution of precise developmental decisions (irreversible). • Cis-acting control elements are located many kb away from the start site. • Promoter region is poorly characterized.

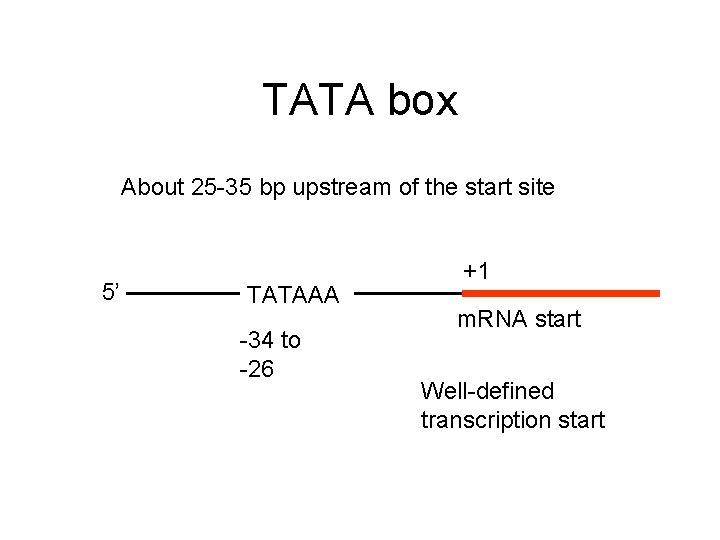
TATA box About 25 -35 bp upstream of the start site 5’ TATAAA -34 to -26 +1 m. RNA start Well-defined transcription start

Initiator • Instead of a TATA box, some eukaryotic gene contain an alternative promoter element, called an initiator. • Initiator is highly degenerative. +1 5’ Y Y A N T/A Y Y = pyrimidine (C or T) N = any

Cp. G island • Genes coding for intermediary metabolism are transcribed at low rates, and do not contain a TATA box or initiator. • Most genes of this type contain a CG-rich stretch of 2050 nt within ~100 bp upstream of the start site region. • A transcription factor called SP 1 recognizes these CGrich region. • Gives multiple alternative m. RNA start sites. ~100 bp Cp. G island m. RNA Multiple 5’-start sites

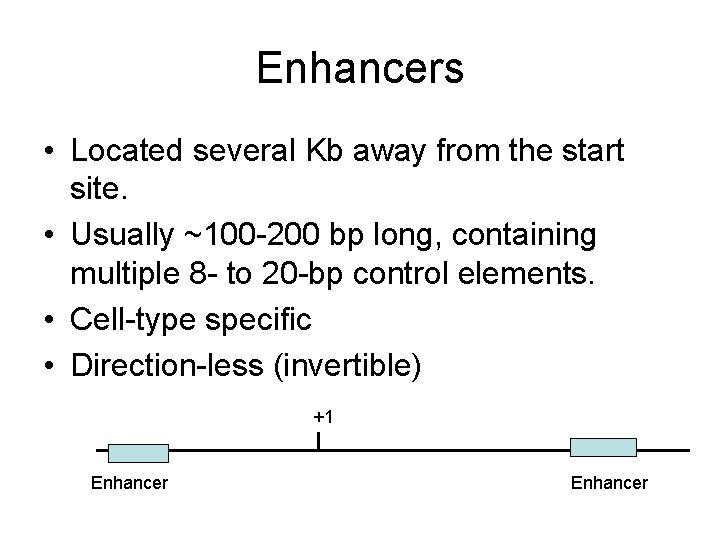
Enhancers • Located several Kb away from the start site. • Usually ~100 -200 bp long, containing multiple 8 - to 20 -bp control elements. • Cell-type specific • Direction-less (invertible) +1 Enhancer

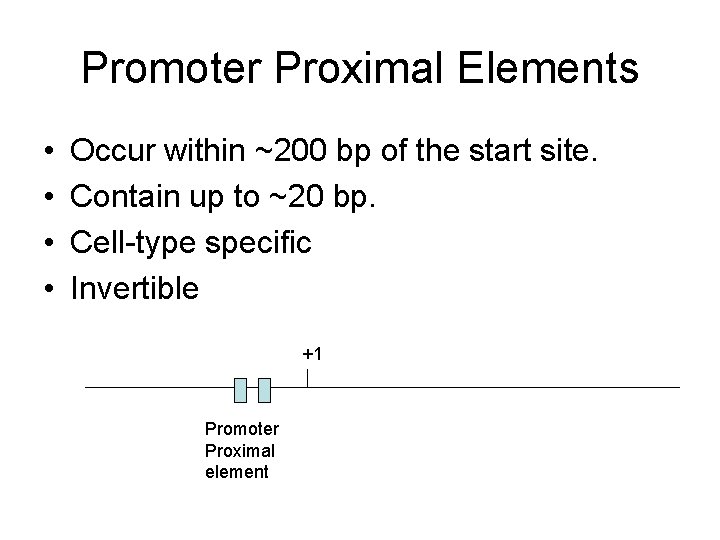
Promoter Proximal Elements • • Occur within ~200 bp of the start site. Contain up to ~20 bp. Cell-type specific Invertible +1 Promoter Proximal element

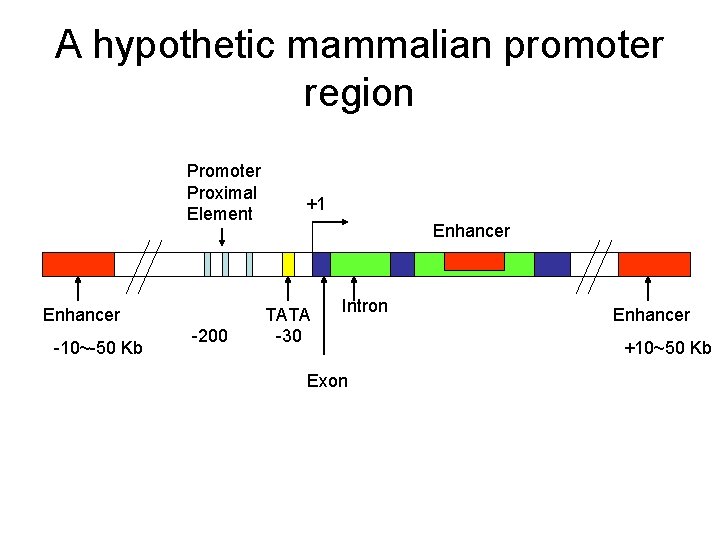
A hypothetic mammalian promoter region Promoter Proximal Element Enhancer -10~-50 Kb -200 +1 Enhancer TATA -30 Intron Exon Enhancer +10~50 Kb

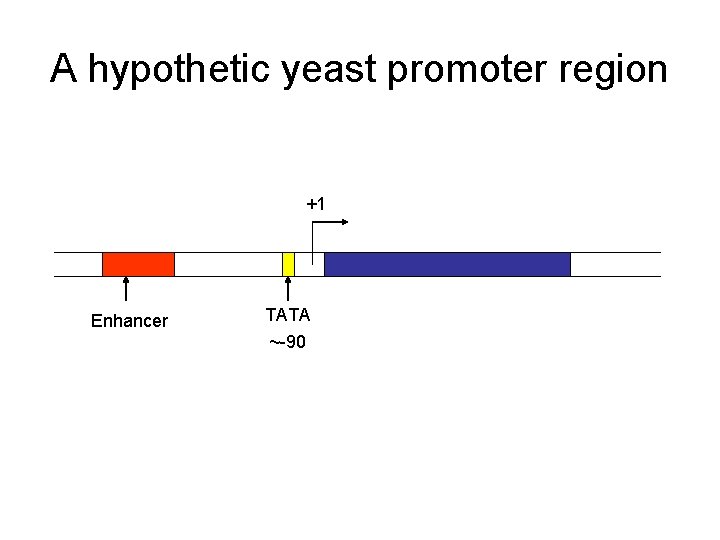
A hypothetic yeast promoter region +1 Enhancer TATA ~-90

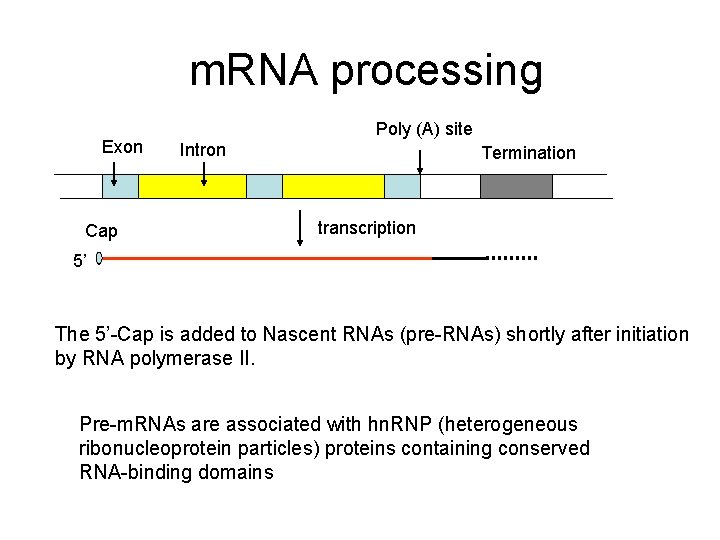
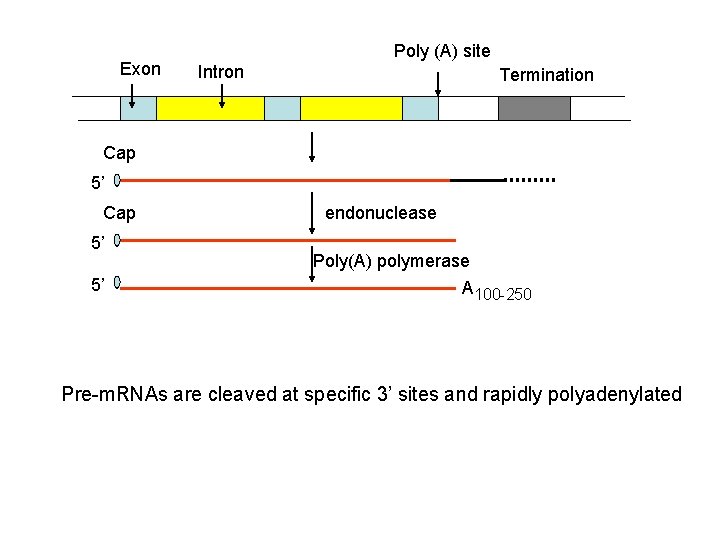
m. RNA processing Exon Cap Poly (A) site Intron Termination transcription 5’ The 5’-Cap is added to Nascent RNAs (pre-RNAs) shortly after initiation by RNA polymerase II. Pre-m. RNAs are associated with hn. RNP (heterogeneous ribonucleoprotein particles) proteins containing conserved RNA-binding domains

Exon Poly (A) site Intron Termination Cap 5’ 5’ endonuclease Poly(A) polymerase A 100 -250 Pre-m. RNAs are cleaved at specific 3’ sites and rapidly polyadenylated

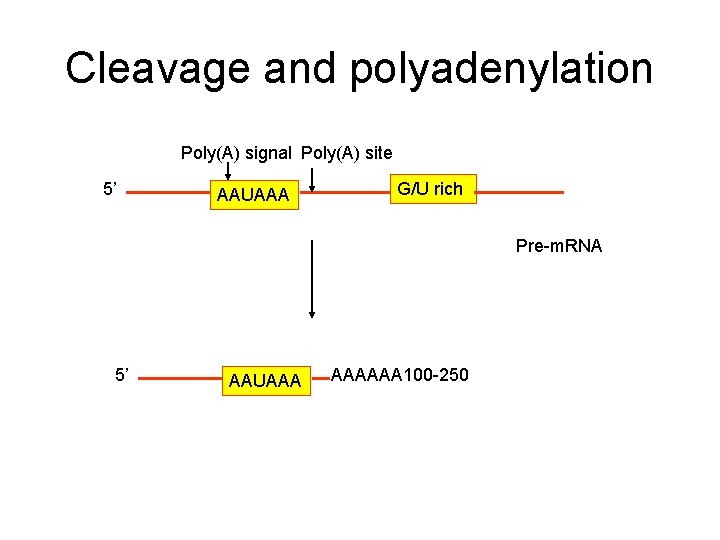
Cleavage and polyadenylation Poly(A) signal Poly(A) site 5’ AAUAAA G/U rich Pre-m. RNA 5’ AAUAAA AAAAAA 100 -250

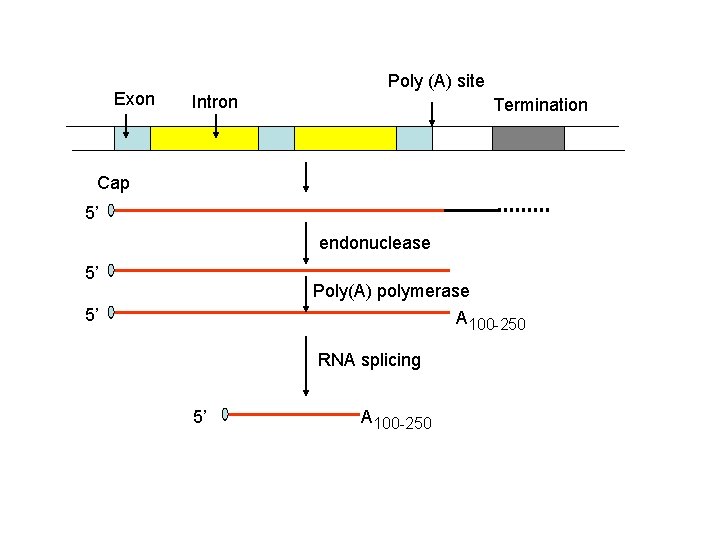
Exon Poly (A) site Intron Termination Cap 5’ endonuclease 5’ Poly(A) polymerase A 100 -250 5’ RNA splicing 5’ A 100 -250
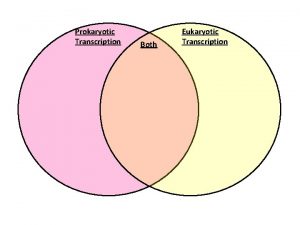
 Prokaryotic and eukaryotic venn diagram
Prokaryotic and eukaryotic venn diagram Prokaryote vs eukaryote cells
Prokaryote vs eukaryote cells Eukaryotic transcription
Eukaryotic transcription Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Slidetodoc
Slidetodoc Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Gấu đi như thế nào
Gấu đi như thế nào Chụp tư thế worms-breton
Chụp tư thế worms-breton Alleluia hat len nguoi oi
Alleluia hat len nguoi oi Các môn thể thao bắt đầu bằng tiếng đua
Các môn thể thao bắt đầu bằng tiếng đua Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Cong thức tính động năng
Cong thức tính động năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ