SFSU Center for Computing for Life Sciences CCLS















- Slides: 35

SFSU Center for Computing for Life Sciences - CCLS Dragutin Petkovic 1, Chris Smith 2, 3, Mike Wong 1, 3 1 - SFSU Department of Computer Science 2 - SFSU Department of Biology 3 - SFSU Center for Computing for Life Sciences

Outline • About Center for Computing for Life Sciences (CCLS) at SFSU – cs. sfsu. edu/ccls/index. html • What is computing for life sciences? • CCLS Dell Cluster Computer and its usage • Chris Smith - Turning Processor Cycles into Research-Based Teaching

cs. sfsu. edu/ccls/index. html

Mission • CCLS addresses the emerging trend of integration of life sciences and computational and mathematical sciences. It involves faculty, researchers, and students from the SFSU departments of Biology, Biochemistry, Computer Science, Mathematics, and Physics and other SFSU departments. • The broad research program of the center emphasizes investigations in topics varying from Bioinformatics and Computational Drug Discovery to complex data visualization and development of new paradigms for data modeling, user interfaces and web-engineering in contexts involving life sciences. • The CCLS provides an environment for faculty to cooperate, for students to work on multidisciplinary projects including those involving culmination degrees and for collaboration with industrial and academic partners. The center also hosts a number of external advisors and collaborators. ccls. lab. sfsu. edu

CCLS: An Interdisciplinary Collaboration Space 5

Areas addressed by CCLS projects – they are broad by design Bioinformatics but also… • • • Use of Machine Learning or analysis and classifications of genotypes Data management for biology and drug development Data visualization Mathematical modeling of genetic structures Advanced WWW applications and user interfaces Serious games in health education Sensor networks for biological and environmental applications Data mining of biological data High performance clusters and SW tools for computing for life sciences ……whatever is next

Range of Activities • Broad, but focused on fostering research, not direct teaching – – – – Projects and theses Research and publications External grants Collaboration with industry and academia Hosting seminar visitors Helping faculty with grants and travel IT and high performance computing support Also incubators for high tech

• 20+ faculty CCLS Accomplishments – COSE (CS. Math, Biology, Chemistry and Biochemistry) – Health and Human Services, Industrial design, Philosophy • Over 19 MS Theses in CCLS area since 2004 • Over 28 refereed publications in CCLS area since 2003. – One best paper award & one second best paper award – Several top awards at COSE science fairs • NSF Career Grant to Prof. R. Singh for proposed research in CCLS area – Data management and search for chemical data • Funding: – External sources - NSF, CSUPERB, Microsoft, Sun/Agilent, NIH – Support for CCLS investigators - Three rounds of mini grant and travel grants funded by CCLS: • 30+ faculty and students funded (about $ 150 K in three years) • External Collaborators: UCSF, UC Davis, SUN/Agilent, Microsoft, Washington University Genome Center, Lawrence Berkeley National Lab • Core Computing Resources – Dell High performance cluster computer – Teaching Cluster & shared application servers – Climate and power control for independent research groups

CCLS Computing Resources • A cluster is – Multiple computers offer high compute power – Work closely together such that they can be viewed as a single computer. – Network/WWW accessible – Small footprint • Applications include – Predicting molecular structure (e. g. protein folding) – Gene sequence searches (e. g. BLAST searches) – Genetic similarity comparison between species (e. g. PAUP phylogentics analysis). – Predicting RNA secondary structures CCLS DELL Power. Edge 1955 Quad-Processor Compute Nodes Mike Wong M. S. Researcher Programmer

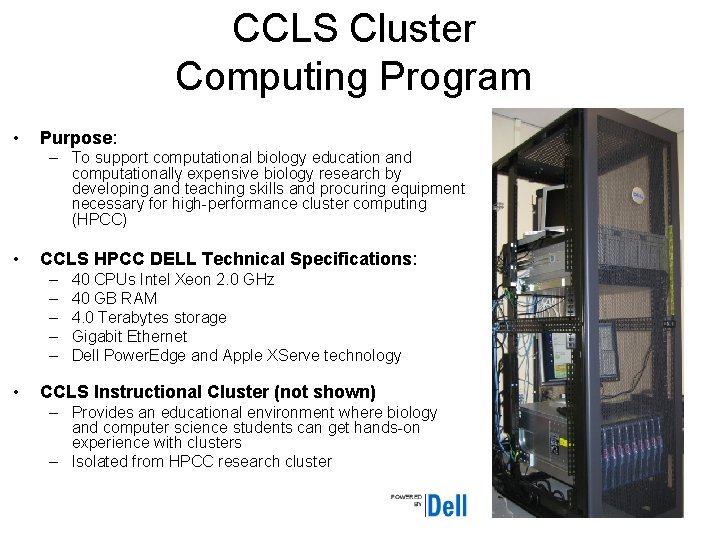
CCLS Cluster Computing Program • Purpose: – To support computational biology education and computationally expensive biology research by developing and teaching skills and procuring equipment necessary for high-performance cluster computing (HPCC) • CCLS HPCC DELL Technical Specifications: – – – • 40 CPUs Intel Xeon 2. 0 GHz 40 GB RAM 4. 0 Terabytes storage Gigabit Ethernet Dell Power. Edge and Apple XServe technology CCLS Instructional Cluster (not shown) – Provides an educational environment where biology and computer science students can get hands-on experience with clusters – Isolated from HPCC research cluster

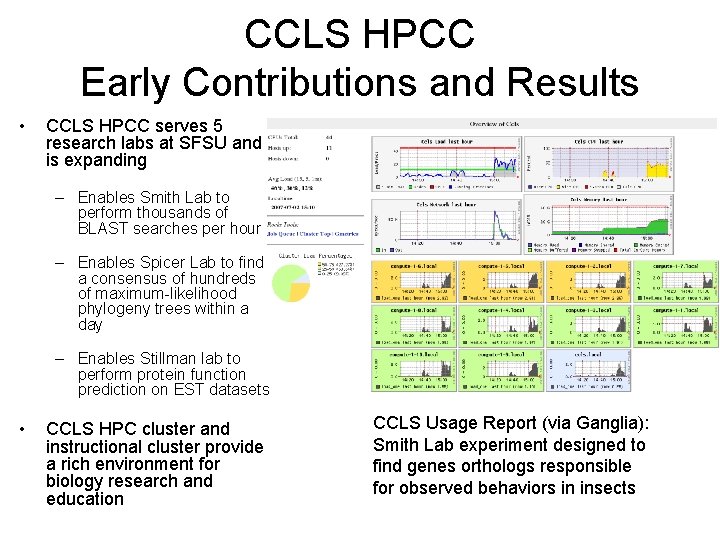
CCLS HPCC Early Contributions and Results • CCLS HPCC serves 5 research labs at SFSU and is expanding – Enables Smith Lab to perform thousands of BLAST searches per hour – Enables Spicer Lab to find a consensus of hundreds of maximum-likelihood phylogeny trees within a day – Enables Stillman lab to perform protein function prediction on EST datasets • CCLS HPC cluster and instructional cluster provide a rich environment for biology research and education CCLS Usage Report (via Ganglia): Smith Lab experiment designed to find genes orthologs responsible for observed behaviors in insects

Summary • CS and math are becoming a critical tool for future advances in biotechnology and is exciting area for research and teaching • CSU must address this area adequately • CCLS at SFSU is one example of a working model of Biology/Chemistry/CS/Math/Life Sciences collaboration • CCLS advocates addressing this area very broadly, NOT only as bioinformatics • Critical need for infrastructure support: technical support, admin, people, space, networking, SW, HW (NOT ONLY HW)


Turning Processor Cycles into Research-Based Teaching • Annotation Background • Genomics Education Partnership • CCLS Genome Annotation Pipeline • Biol 638/738: Student Genome Annotation © Smith. Lab 2007

Given Some Raw Sequence? G GTATCTTATTCGCCATCGAAGCGGTCACACTGGGTGCCGCCGCCAACTTCACTCTTTCCGT TCTGTGAGCGAAAACCGAAAAGTCTGTGCTTTGGTAAGTGTTGCTAAAAGTTCGGAATAAT GTTGCATCCCGAGCATTTTCGGGTACATAACTGTTCCACGGCGGTGGTCCAGCAAAGACT AATCGTTATCACGCCTTTCGCAGTTCTTAAATTCACCCGACGAGTCCCTAATACACAATTAA AATGGTTAGGGAGAACAAGGCAGCGTGGAAGGCTCAGTACTTCATCAAGGTTGTGGTAAG TATAGAACCTTATAGAATTCGCTCACTAGCTGGCGCCTGGCTTATGCTGTTAACTGATCCC TCCTCCAGGAACTGTTCGATGAGTTCCCAAAGTGCTTCATCGTGGGCGCCGACAACGTGG GCTCCAAGCAGATGCAGAACATCCGTACCAGCCTGCGTGGACTGGCCGTCGTGCTTATGG GCAAGAACACCATGATGCGCAAGGCCATCCGCGGTCATCTGGAGAACAACCCGCAGCTG GAGAAGCTGCTACCCCACATCAAGGGCAACGTGGGATTCGTGTTCACCAAGGGCGATCTC GCCGAGGTGCGCGACAAGCTGCTGGAGTCCAAGGTGCGCGCCCCCGTCCCGGCG CTATTGCCCCTCTGCACGTCATCATCCCGGCGCAGAACACCGGCTTGGGACCCGAGAAGA CCAGTTTCTTCCAGGCCCTGTCCATCCCGACCAAAATTTCCAAGGGAACAATTGAAATCAT CAACGATGTGCCCATCCTGAAGCCTGGCGACAAGGTCGGCGCCTCCGAGGCGACACTGC TCAACATGTTGAACATCTCGCCCTTCTCGTACGGTCTGATTGTCAACCAGGTCTACGACTC CGGCTCGATCTTTTCGCCGGAGATCCTGGACATCAAGCCCGAGGATCTGCGCGCCAAGTT CCAACAGGGAGTGGCCAACTTGGCCGCCGTTTGTCCGTGGGCTACCCCACCATCGC CTCGGCCCCGCACAGCATTGCCAACGGATTCAAGAATCTGCTGGCCATTGCTGCCACCAC CGAGGTGGAGTTCAAGGAGGCGACCACCATCAAGGAGTACATCAAGGACCCCAGCAAGTT CGCCGCAGCTGCTTCGGCTGCCCCCGCGGCGGAGCTACCGAGAAGAAG GAGGAGGCCAAGAAGCCCGAGTCCGAATCAGAGGAGGAGGACGATGATATGGGTTTCGG TCTGTTCGACTAAGCTGGATCCCGATTGCAGAATGCCCTCTGCGGCGCCCGCGAACCATC GCTTCCGCTTTCGGCGTTTACCCACTAAGACCCTTTGTTATGTT

What Does The Sequence Encode? G GTATCTTATTCGCCATCGAAGCGGTCACACTGGGTGCCGCCGCCAACTTCACTCTTTCCG TTCTGTGAGCGAAAACCGAAAAGTCTGTGCTTTGGTAAGTGTTGCTAAAAGTTCGGAATAA TGTTGCATCCCGAGCATTTTCGGGTACATAACTGTTCCACGGCGGTGGTCCAGCAAAGAC TAATCGTTATCACGCCTTTCGCAGTTCTTAAATTCACCCGACGAGTCCCTAATACACAATT AAAATGGTTAGGGAGAACAAGGCAGCGTGGAAGGCTCAGTACTTCATCAAGGTTGTGGT AAGTATAGAACCTTATAGAATTCGCTCACTAGCTGGCGCCTGGCTTATGCTGTTAACTGAT CCCTCCTCCAGGAACTGTTCGATGAGTTCCCAAAGTGCTTCATCGTGGGCGCCGACAAC GTGGGCTCCAAGCAGATGCAGAACATCCGTACCAGCCTGCGTGGACTGGCCGTCGTGC TTATGGGCAAGAACACCATGATGCGCAAGGCCATCCGCGGTCATCTGGAGAACAACCC GCAGCTGGAGAAGCTGCTACCCCACATCAAGGGCAACGTGGGATTCGTGTTCACCAAG GGCGATCTCGCCGAGGTGCGCGACAAGCTGCTGGAGTCCAAGGTGCGCGCCCCCGCCC GTCCCGGCGCTATTGCCCCTCTGCACGTCATCATCCCGGCGCAGAACACCGGCTTGGGA CCCGAGAAGACCAGTTTCTTCCAGGCCCTGTCCATCCCGACCAAAATTTCCAAGGGAAC AATTGAAATCATCAACGATGTGCCCATCCTGAAGCCTGGCGACAAGGTCGGCGCCTCCG AGGCGACACTGCTCAACATGTTGAACATCTCGCCCTTCTCGTACGGTCTGATTGTCAACC AGGTCTACGACTCCGGCTCGATCTTTTCGCCGGAGATCCTGGACATCAAGCCCGAGGAT CTGCGCGCCAAGTTCCAACAGGGAGTGGCCAACTTGGCCGCCGTTTGTCCGTGGG CTACCCCACCATCGCCTCGGCCCCGCACAGCATTGCCAACGGATTCAAGAATCTGCTGG CCATTGCTGCCACCACCGAGGTGGAGTTCAAGGAGGCGACCACCATCAAGGAGTACAT CAAGGACCCCAGCAAGTTCGCCGCAGCTGCTTCGGCTGCCCCCGCGGC GGAGCTACCGAGAAGAAGGAGGAGGCCAAGAAGCCCGAGTCCGAATCAGAGGAGGAG GACGATGATATGGGTTTCGGTCTGTTCGACTAAGCTGGATCCCGATTGCAGAATGCCCTC TGCGGCGCCCGCGAACCATCGCTTCCGCTTTCGGCGTTTACCCACTAAGACCCTTTGTTA TGTT 5’ UTR START CODING EXON STOP 3’UTR

Genome Annotation The Problem: Too many genomes, not many reliably annotated. Bad gene models make it harder to clone genes and use genomic data in the lab. Reliance on automated annotations means that many analyses are ‘quick & dirty’ © Smith. Lab 2007 www. genomesonline. org/ Liolios et al. NAR 2006 (DOI: 10. 1093/NARGKJ 145)

The World is Filled with Non-Model Organisms • • Only a few model organisms annotated, only 5 done ‘well’ Most new genomes are automatically annotated, if at all Human curation is poorly funded or not funded Little infrastructure exists for normal people to do bioinformatics analyses in their own organisms © Smith. Lab 2007

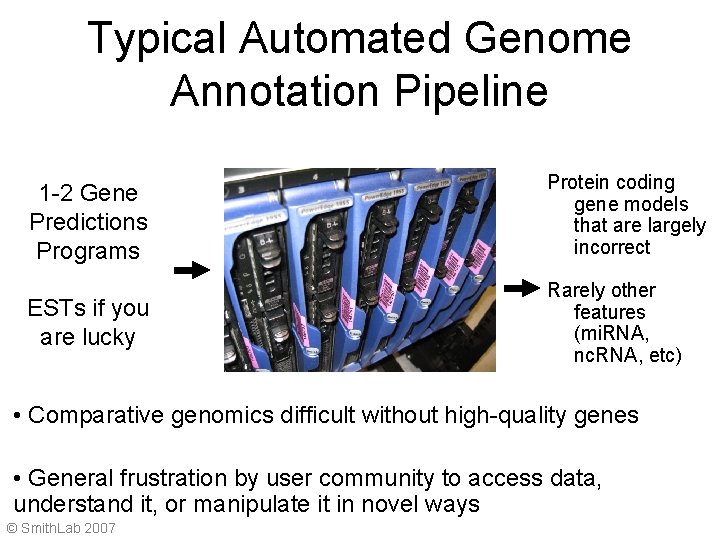
Typical Automated Genome Annotation Pipeline 1 -2 Gene Predictions Programs Protein coding gene models that are largely incorrect ESTs if you are lucky Rarely other features (mi. RNA, nc. RNA, etc) • Comparative genomics difficult without high-quality genes • General frustration by user community to access data, understand it, or manipulate it in novel ways © Smith. Lab 2007

Automated Annotation is Better Than Some Methods*… theredsrocket. blogspot. com/2007/04/finals. html * Methods have not been actually tested • Web tools & common formats enable distributed annotation • Easier technology puts annotation in grasp of students © Smith. Lab 2007

• Student Driven Community Genome Annotation • Collaborators (34 US Universities) • • Smith Lab @ SFSU Jim Youngblom @ CSU Stanislaus Anya Goodman @ California Poly Catherine Coyle-Thompson @ CSU Northridge • Use real research data as a teaching tool © Smith. Lab 2007

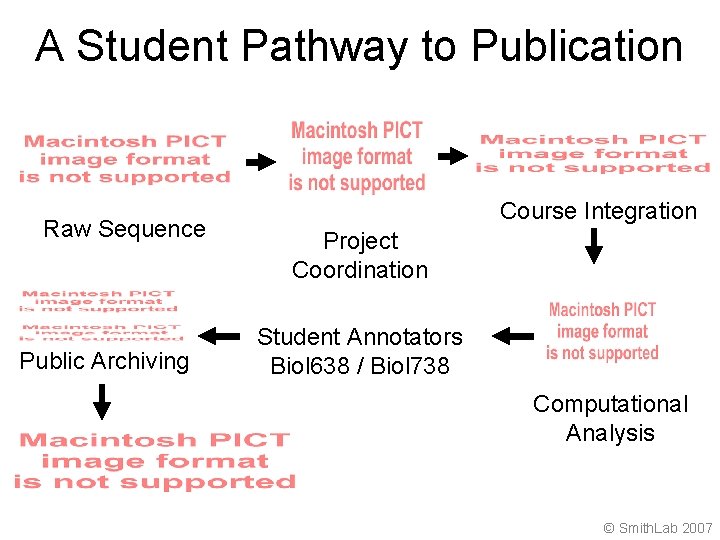
A Student Pathway to Publication Raw Sequence Public Archiving Course Integration Project Coordination Student Annotators Biol 638 / Biol 738 Computational Analysis © Smith. Lab 2007

Students perform analysis and annotation in class • Biol 638/Biol 738 – Paired Undergraduate/Graduate Genome Annotation Workshop – 1 Semester, 4 units – Fall 2007 20 enrolled, 14 finished – Taught in SFSU SEGA Teaching Lab • 20 i. Mac G 4’s • Students can also use their own computer All subjects & ages under • Each student annotates 50 kb of sequence one roof – Finds repeat, genes, protein functions, promoters – Learn basic UNIX, command-line programs © Smith. Lab 2007 • Pre- and Post-Course Assessment Surveys

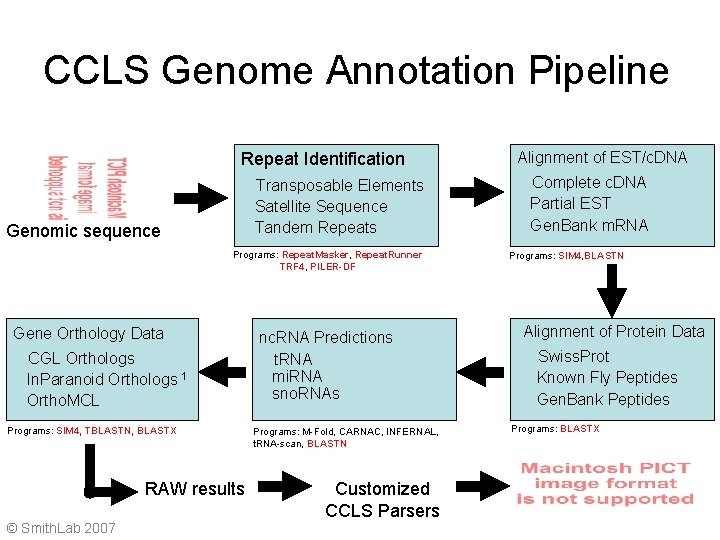
CCLS Genome Annotation Pipeline Repeat Identification Transposable Elements Satellite Sequence Tandem Repeats Genomic sequence Programs: Repeat. Masker, Repeat. Runner TRF 4, PILER-DF Gene Orthology Data CGL Orthologs In. Paranoid Orthologs 1 Ortho. MCL Programs: SIM 4, TBLASTN, BLASTX RAW results © Smith. Lab 2007 nc. RNA Predictions t. RNA mi. RNA sno. RNAs Programs: M-Fold, CARNAC, INFERNAL, t. RNA-scan, BLASTN Customized CCLS Parsers Alignment of EST/c. DNA Complete c. DNA Partial EST Gen. Bank m. RNA Programs: SIM 4, BLASTN Alignment of Protein Data Swiss. Prot Known Fly Peptides Gen. Bank Peptides Programs: BLASTX

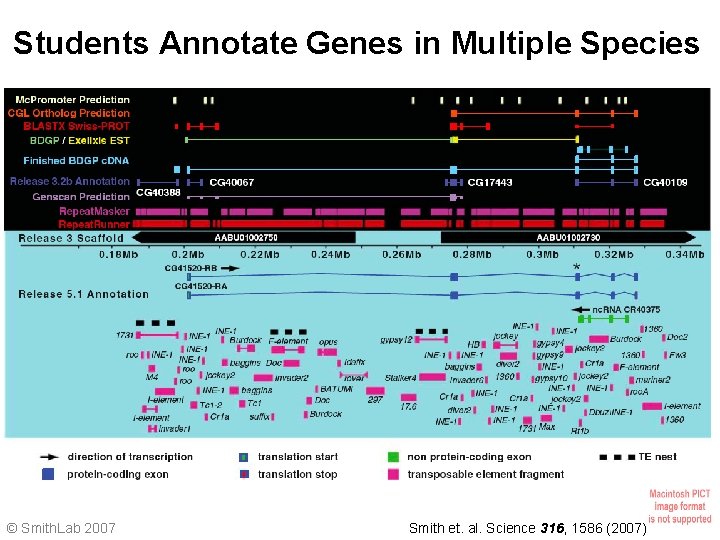
Students Annotate Genes in Multiple Species Release 5. 1 Annotation © Smith. Lab 2007 Smith et. al. Science 316, 1586 (2007)

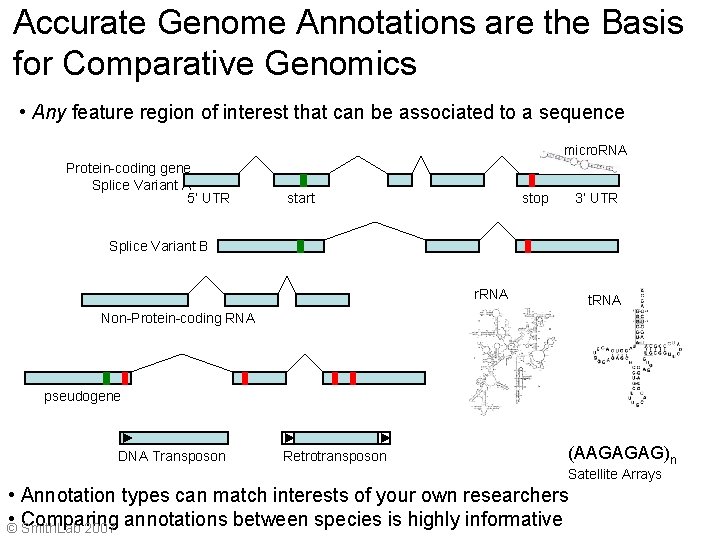
Accurate Genome Annotations are the Basis for Comparative Genomics • Any feature region of interest that can be associated to a sequence micro. RNA Protein-coding gene Splice Variant A 5’ UTR start stop 3’ UTR Splice Variant B r. RNA t. RNA Non-Protein-coding RNA pseudogene DNA Transposon Retrotransposon (AAGAGAG)n Satellite Arrays • Annotation types can match interests of your own researchers • Comparing annotations between species is highly informative © Smith. Lab 2007

Multiple Fly Genomes Give Student Access to Cutting Edge Research Data • Currently 12 Drosophlid genomes • Several more insect genomes • Possible to do in-depth comparative genomic analyses – – Conserved promoters Rates of gene evolution New/Lost genes Much more… © Smith. Lab 2007 229 Annotation authors including C. D. Smith Nature 2007 450 (8), 25 -40.

Comparative Genomic Analysis D. erecta D. melanogaster Bad D. mojavensis annotation! © Smith. Lab 2007 From Biol 738 Final Report of Jennifer Placek

RNA Structure Motifs Conserved Across Species Are Candidates for Further Study © Smith. Lab 2007 From Biol 638 Report by Lucas Hanscom Spring 2007

CCLS Injects Computing in Biology Courses • Standardized core facilities that are actively maintained • Advanced software installation and support • Custom software development for individual researchers • Access to faculty and students from other disciplines • Comfortable collaborative meeting space • Engaged staff who meets the needs of researchers

CCLS People Make the Difference Pre-CCLS Code screenshot #!/bin/csh set query = /home/cdsmith/results cd $query foreach file (*fst) current directory Conclusions blastx Pfam-A. fasta $file > $query. results end 1) I write hacky error ridden code 2) CCLS Mike Wong fixes my code Adapted to cluster Error handling Scalability New analyses & features © Smith. Lab 2007 Post-CCLS Code screenshot #!/share/apps/bin/perl -w use Datastore: : MD 5; use File: : Path qw( mkpath ); use Statistics: : Descriptive; use Proc: : Daemon; Proc: : Daemon: : Init; # This script will continue running after you log out # ===== INITIALIZE VARIABLES # It's important to use absolute paths; Proc: : Daemon: : Init requires it our $prefix = "/home/mikewong/research/stillmanlab"; # CHANGE THIS VARIABLE our $path = { results => "$prefix/JGI_Project/results", queries => "$prefix/JGI_Project/queries", }; my $job_name = 'anu_blast'; my $species = '/share/apps/data/blastdb/Gen. Bank_v 159_aa. fasta'; my $datastore = new Datastore: : MD 5( root => $path->{ results }, depth => 2 ); mkdir $path->{ results } unless -e $path->{ results }; # ===== READ THE QUERY DIRECTORY FOR FST FILES opendir DIR, $path->{ queries }; my @files = sort grep { /. fst$/ } readdir DIR; closedir DIR; my $job_processing_times = new Statistics: : Descriptive: : Full(); open LOG, ">>$prefix/JGI_Project/log"; # ===== GENERATE THE COMMAND FOR EACH FILE/SPECIES COMBINATION foreach my $file (@files) { my $results_path = $datastore->id_to_dir( $file ); mkpath $results_path unless -e $results_path; my $db = $species;

Acknowledgements • Bioinformatics & Genome Annotation Class Fall 2007 – Tobias Sayre (Graduate Assistant) • • • Ari A. Ramsey M. Amy S. Joseph B. Vy N. Elinor V. Eugenel E. Jennifer P. Tyler W. Henry H. Bhamini P. Mike W. Jay K. Marvin S. Lucas H. (S 07) CCLS Pipeline - Mike Wong SFSU COSE Hardware Support - Alan Der SFSU COSE Network Support - Tina Easter © Smithlab 2007

fin

Using the Semantic Web to Link Genes and Behaviors • Took 200 known behavior genes from flies • Used CCLS cluster to identify orthologs in ants and bees • Designed primers to find in new ant species • Created networks of genes linked to behaviors © Smith. Lab 2007

1 -Student, 1 -Gene Independent Project • Romeo-Smith HIV Project – HIV is known to suppress host immune system genes – HIV Tar RNA secondary structure may act to inhibit through RNAi • Screen all human genes & genome for novel Tar targets – Human genes may also adopt Tarlike shapes • Use CCLS cluster + RNA folding tools to fold all 30, 000 human genes © Smith. Lab 2007 www. mcld. co. uk