CS 173 Lecture 6 NON protein coding genes


![“non coding” RNAs (nc. RNA) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 4 “non coding” RNAs (nc. RNA) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 4](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-4.jpg)
![Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 23 Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 23](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-23.jpg)
![Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 24 Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 24](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-24.jpg)
![Other Non Coding Transcripts http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 27 Other Non Coding Transcripts http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 27](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-27.jpg)
![lnc. RNAs come in many flavors http: //cs 173. stanford. edu [Bejerano. Winter 12/13] lnc. RNAs come in many flavors http: //cs 173. stanford. edu [Bejerano. Winter 12/13]](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-29.jpg)
![Or are they? http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 33 Or are they? http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 33](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-33.jpg)
- Slides: 42

CS 173 Lecture 6: NON protein coding genes MW 11: 00 -12: 15 in Beckman B 302 Prof: Gill Bejerano TAs: Jim Notwell & Harendra Guturu http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 1

Announcements • HW 1 due in one week. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 2

ATATTGAATTTTCAAAAATTCTTACTTTTTGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA TATCCATATCTAATCTTATATGTTGTGGAAATGTAAAGAGCCCCATTATCTTAGCCTAAAAAAACCTTCTCTTTGGAACTTTC TAATACGCTTAACTGCTCATTGCTATATTGAAGTACGGATTAGAAGCCGCCGAGCGGGCGACAGCCCTCCGACGGAAGACTCTCCTC TGCGTCCTCGTCTTCACCGGTCGCGTTCCTGAAACGCAGATGTGCCTCGCGCCGCACTGCTCCGAACAATAAAGATTCTACAATACT CTTTTATGGTTATGAAGAGGAAAAATTGGCAGTAACCTGGCCCCACAAACCTTCAAATTAACGAATCAAATTAACAACCATAGGATG AATGCGATTAGTTTTTTAGCCTTATTTCTGGGGTAATCAGCGATGATTTTTGATCTATTAACAGATATATAAATGGAA GCTGCATAACCACTTTAACTAATACTTTCAACATTTTCAGTTTGTATTACTTCTTATTCAAATGTCATAAAAGTATCAACAAAAAAT TTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAATGACTAAATCTCATTCAGAAGAAGTGATTGTACCTGAGTTCAA CTAGCGCAAAGGAATTACCAAGACCATTGGCCGAAAAGTGCCCGAGCATAATTAAGAAATTTATAAGCGCTTATGATGCTAAACCGG TTTGTTGCTAGATCGCCTGGTAGAGTCAATCTAATTGGTGAACATATTGATTATTGTGACTTCTCGGTTTTACCTTTAGCTATTGATATGCTTTGCGCCGTCAAAGTTTTGAACGATGAGATTTCAAGTCTTAAAGCTATATCAGAGGGCTAAGCATGTGTATTCTGAAT TTAAGAGTCTTGAAGGCTGTGAAATTAATGACTACAGCGAGCTTTACTGCCGACGAAGACTTTTTCAAGCAATTTGGTGCCTTGATG CGAGTCTCAAGCTTCTTGCGATAAACTTTACGAATGTTCTTGTCCAGAGATTGACAAAATTTGTTCCATTGCTTTGTCAAATGGATC ATGGTTCCCGTTTGACCGGAGCTGGGGTGGTTGTACTGTTCACTTGGTTCCAGGGGGCCCAAATGGCAACATAGAAAAGGTAA GAAGCCCTTGCCAATGAGTTCTACAAGGTCAAGTACCCTAAGATCACTGATGCTGAGCTAGAAAATGCTATCATCGTCTCTAAACCA ATTGGGCAGCTGTCTATATGAATTAGTCAAGTATACTTCTTTTACTTTGTTCAGAACAACTTCTCATTTTTTTCTACTCATAA TTAGCATCACAAAATACGCAATAATAACGAGTAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGA ATGTTTTCAATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTT ATACCTATTCTTGACATGATATGACTACCATTTTGTTATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTTGCGAAGTT TGGCAAGTTGCCAACTGACGAGATGCAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGT TCAATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTTGGATAC ATTCTTGACATGATATGACTACCATTTTGTTATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTCATTTGCGAAGTTCT GCAAGTTGCCAACTGACGAGATGCAGTTTCCTACGCATAATAAGAATAGGAGGGAATATCAAGCCAGACAATCTATCATTACATTTA CGGCTCTTCAAAAAGATTGAACTCTCGCCAACTTATGGAATCTTCCAATGAGACCTTTGCGCCAAATAATGTGGATTTGGAAAAAGA ATAAGTCATCTCAGAGTAATATAACTACCGAAGTTTATGAGGCATCGAGCTTTGAAGAAAAAGTAAGCTCAGAAAAACCTCAATACA TCATTCTGGAAGAAAATCTATTATGAATATGTGGTCGTTGACAAATCTTGGGTGTTTCTATTCTGGATTCATTTATGTACAAC GGACTTGAAGCCCGTCGAAAAAGGCGGGTTTGGTCCTGGTACAATTATTGTTACTTCTGGCTTGCTGAATGTTTCAATATCAA CTTGGCAAATTGCAGCTACAGGTCTACAACTGGGTCTAAATTGGTGGCAGTGTTGGATAACAATTTGGATTGGGTACGGTTTCGTTG GCTTTTGTTGTTTTGGCCTCTAGAGTTGGATCTGCTTATCATTTGTCATTCCCTATATCATCTAGAGCATCATTCGGTATTTTCTTC TTTATGGCCCGTTATTAACAGAGTCGTCATGGCCATCGTTTGGTATAGTGTCCAAGCTTATATTGCGGCAACTCCCGTATCATTAAT TGAAATCTTTGGAAAAGATTTACAATGATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTCATTTGCGAAGTTCT GCAAGTTGCCAACTGACGAGATGCAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTT AATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTTGGATACCT TCTTGACATGATATGACTACCATTTTGTTATTGTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTT AATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTA CTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTT ACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 3 AATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGT
![non coding RNAs nc RNA http cs 173 stanford edu Bejerano Winter 1213 4 “non coding” RNAs (nc. RNA) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 4](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-4.jpg)
“non coding” RNAs (nc. RNA) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 4

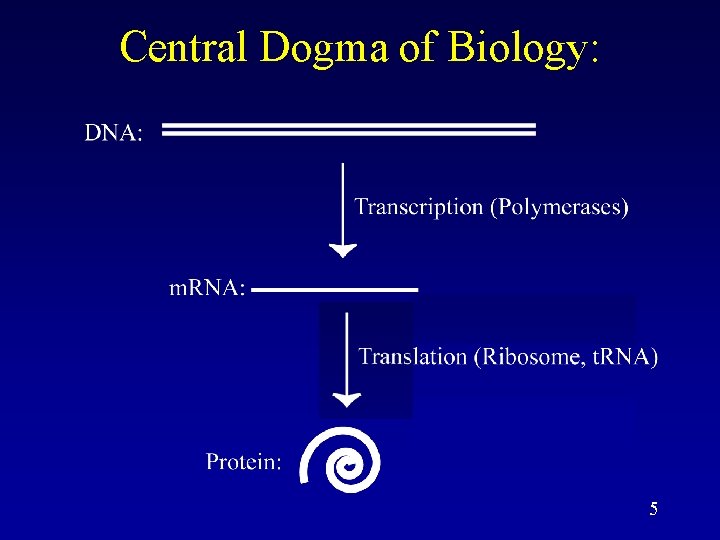
Central Dogma of Biology: 5

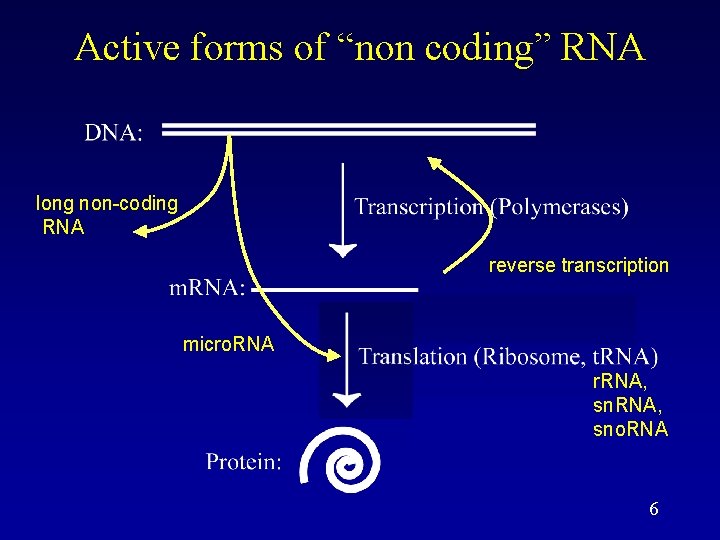
Active forms of “non coding” RNA long non-coding RNA reverse transcription micro. RNA r. RNA, sno. RNA 6

What is nc. RNA? • Non-coding RNA (nc. RNA) is an RNA that functions without being translated to a protein. • Known roles for nc. RNAs: – RNA catalyzes excision/ligation in introns. – RNA catalyzes the maturation of t. RNA. – RNA catalyzes peptide bond formation. – RNA is a required subunit in telomerase. – RNA plays roles in immunity and development (RNAi). – RNA plays a role in dosage compensation. – RNA plays a role in carbon storage. – RNA is a major subunit in the SRP, which is important in protein trafficking. – RNA guides RNA modification. – RNA can do so many different functions, it is thought in the beginning there was an RNA World, where RNA was both the information carrier and active molecule. 7

“non coding” RNAs (nc. RNA) Small structural RNAs (ss. RNA) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 8

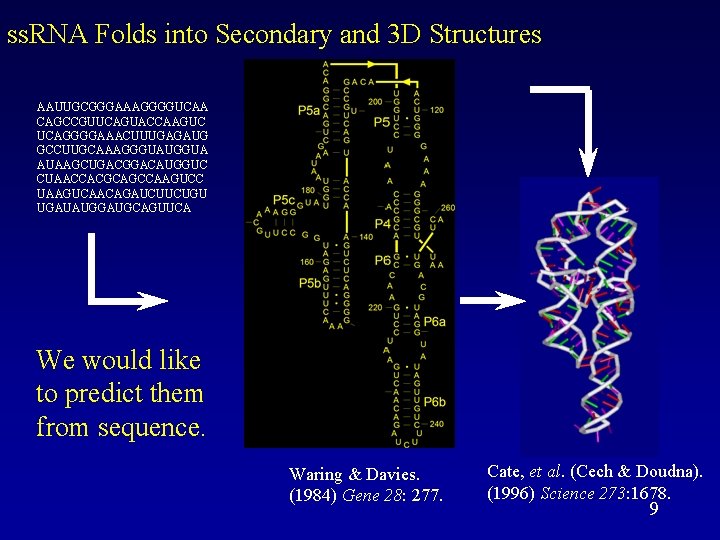
ss. RNA Folds into Secondary and 3 D Structures AAUUGCGGGAAAGGGGUCAA CAGCCGUUCAGUACCAAGUC UCAGGGGAAACUUUGAGAUG GCCUUGCAAAGGGUAUGGUA AUAAGCUGACGGACAUGGUC CUAACCACGCAGCCAAGUCC UAAGUCAACAGAUCUUCUGU UGAUAUGGAUGCAGUUCA We would like to predict them from sequence. Waring & Davies. (1984) Gene 28: 277. Cate, et al. (Cech & Doudna). (1996) Science 273: 1678. 9

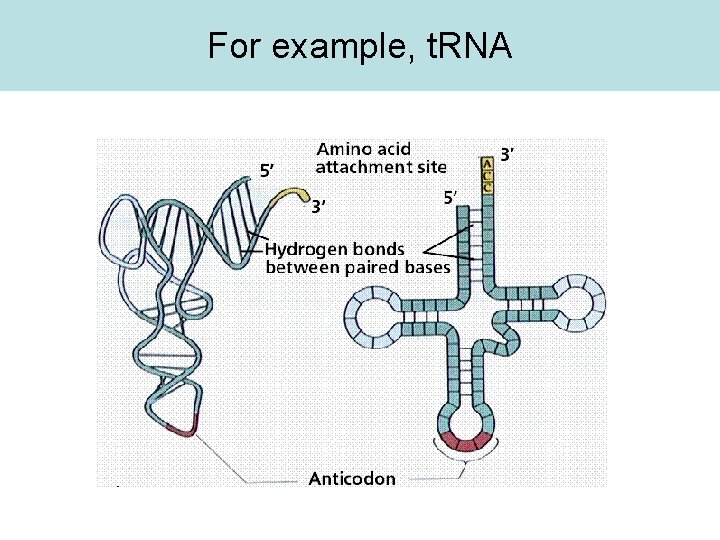
For example, t. RNA

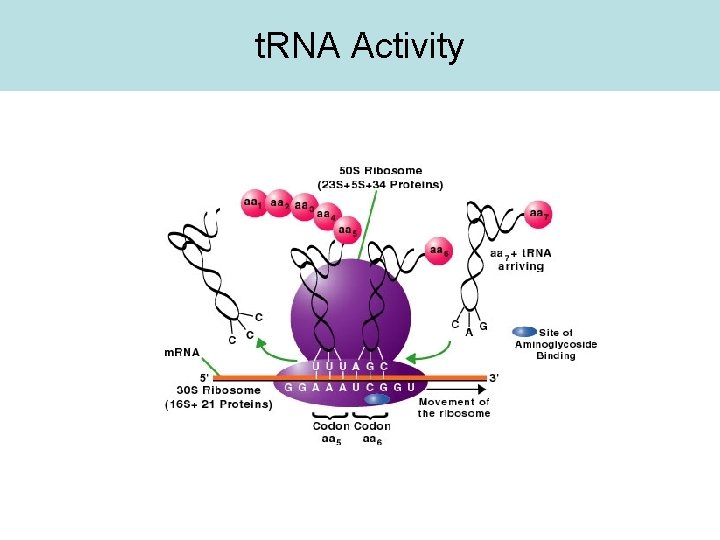
t. RNA Activity

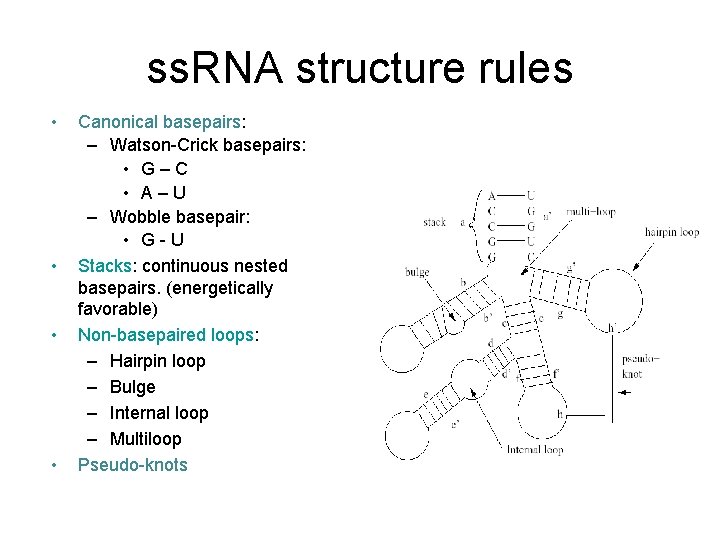
ss. RNA structure rules • • Canonical basepairs: – Watson-Crick basepairs: • G – C • A – U – Wobble basepair: • G - U Stacks: continuous nested basepairs. (energetically favorable) Non-basepaired loops: – Hairpin loop – Bulge – Internal loop – Multiloop Pseudo-knots

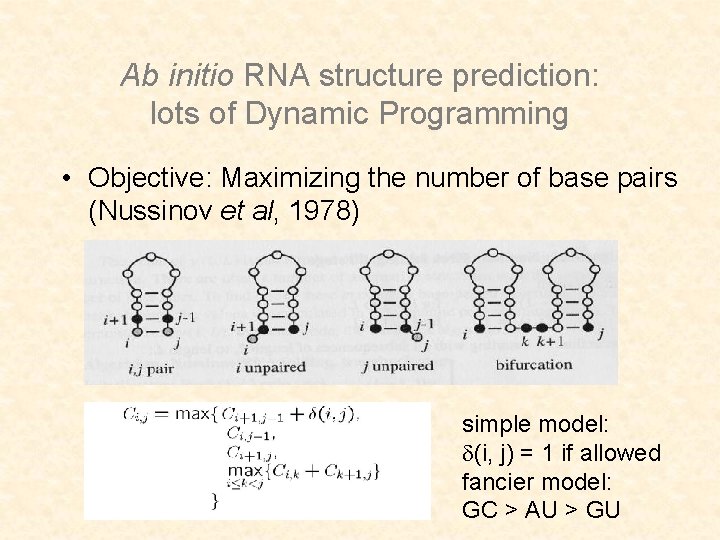
Ab initio RNA structure prediction: lots of Dynamic Programming • Objective: Maximizing the number of base pairs (Nussinov et al, 1978) simple model: (i, j) = 1 if allowed fancier model: GC > AU > GU

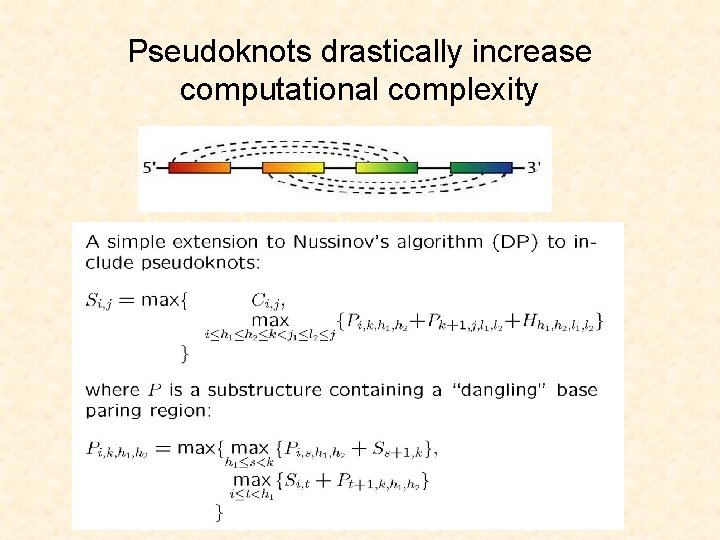
Pseudoknots drastically increase computational complexity

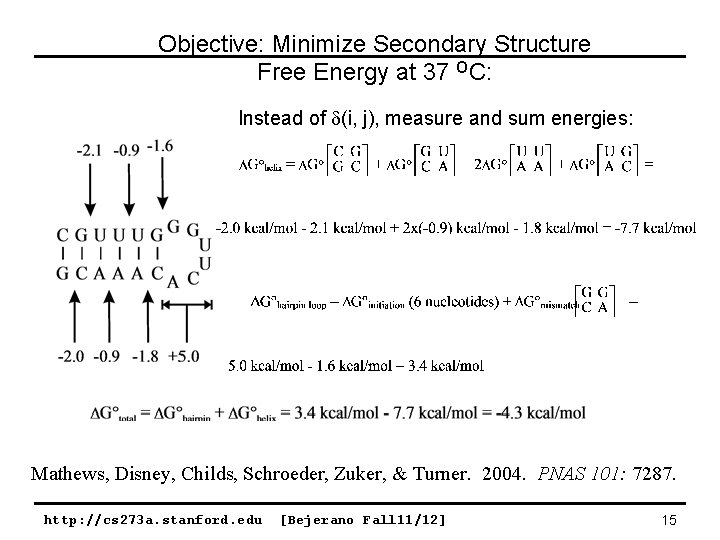
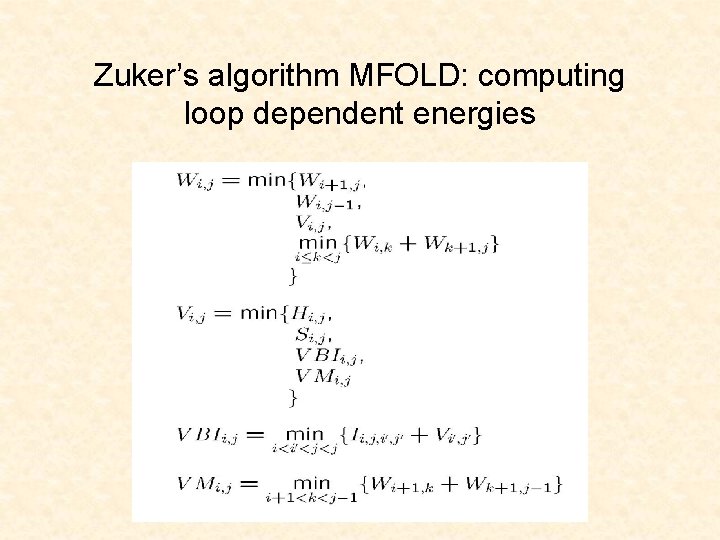
Objective: Minimize Secondary Structure Free Energy at 37 OC: Instead of (i, j), measure and sum energies: Mathews, Disney, Childs, Schroeder, Zuker, & Turner. 2004. PNAS 101: 7287. http: //cs 273 a. stanford. edu [Bejerano Fall 11/12] 15

Zuker’s algorithm MFOLD: computing loop dependent energies

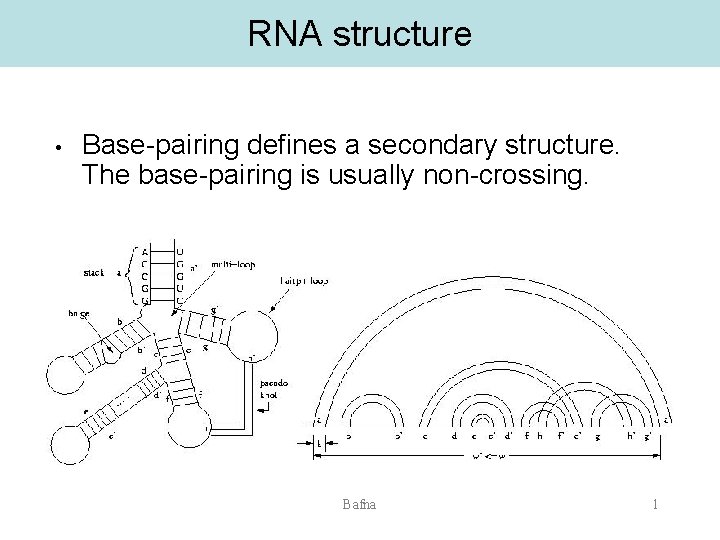
RNA structure • Base-pairing defines a secondary structure. The base-pairing is usually non-crossing. Bafna 1

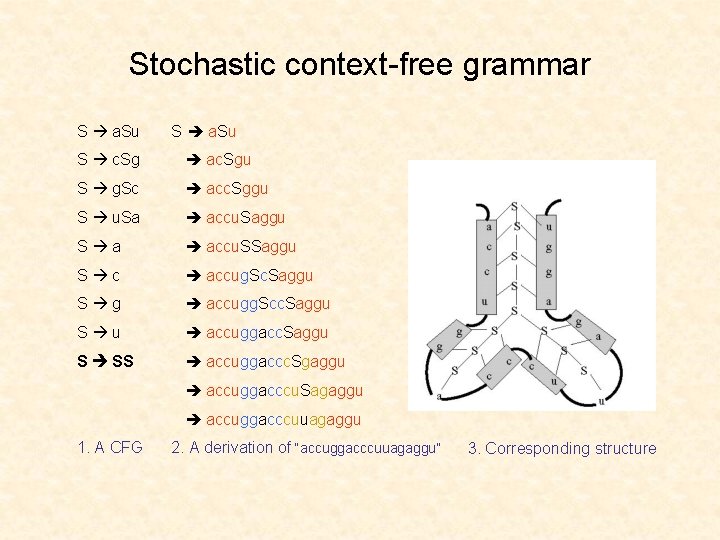
Stochastic context-free grammar S a. Su S c. Sg ac. Sgu S g. Sc acc. Sggu S u. Sa accu. Saggu S a accu. SSaggu S c accug. Sc. Saggu S g accugg. Scc. Saggu S u accuggacc. Saggu S SS accuggaccc. Sgaggu accuggacccu. Sagaggu accuggacccuuagaggu 1. A CFG 2. A derivation of “accuggacccuuagaggu” 3. Corresponding structure

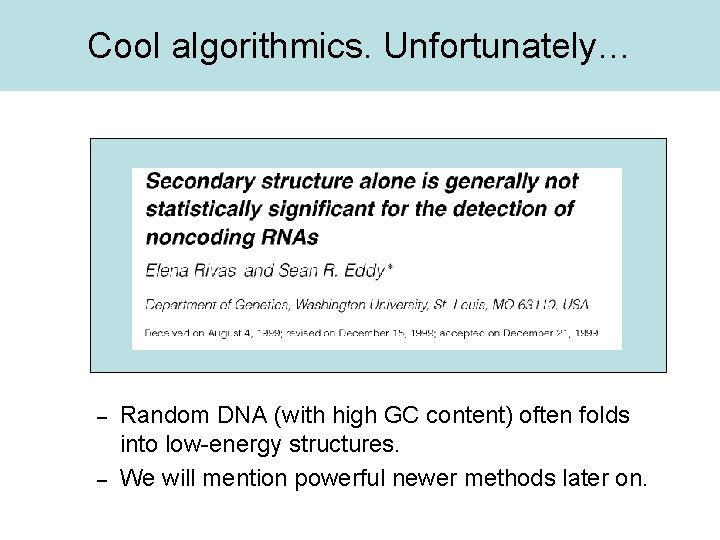
Cool algorithmics. Unfortunately… – – Random DNA (with high GC content) often folds into low-energy structures. We will mention powerful newer methods later on.

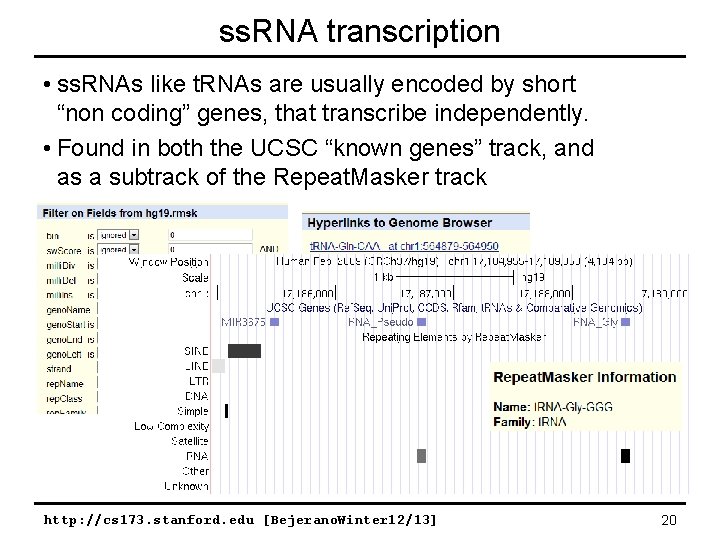
ss. RNA transcription • ss. RNAs like t. RNAs are usually encoded by short “non coding” genes, that transcribe independently. • Found in both the UCSC “known genes” track, and as a subtrack of the Repeat. Masker track http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 20

“non coding” RNAs (nc. RNA) micro. RNAs (mi. RNA/mi. R) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 21

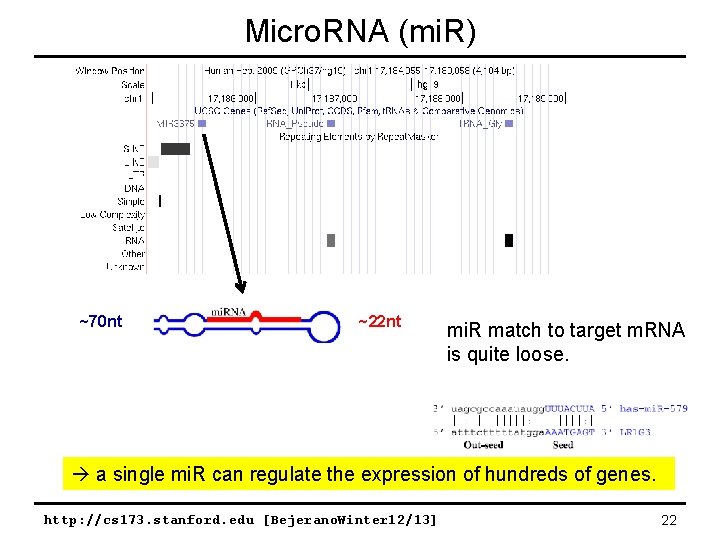
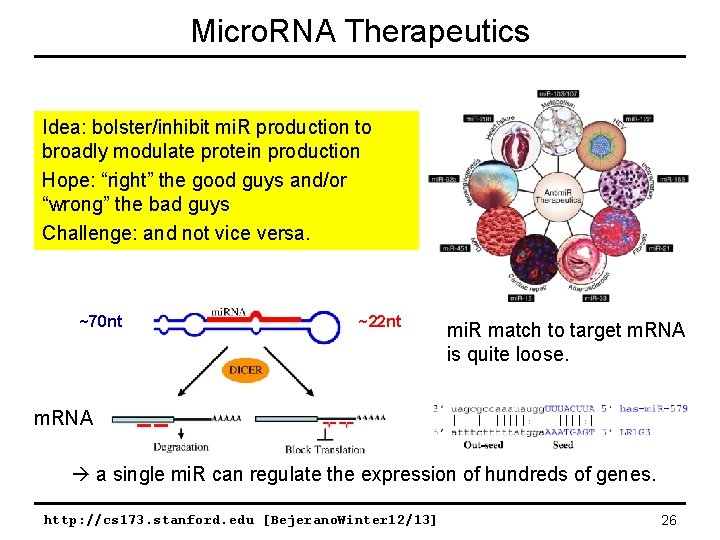
Micro. RNA (mi. R) ~70 nt ~22 nt mi. R match to target m. RNA is quite loose. m. RNA a single mi. R can regulate the expression of hundreds of genes. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 22
![Micro RNA Transcription m RNA http cs 173 stanford edu Bejerano Winter 1213 23 Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 23](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-23.jpg)
Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 23
![Micro RNA Transcription m RNA http cs 173 stanford edu Bejerano Winter 1213 24 Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 24](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-24.jpg)
Micro. RNA Transcription m. RNA http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 24

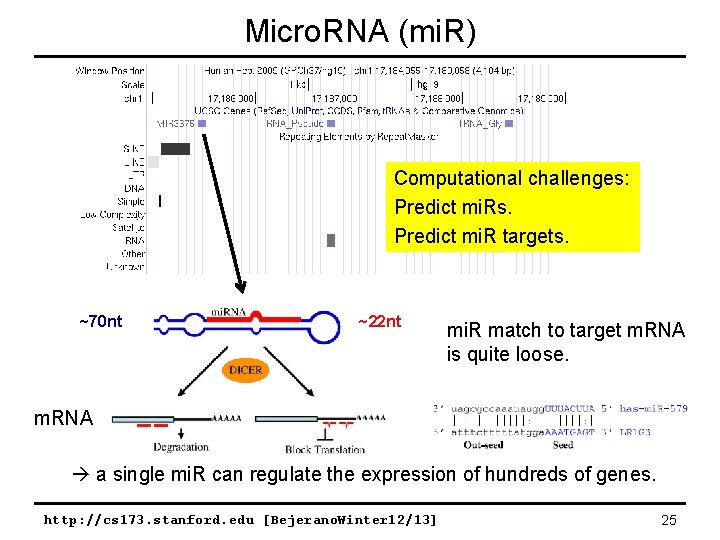
Micro. RNA (mi. R) Computational challenges: Predict mi. Rs. Predict mi. R targets. ~70 nt ~22 nt mi. R match to target m. RNA is quite loose. m. RNA a single mi. R can regulate the expression of hundreds of genes. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 25

Micro. RNA Therapeutics Idea: bolster/inhibit mi. R production to broadly modulate protein production Hope: “right” the good guys and/or “wrong” the bad guys Challenge: and not vice versa. ~70 nt ~22 nt mi. R match to target m. RNA is quite loose. m. RNA a single mi. R can regulate the expression of hundreds of genes. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 26
![Other Non Coding Transcripts http cs 173 stanford edu Bejerano Winter 1213 27 Other Non Coding Transcripts http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 27](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-27.jpg)
Other Non Coding Transcripts http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 27

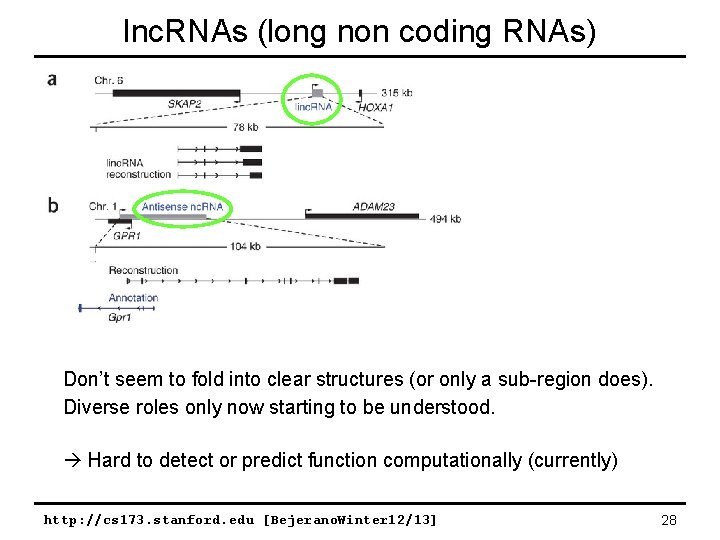
lnc. RNAs (long non coding RNAs) Don’t seem to fold into clear structures (or only a sub-region does). Diverse roles only now starting to be understood. Hard to detect or predict function computationally (currently) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 28
![lnc RNAs come in many flavors http cs 173 stanford edu Bejerano Winter 1213 lnc. RNAs come in many flavors http: //cs 173. stanford. edu [Bejerano. Winter 12/13]](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-29.jpg)
lnc. RNAs come in many flavors http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 29

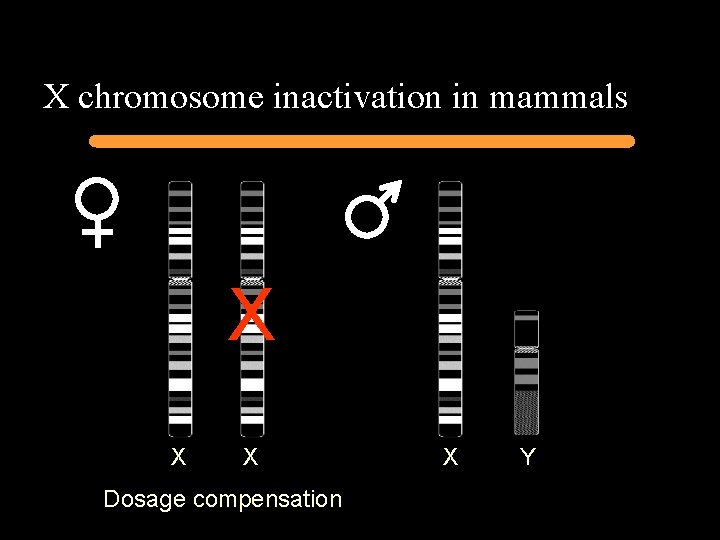
X chromosome inactivation in mammals X X X Dosage compensation X Y

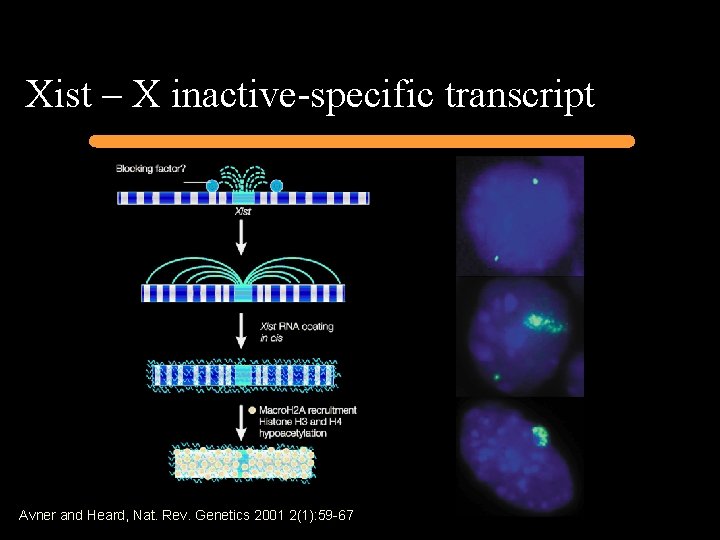
Xist – X inactive-specific transcript Avner and Heard, Nat. Rev. Genetics 2001 2(1): 59 -67

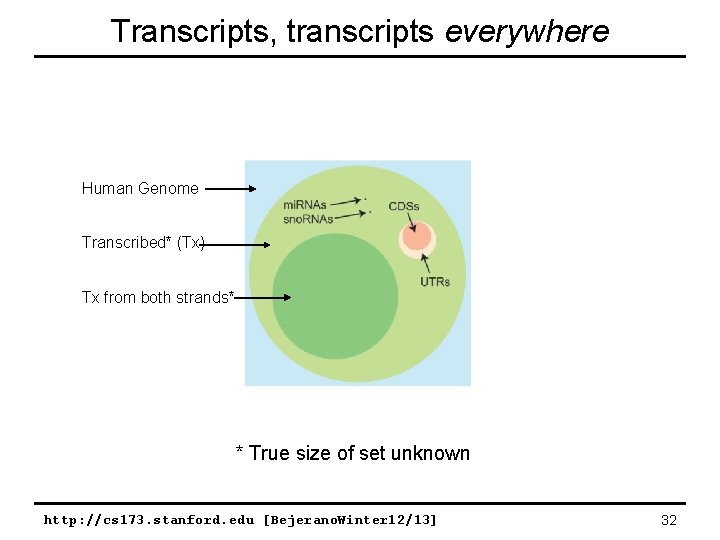
Transcripts, transcripts everywhere Human Genome Transcribed* (Tx) Tx from both strands* * True size of set unknown http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 32
![Or are they http cs 173 stanford edu Bejerano Winter 1213 33 Or are they? http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 33](https://slidetodoc.com/presentation_image/049edb2cc8ac1bdefe7b02372d91db5c/image-33.jpg)
Or are they? http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 33

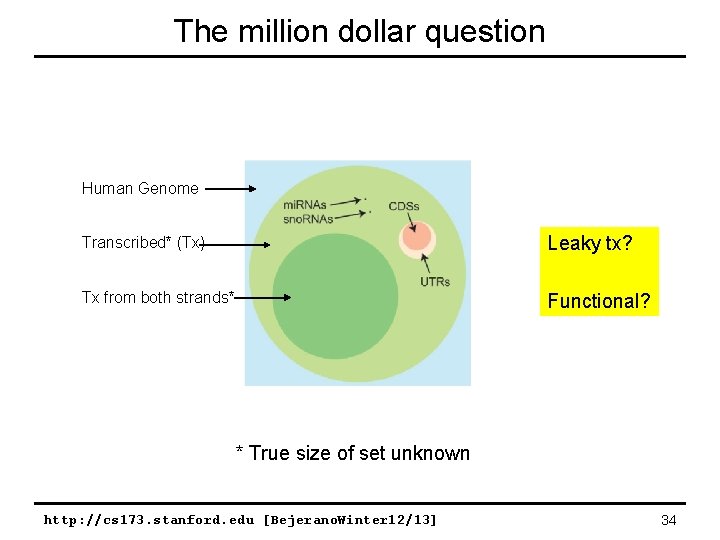
The million dollar question Human Genome Transcribed* (Tx) Leaky tx? Tx from both strands* Functional? * True size of set unknown http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 34

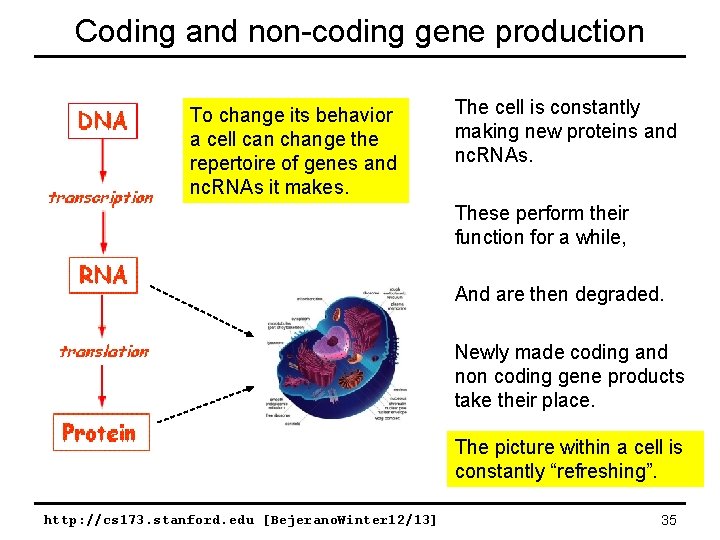
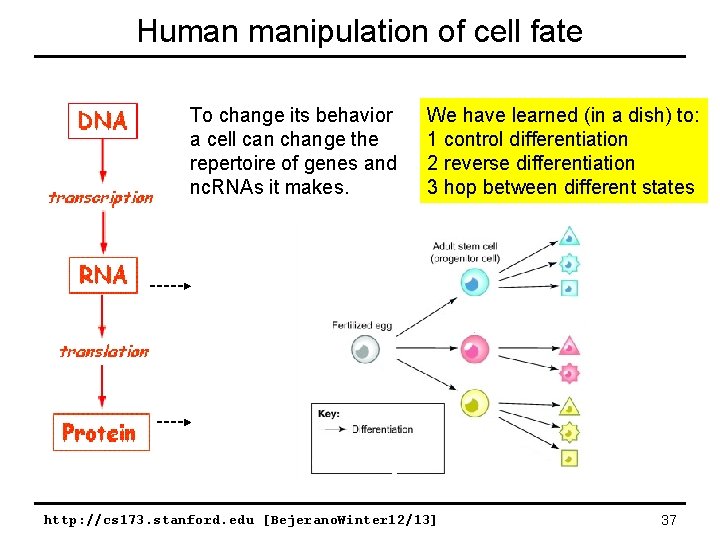
Coding and non-coding gene production To change its behavior a cell can change the repertoire of genes and nc. RNAs it makes. The cell is constantly making new proteins and nc. RNAs. These perform their function for a while, And are then degraded. Newly made coding and non coding gene products take their place. The picture within a cell is constantly “refreshing”. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 35

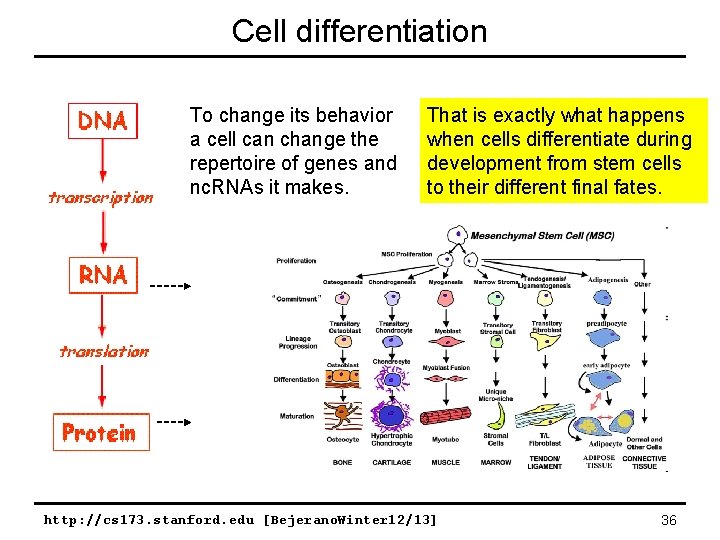
Cell differentiation To change its behavior a cell can change the repertoire of genes and nc. RNAs it makes. That is exactly what happens when cells differentiate during development from stem cells to their different final fates. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 36

Human manipulation of cell fate To change its behavior a cell can change the repertoire of genes and nc. RNAs it makes. We have learned (in a dish) to: 1 control differentiation 2 reverse differentiation 3 hop between different states http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 37

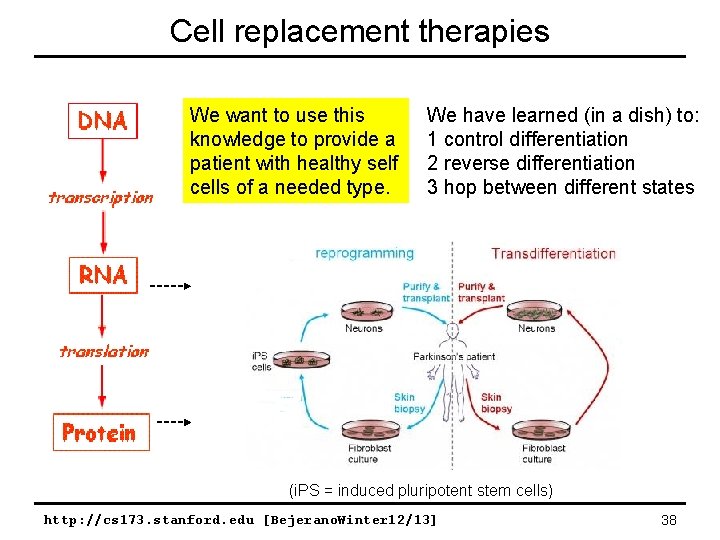
Cell replacement therapies We want to use this knowledge to provide a patient with healthy self cells of a needed type. We have learned (in a dish) to: 1 control differentiation 2 reverse differentiation 3 hop between different states (i. PS = induced pluripotent stem cells) http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 38

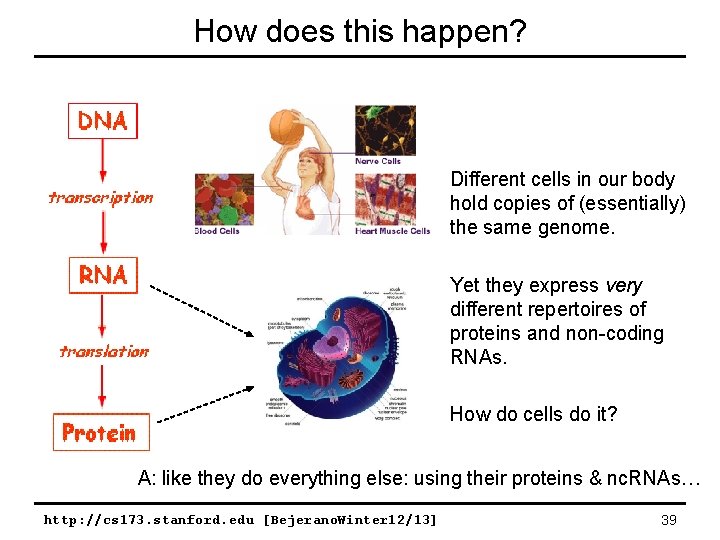
How does this happen? Different cells in our body hold copies of (essentially) the same genome. Yet they express very different repertoires of proteins and non-coding RNAs. How do cells do it? A: like they do everything else: using their proteins & nc. RNAs… http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 39

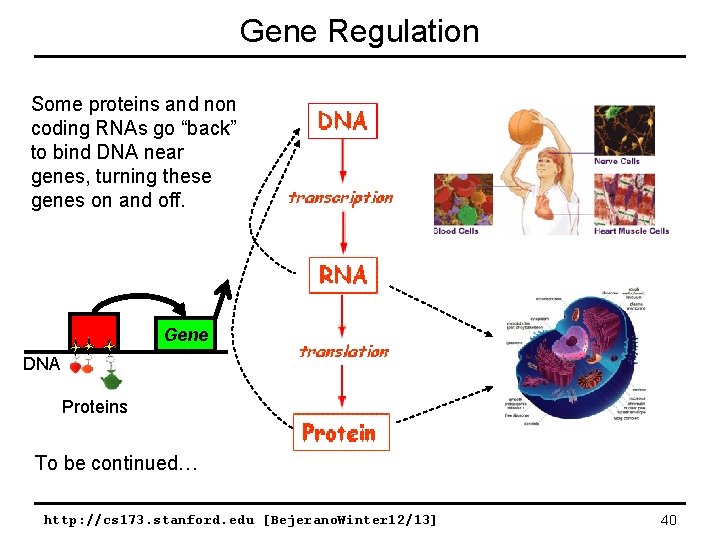
Gene Regulation Some proteins and non coding RNAs go “back” to bind DNA near genes, turning these genes on and off. Gene DNA Proteins To be continued… http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 40

Review Lecture 6 • Central dogma recap – Genes, proteins and non coding RNAs • RNA world hypothesis • Small structural RNAs – Sequence, structure, function – Structure prediction – Transcription mode • Micro. RNAs – Functions – Modes of transcription • lnc. RNAs – Xist • Genome wide (and context wide) transcription – How much? – To what goals? • Gene transcription and cell identity – Cell differentiation – Human manipulation of cell fates • Gene regulation control http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 41

(On Mondays) ask students to stack the chairs without wheels at the back of the room at the end of class. http: //cs 173. stanford. edu [Bejerano. Winter 12/13] 42