Computational Proteomics of Protein Complexes Talk at NIH




- Slides: 40

Computational Proteomics of Protein Complexes Talk at NIH 2003. 04. 07 Do not reproduce without permission 2 2 Gerstein. info/talks (c) 2003 Mark B Gerstein Yale U

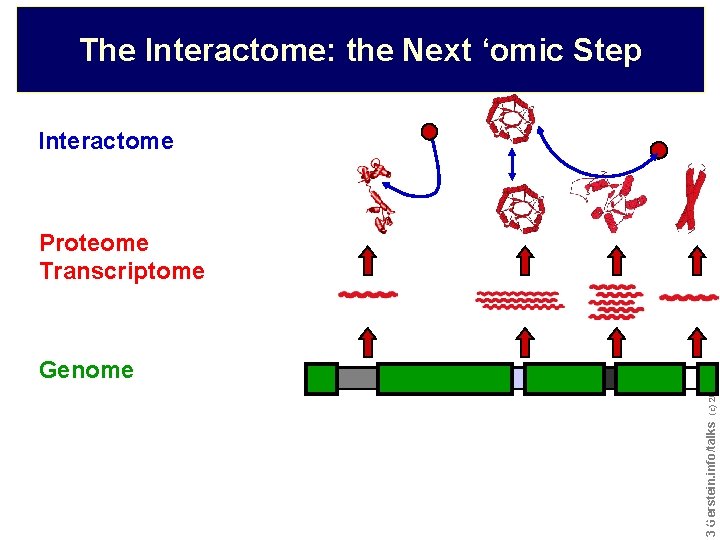
The Interactome: the Next ‘omic Step Interactome Proteome Transcriptome Do not reproduce without permission 3 3 Gerstein. info/talks (c) 2003 Genome

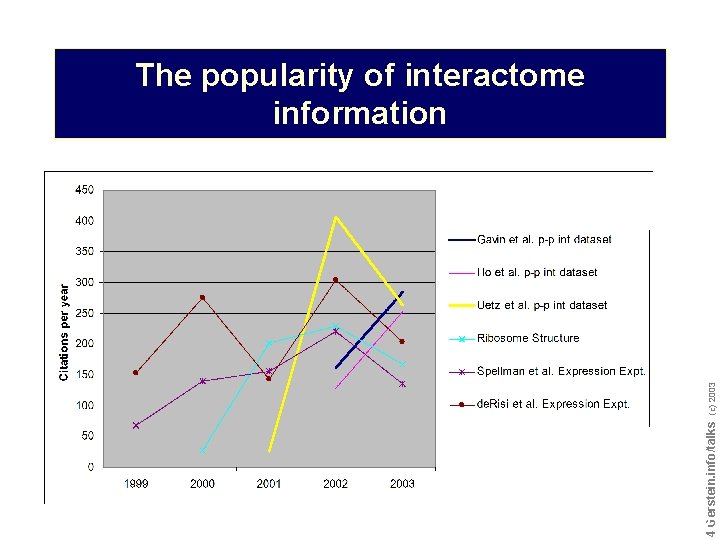
Do not reproduce without permission 4 4 Gerstein. info/talks (c) 2003 The popularity of interactome information

Do not reproduce without permission 5 5 Gerstein. info/talks 1. Interactions provide a systematic way of defining protein function on a genomic scale 2. Known complexes provide a benchmark to validate and integrate genome-wide interaction experiments, providing a more accurate interactome 3. Known complexes provide a focus for the intergration of (non-interaction) genomic information – e. g. expression data 4. Extrapolating from known complexes, one can predict protein complexes on a genome-scale via integrating experimental interactions and noninteraction information (combining #1 and #2) (c) 2003 Computational Proteomics of Complexes

Do not reproduce without permission 6 6 Gerstein. info/talks (c) 2003 Circumscribing Protein Function in terms of Interactions

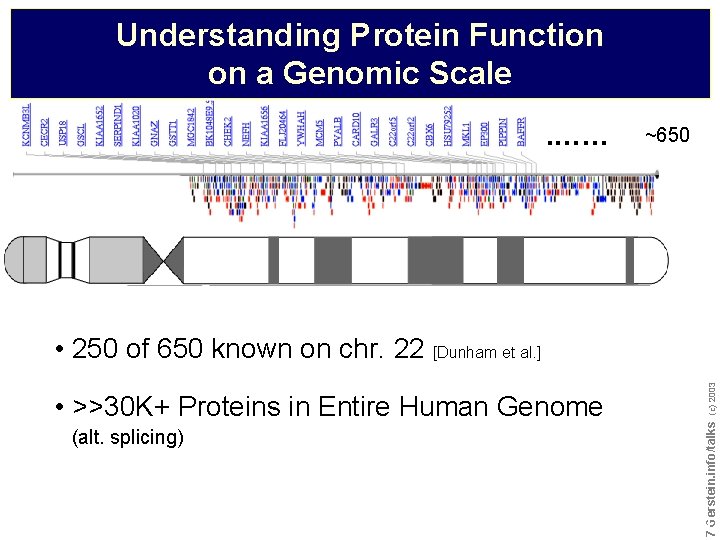
Understanding Protein Function on a Genomic Scale. …… ~650 (alt. splicing) Do not reproduce without permission 7 7 Gerstein. info/talks • >>30 K+ Proteins in Entire Human Genome (c) 2003 • 250 of 650 known on chr. 22 [Dunham et al. ]

Issues in defining protein function on a genomic scale • Multi-functionality: 2 functions/protein (also 2 proteins/function) • Role Conflation: molecular, cellular, phenotypic • Fun terms… but do they scale? • Starry night • For now, definable aspects of function: interactions, location, [Babbit] enzymatic rxn. Do not reproduce without permission 8 8 Gerstein. info/talks (c) 2003 • Sarah (affects female fertility); Sonic; Darkener of apricot & suppressor of white apricot; Redtape, gridlock, roadblock (when mutated block transport along axons); ROP vs ROM ("Regulator of Copy Number" or RNA-I-II-complex-binding-protein)

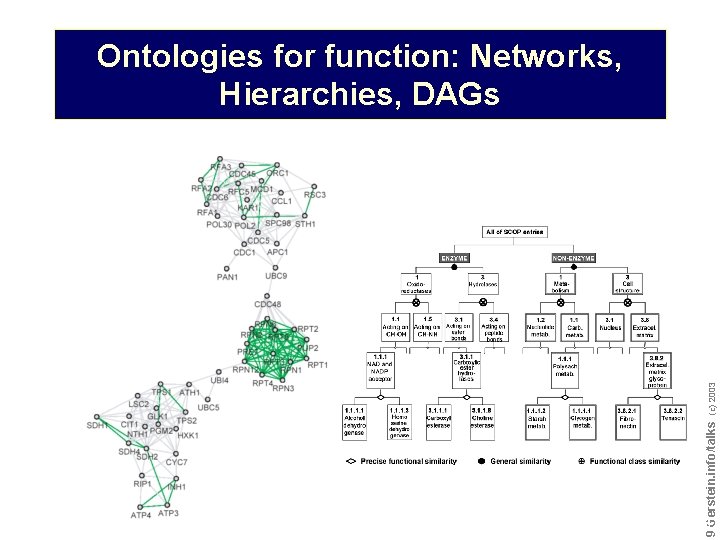
Do not reproduce without permission 9 9 Gerstein. info/talks (c) 2003 Ontologies for function: Networks, Hierarchies, DAGs

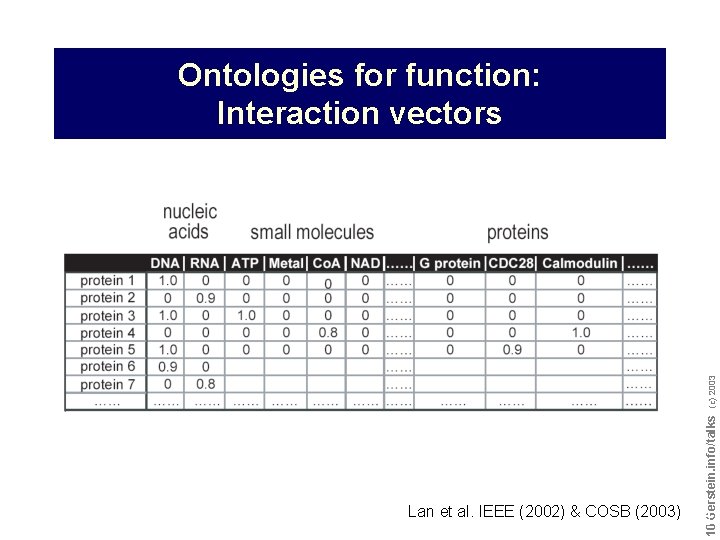
Lan et al. IEEE (2002) & COSB (2003) Do not reproduce without permission 1010 Gerstein. info/talks (c) 2003 Ontologies for function: Interaction vectors

Do not reproduce without permission 1111 Gerstein. info/talks (c) 2003 Validating and Integrating Genomic Protein-Protein Interaction Datasets with Known Complexes

Protein interaction data • Databases (BIND, DIP, MIPS etc. ) à literature • High-throughput datasets à in vivo pull down à yeast two-hybrid • Computational predictions à Tangential genomic data Do not reproduce without permission 1212 Gerstein. info/talks (c) 2003 • Expression data • Phenotypic data • Localization Data

Combining interaction data • High-throughput data is less reliable than more careful, smaller scale experiments à Orthogonal datasets • Combining data increases à How to weight the different data sources? à General classification problem (machine learning) à Bayesian networks: probabilistic Do not reproduce without permission 1313 Gerstein. info/talks • How to do this in a quantitative way? (c) 2003 à accuracy à coverage

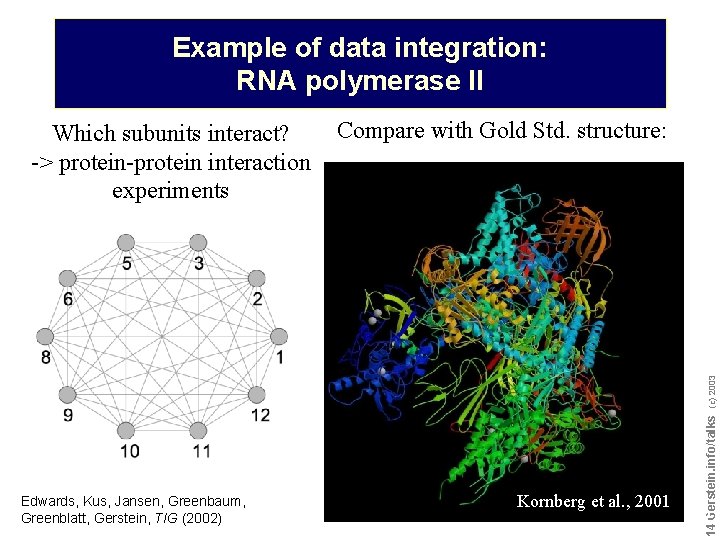
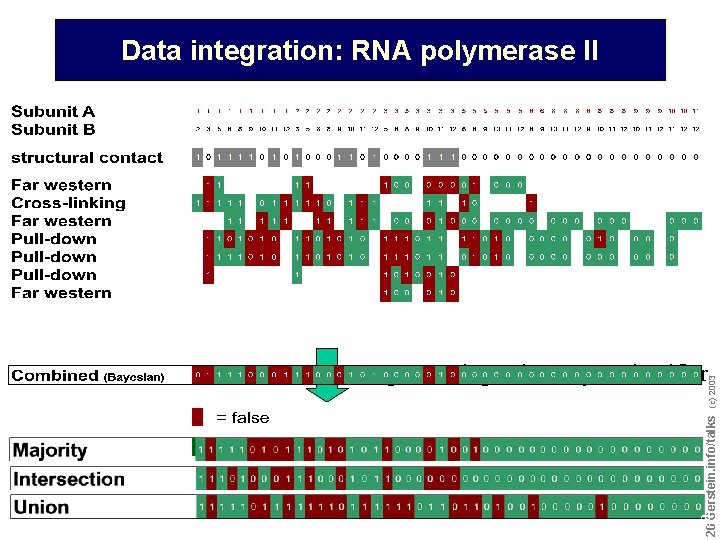
Example of data integration: RNA polymerase II Compare with Gold Std. structure: Edwards, Kus, Jansen, Greenbaum, Greenblatt, Gerstein, TIG (2002) Kornberg et al. , 2001 Do not reproduce without permission 1414 Gerstein. info/talks (c) 2003 Which subunits interact? -> protein-protein interaction experiments

Do not reproduce without permission 1515 Gerstein. info/talks (c) 2003 Data integration: RNA polymerase II

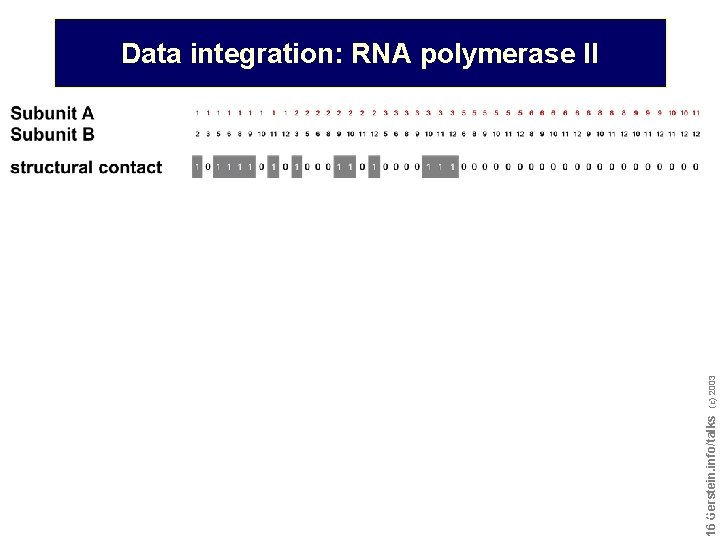
Do not reproduce without permission 1616 Gerstein. info/talks (c) 2003 Data integration: RNA polymerase II

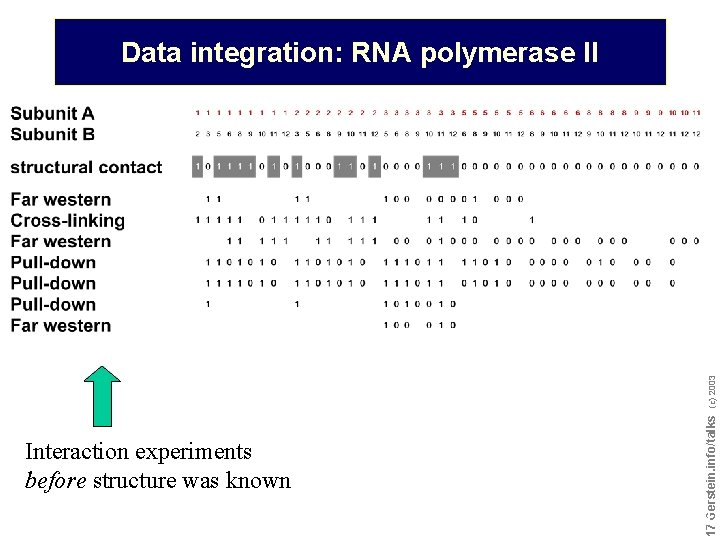
Interaction experiments before structure was known Do not reproduce without permission 1717 Gerstein. info/talks (c) 2003 Data integration: RNA polymerase II

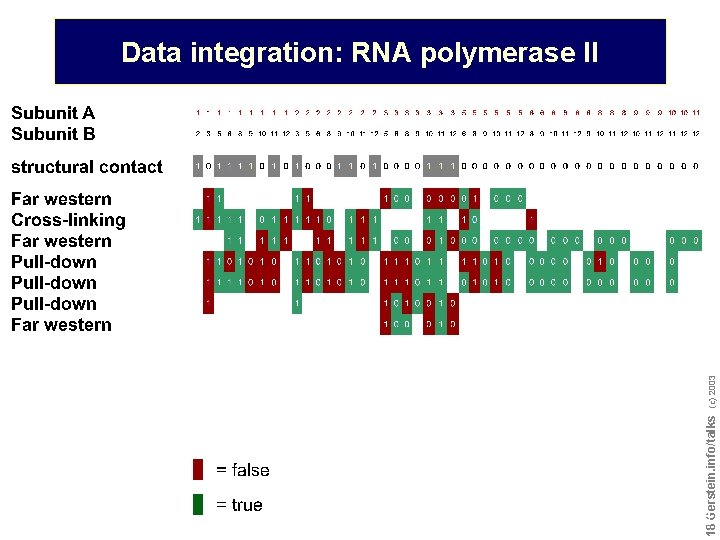
Do not reproduce without permission 1818 Gerstein. info/talks (c) 2003 Data integration: RNA polymerase II

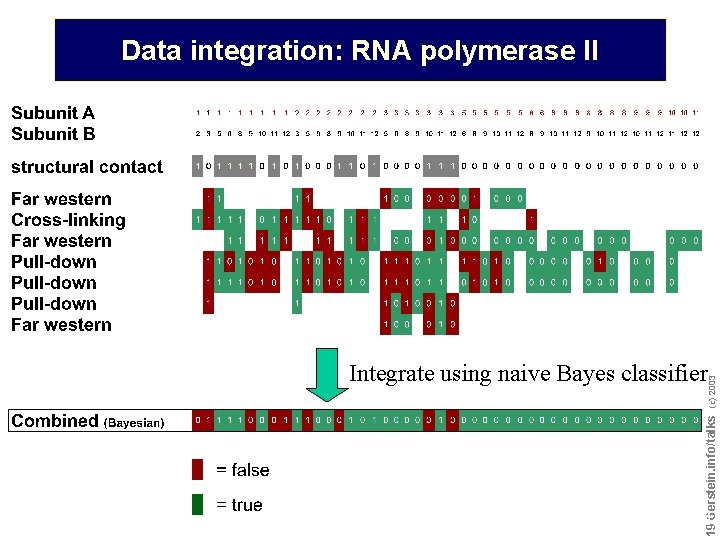
Data integration: RNA polymerase II Do not reproduce without permission 1919 Gerstein. info/talks (c) 2003 Integrate using naive Bayes classifier

Data integration: RNA polymerase II Do not reproduce without permission 2020 Gerstein. info/talks (c) 2003 Integrate using naive Bayes classifier

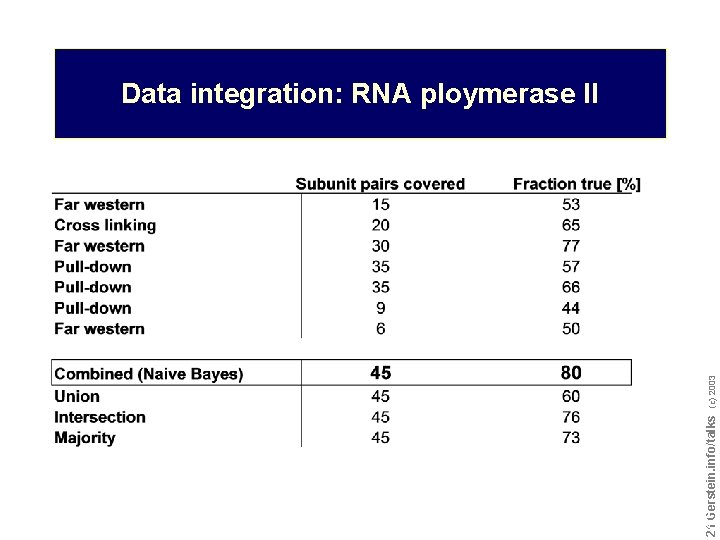
Do not reproduce without permission 2121 Gerstein. info/talks (c) 2003 Data integration: RNA ploymerase II

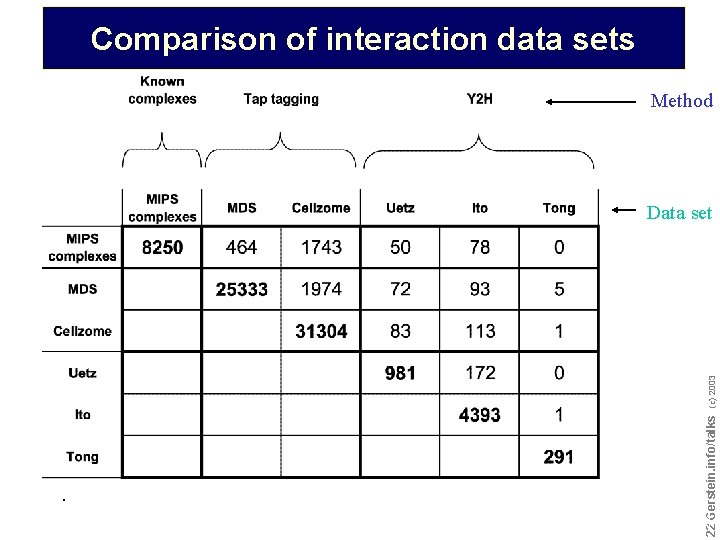
Comparison of interaction data sets Method . Do not reproduce without permission 2222 Gerstein. info/talks (c) 2003 Data set

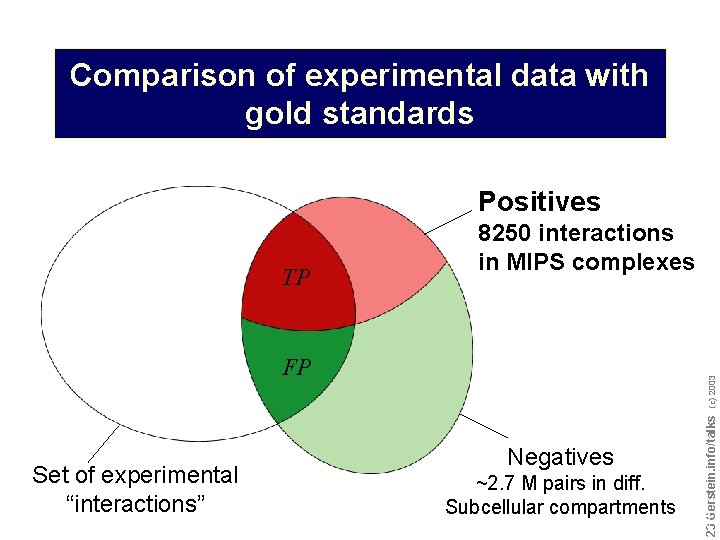
Comparison of experimental data with gold standards Positives Set of experimental “interactions” (c) 2003 FP Negatives ~2. 7 M pairs in diff. Subcellular compartments Do not reproduce without permission 2323 Gerstein. info/talks TP 8250 interactions in MIPS complexes

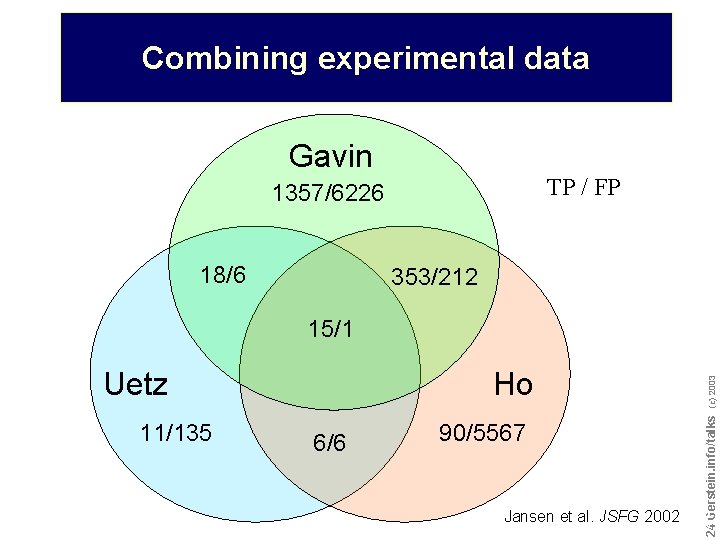
Combining experimental data Gavin TP / FP 1357/6226 18/6 353/212 15/1 11/135 6/6 (c) 2003 Ho 90/5567 Jansen et al. JSFG 2002 Do not reproduce without permission 2424 Gerstein. info/talks Uetz

Do not reproduce without permission 2525 Gerstein. info/talks (c) 2003 Integrating Structural Complexes with Non-interaction Genomic Information: Using them to Interpret Gene Expression data

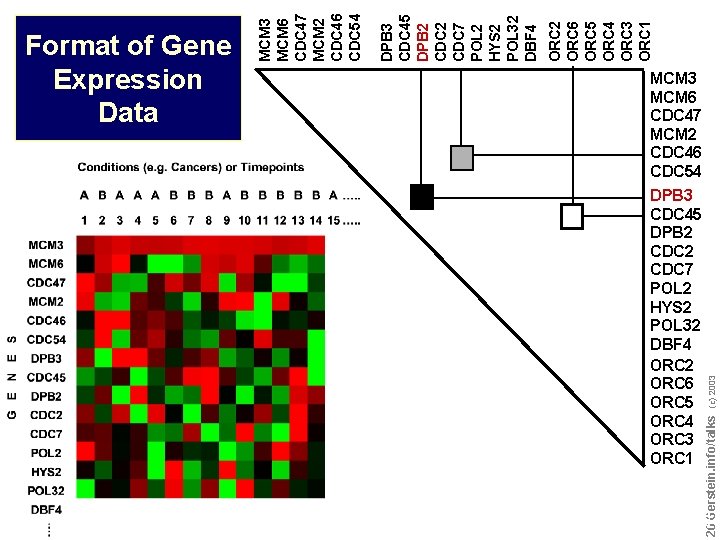
Do not reproduce without permission 2626 Gerstein. info/talks MCM 3 MCM 6 CDC 47 MCM 2 CDC 46 CDC 54 DPB 3 CDC 45 DPB 2 CDC 7 POL 2 HYS 2 POL 32 DBF 4 ORC 2 ORC 6 ORC 5 ORC 4 ORC 3 ORC 1 (c) 2003 ORC 2 ORC 6 ORC 5 ORC 4 ORC 3 ORC 1 DPB 3 CDC 45 DPB 2 CDC 7 POL 2 HYS 2 POL 32 DBF 4 MCM 3 MCM 6 CDC 47 MCM 2 CDC 46 CDC 54 Format of Gene Expression Data

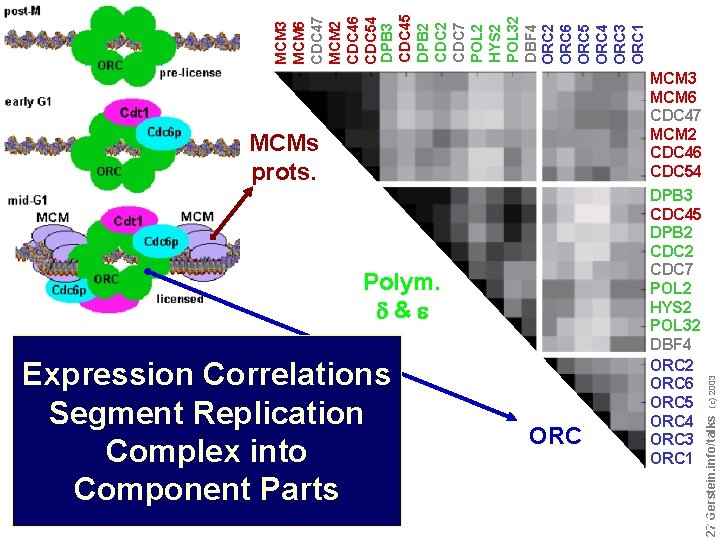
Expression Correlations Segment Replication Complex into Component Parts ORC Do not reproduce without permission 2727 Gerstein. info/talks Polym. d&e (c) 2003 MCM 6 CDC 47 MCM 2 CDC 46 CDC 54 DPB 3 CDC 45 DPB 2 CDC 7 POL 2 HYS 2 POL 32 DBF 4 ORC 2 ORC 6 ORC 5 ORC 4 ORC 3 ORC 1 MCMs prots. MCM 3 MCM 6 CDC 47 MCM 2 CDC 46 CDC 54 DPB 3 CDC 45 DPB 2 CDC 7 POL 2 HYS 2 POL 32 DBF 4 ORC 2 ORC 6 ORC 5 ORC 4 ORC 3 ORC 1

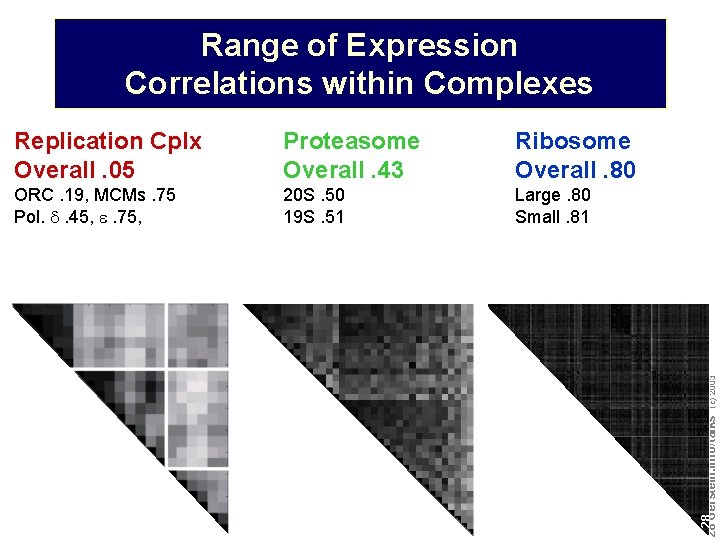
Range of Expression Correlations within Complexes Proteasome Overall. 43 Ribosome Overall. 80 ORC. 19, MCMs. 75 Pol. d. 45, e. 75, 20 S. 50 19 S. 51 Large. 80 Small. 81 Do not reproduce without permission 2828 Gerstein. info/talks (c) 2003 Replication Cplx Overall. 05

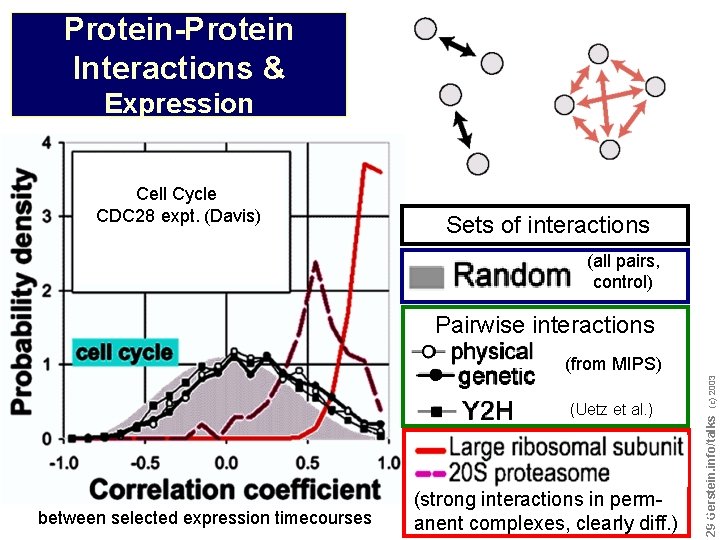
Protein-Protein Interactions & Expression Cell Cycle CDC 28 expt. (Davis) Sets of interactions (all pairs, control) Pairwise interactions between selected expression timecourses (strong interactions in permanent complexes, clearly diff. ) Do not reproduce without permission 2929 Gerstein. info/talks (Uetz et al. ) (c) 2003 (from MIPS)

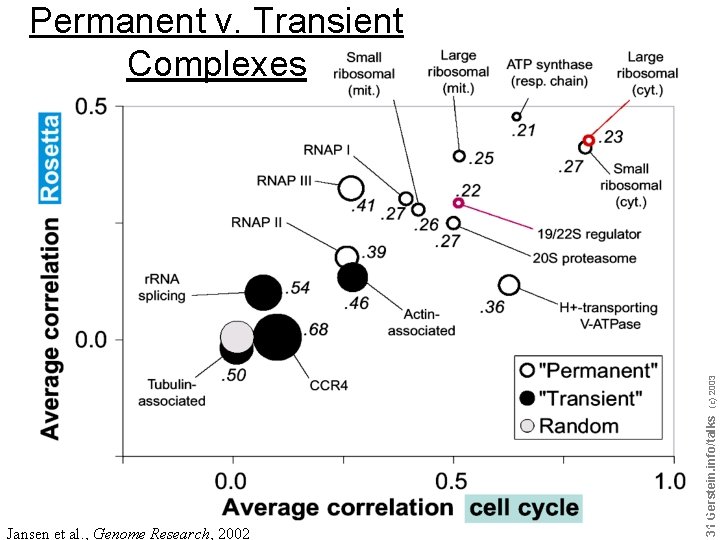
Jansen et al. , Genome Research, 2002 Do not reproduce without permission 3131 Gerstein. info/talks (c) 2003 Permanent v. Transient Complexes

Do not reproduce without permission 3333 Gerstein. info/talks (c) 2003 Genome-wide prediction of protein complexes based on both highthroughput interaction data and noninteraction, genomic information

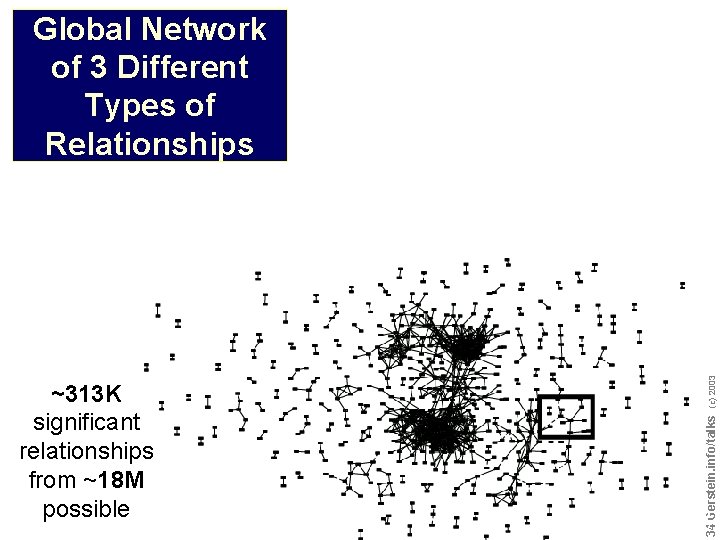
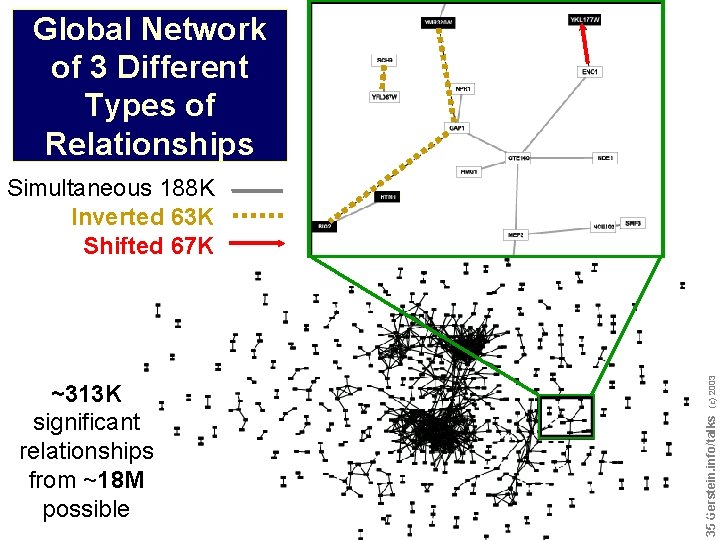
~313 K significant relationships from ~18 M possible Do not reproduce without permission 3434 Gerstein. info/talks (c) 2003 Global Network of 3 Different Types of Relationships

Global Network of 3 Different Types of Relationships ~313 K significant relationships from ~18 M possible Do not reproduce without permission 3535 Gerstein. info/talks (c) 2003 Simultaneous 188 K Inverted 63 K Shifted 67 K

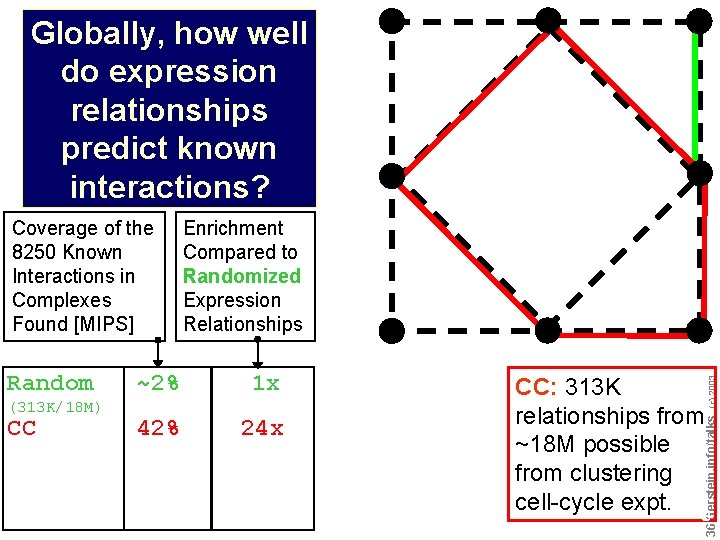
Globally, how well do expression relationships predict known interactions? (313 K/18 M) CC ~2% 1 x 42% 24 x CC: 313 K relationships from ~18 M possible from clustering cell-cycle expt. Do not reproduce without permission (c) 2003 Random Enrichment Compared to Randomized Expression Relationships 3636 Gerstein. info/talks Coverage of the 8250 Known Interactions in Complexes Found [MIPS]

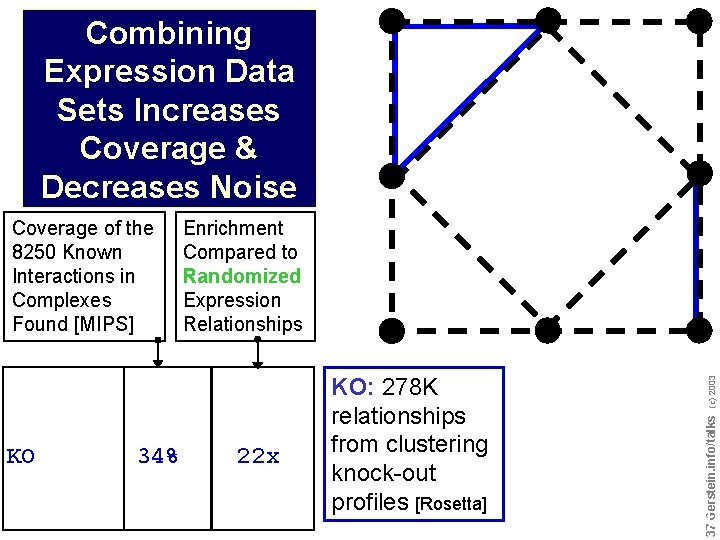
Combining Expression Data Sets Increases Coverage & Decreases Noise KO 34% Enrichment Compared to Randomized Expression Relationships 22 x (c) 2003 KO: 278 K relationships from clustering knock-out profiles [Rosetta] Do not reproduce without permission 3737 Gerstein. info/talks Coverage of the 8250 Known Interactions in Complexes Found [MIPS]

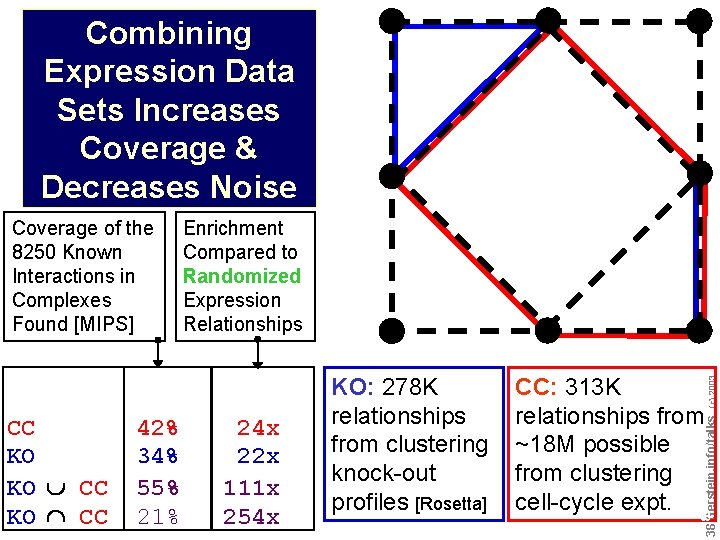
Combining Expression Data Sets Increases Coverage & Decreases Noise CC KO KO v CC KO ^ CC 42% 34% 55% 21% 24 x 22 x 111 x 254 x KO: 278 K relationships from clustering knock-out profiles [Rosetta] CC: 313 K relationships from ~18 M possible from clustering cell-cycle expt. Do not reproduce without permission (c) 2003 Enrichment Compared to Randomized Expression Relationships 3838 Gerstein. info/talks Coverage of the 8250 Known Interactions in Complexes Found [MIPS]

Do not reproduce without permission 3939 Gerstein. info/talks 1. Interactions provide a systematic way of defining protein function on a genomic scale 2. Known complexes provide a benchmark to validate and integrate genome-wide interaction experiments, providing a more accurate interactome 3. Known complexes provide a focus for the intergration of (non-interaction) genomic information – e. g. expression data 4. Extrapolating from known complexes, one can predict protein complexes on a genome-scale via integrating experimental interactions and noninteraction information (combining #1 and #2) (c) 2003 Computational Proteomics of Complexes

Do not reproduce without permission 4040 Gerstein. info/talks • Developing an accurate interactome for the cell, from prediction and through integration of high-throughput information • Development of statistical approaches to combine and integrate information • Development of database technologies to store hetrogeneous and noisy genome-wide interaction datasets • A moderate number of structural complexes are very useful as gold standard data (c) 2003 For the Future

Protein complexes & Structural Genomics • A computational challenge following from the solution of the partslist à Given many monomeric structures produced by structural genomics, predict (or rationalize) the interactome through docking Do not reproduce without permission 4141 Gerstein. info/talks (c) 2003 • Maybe many structures will be only be solved as complexes….

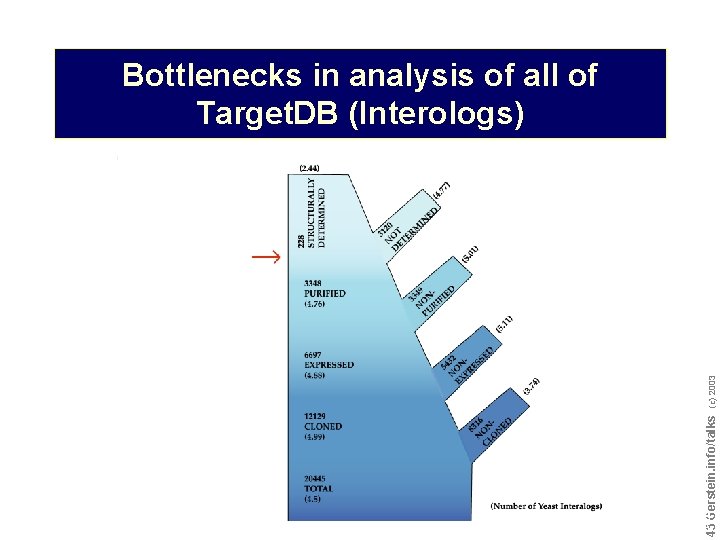
Do not reproduce without permission 4343 Gerstein. info/talks (c) 2003 Bottlenecks in analysis of all of Target. DB (Interologs)

Acknowledgements J Qian, R Jansen, A Drawid, C Wilson, D Greenbaum, C Goh, N Lan, H Hegyi, R Das, S Douglas, B Stenger J Lin, Y Kluger Collaborators A Edwards, B Kus, J Greenblatt NIH Gene. Census. org Do not reproduce without permission 4444 Gerstein. info/talks (c) 2003 M Snyder (A Kumar, H Zhu, …)