Proteomics Informatics Protein Characterization II Protein Interactions Week




- Slides: 60

Proteomics Informatics – Protein Characterization II: Protein Interactions (Week 12)

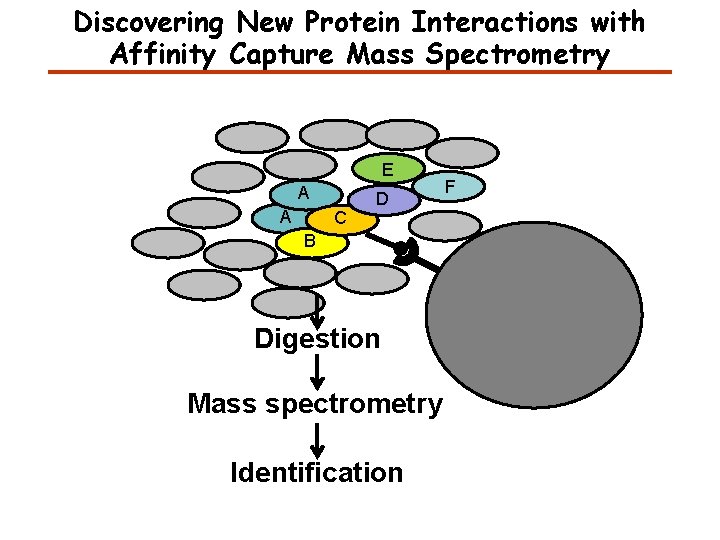
Discovering New Protein Interactions with Affinity Capture Mass Spectrometry E A A C D B Digestion Mass spectrometry Identification F

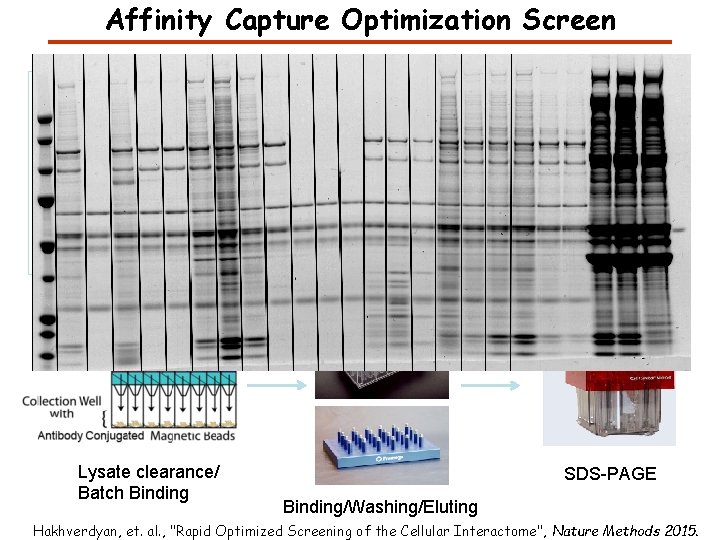
Affinity Capture Optimization Screen Cell extraction More / better quality interactions + Filtration Lysate clearance/ Batch Binding SDS-PAGE Binding/Washing/Eluting Hakhverdyan, et. al. , "Rapid Optimized Screening of the Cellular Interactome", Nature Methods 2015.

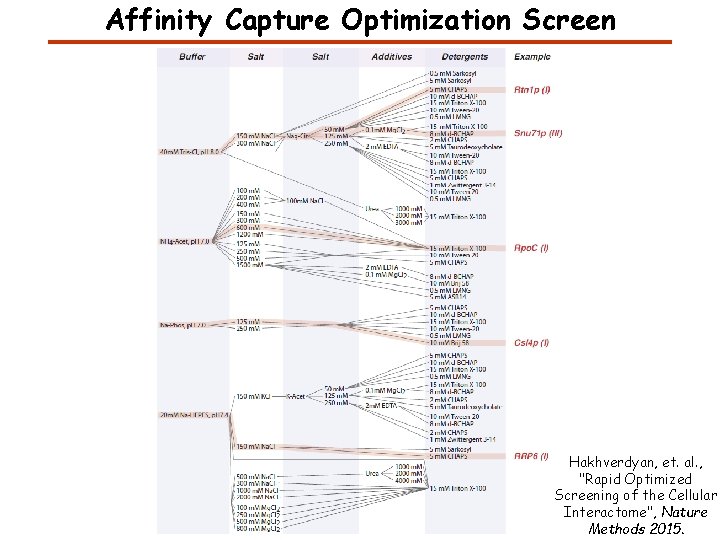
Affinity Capture Optimization Screen Hakhverdyan, et. al. , "Rapid Optimized Screening of the Cellular Interactome", Nature Methods 2015.

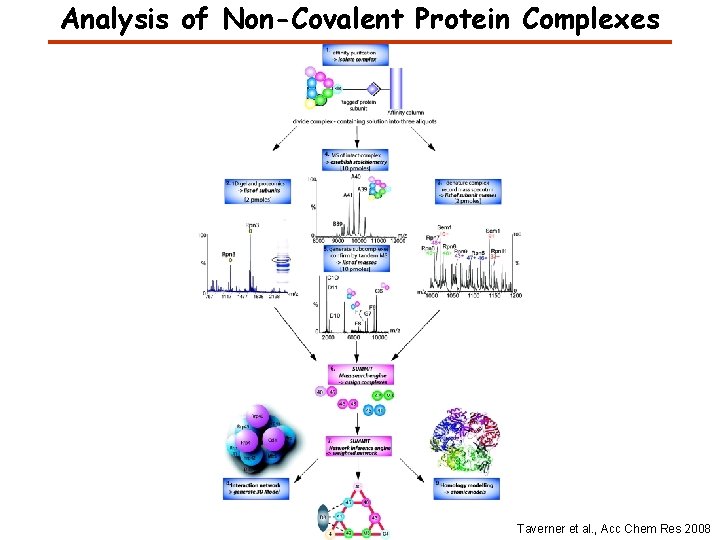
Analysis of Non-Covalent Protein Complexes Taverner et al. , Acc Chem Res 2008

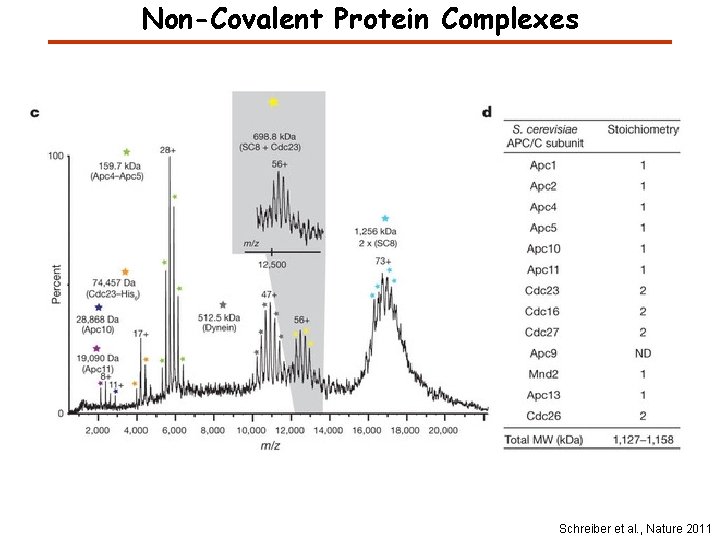
Non-Covalent Protein Complexes Schreiber et al. , Nature 2011

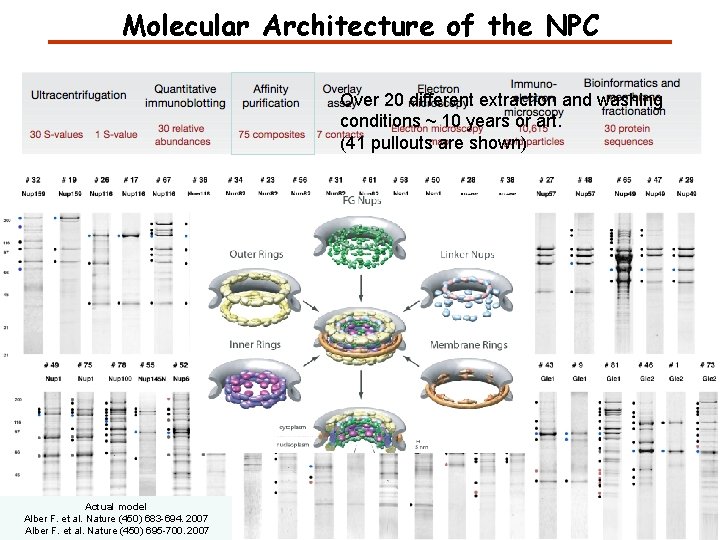
Molecular Architecture of the NPC Over 20 different extraction and washing conditions ~ 10 years or art. (41 pullouts are shown) Actual model Alber F. et al. Nature (450) 683 -694. 2007 Alber F. et al. Nature (450) 695 -700. 2007

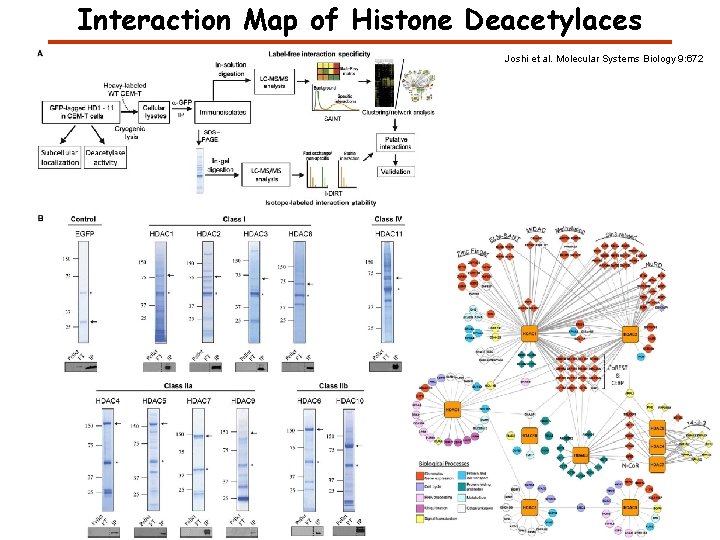
Interaction Map of Histone Deacetylaces Joshi et al. Molecular Systems Biology 9: 672

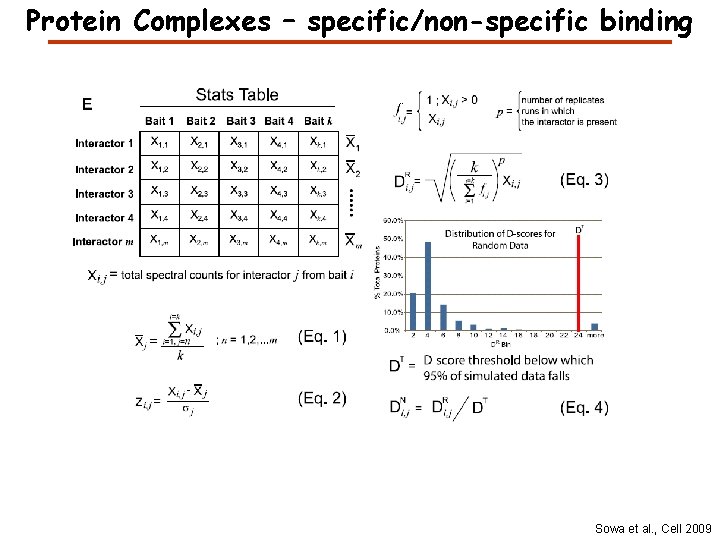
Protein Complexes – specific/non-specific binding Sowa et al. , Cell 2009

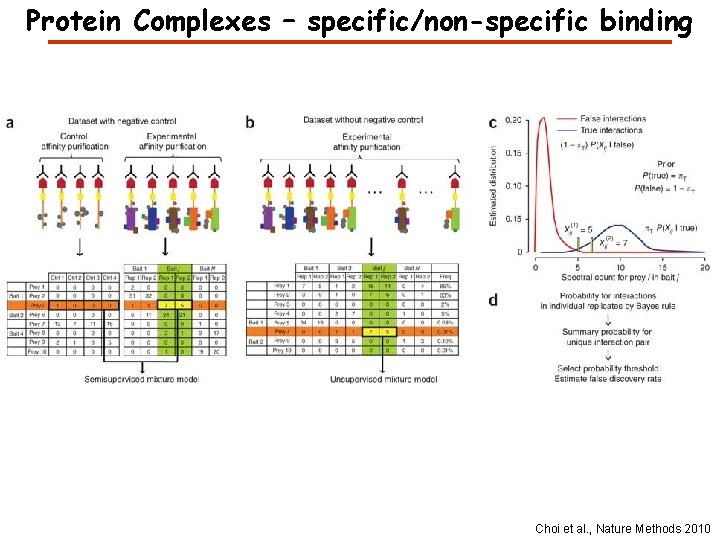
Protein Complexes – specific/non-specific binding Choi et al. , Nature Methods 2010

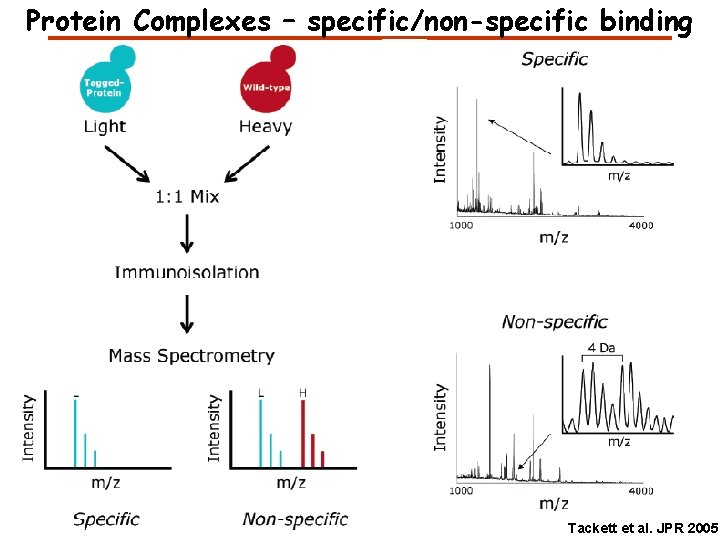
Protein Complexes – specific/non-specific binding Tackett et al. JPR 2005

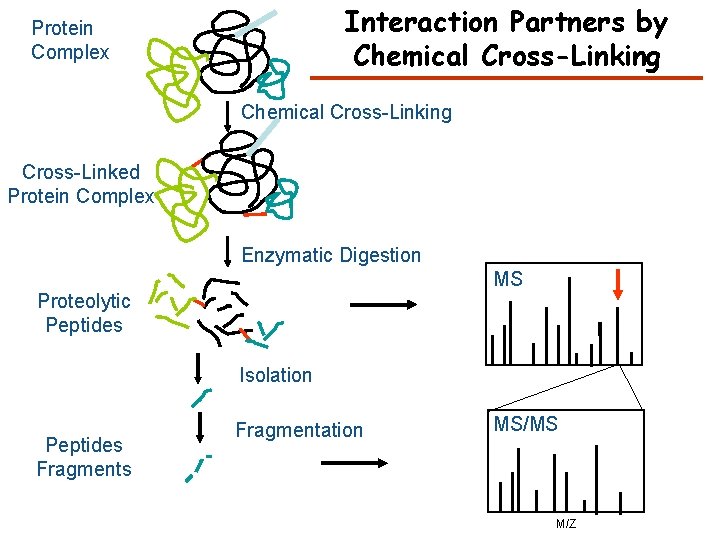
Interaction Partners by Chemical Cross-Linking Protein Complex Chemical Cross-Linking Cross-Linked Protein Complex Enzymatic Digestion MS Proteolytic Peptides Isolation Peptides Fragmentation MS/MS M/Z

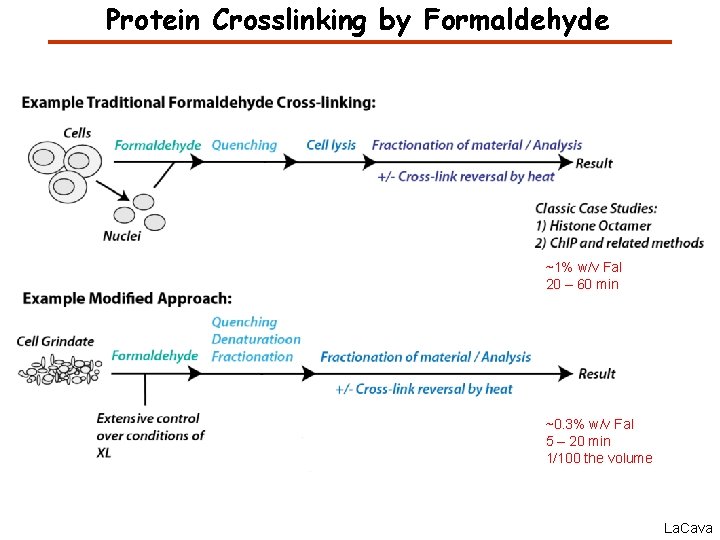
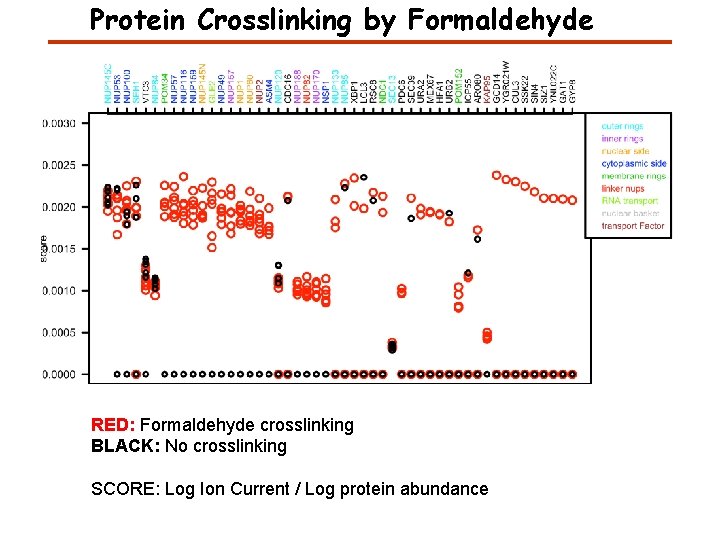
Protein Crosslinking by Formaldehyde ~1% w/v Fal 20 – 60 min ~0. 3% w/v Fal 5 – 20 min 1/100 the volume La. Cava

Protein Crosslinking by Formaldehyde RED: Formaldehyde crosslinking BLACK: No crosslinking SCORE: Log Ion Current / Log protein abundance

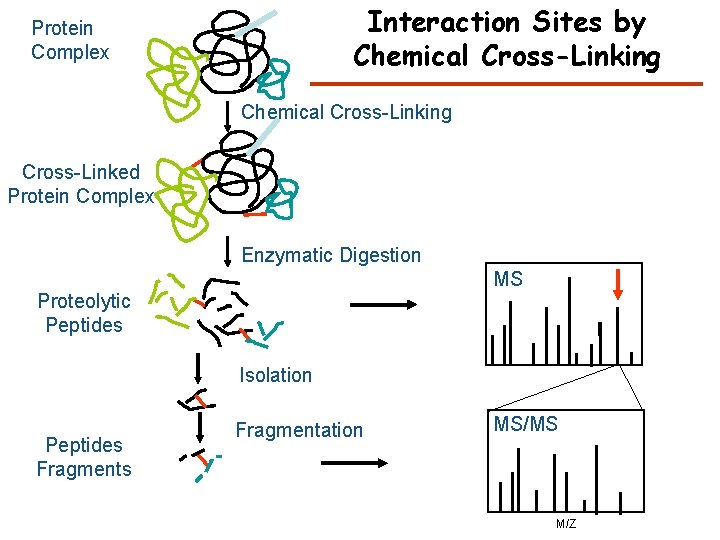
Interaction Sites by Chemical Cross-Linking Protein Complex Chemical Cross-Linking Cross-Linked Protein Complex Enzymatic Digestion MS Proteolytic Peptides Isolation Peptides Fragmentation MS/MS M/Z

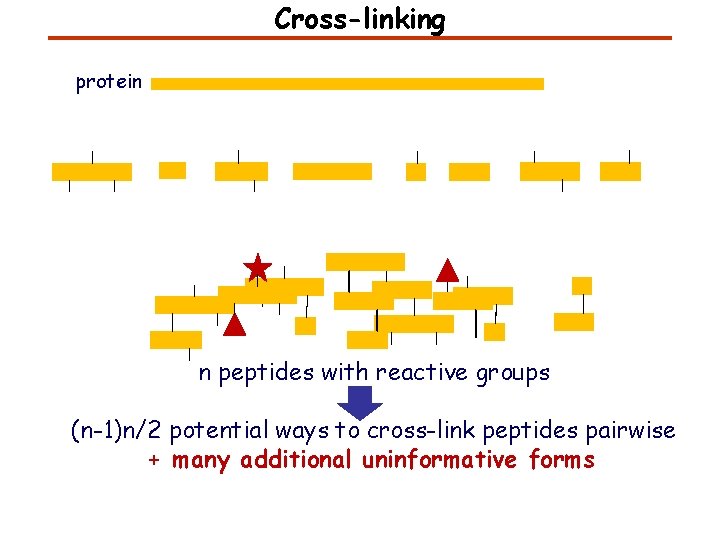
Cross-linking protein n peptides with reactive groups (n-1)n/2 potential ways to cross-link peptides pairwise + many additional uninformative forms Protein A + Ig. G heavy chain 990 possible peptide pairs Yeast NPC 106 possible peptide pairs

Cross-linking Mass spectrometers have a limited dynamic range and it therefore important to limit the number of possible reactions not to dilute the cross-linked peptides. For identification of a cross-linked peptide pair, both peptides have to be sufficiently long and required to give informative fragmentation. High mass accuracy MS/MS is recommended because the spectrum will be a mixture of fragment ions from two peptides. Because the cross-linked peptides are often large, CAD is not ideal, but instead ETD is recommended.

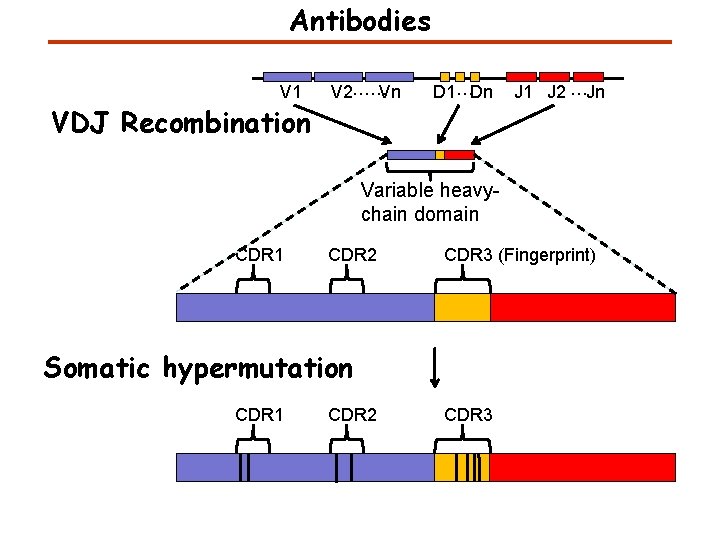
Antibodies V 1 VDJ Recombination V 2……Vn D 1…Dn J 1 J 2 …Jn Variable heavychain domain CDR 1 CDR 2 CDR 3 (Fingerprint) Somatic hypermutation CDR 1 CDR 2 CDR 3

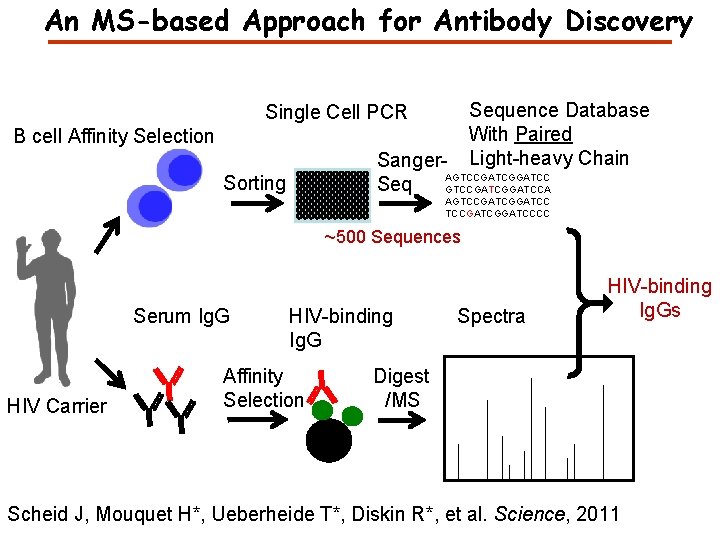
An MS-based Approach for Antibody Discovery Sequence Database With Paired Light-heavy Chain Single Cell PCR B cell Affinity Selection Sanger. AGTCCGATCGGATCC Seq GTCCGATCGGATCCA Sorting AGTCCGATCGGATCCCC ~500 Sequences Serum Ig. G HIV Carrier HIV-binding Ig. G Affinity Selection Spectra HIV-binding Ig. Gs Digest /MS Scheid J, Mouquet H*, Ueberheide T*, Diskin R*, et al. Science, 2011

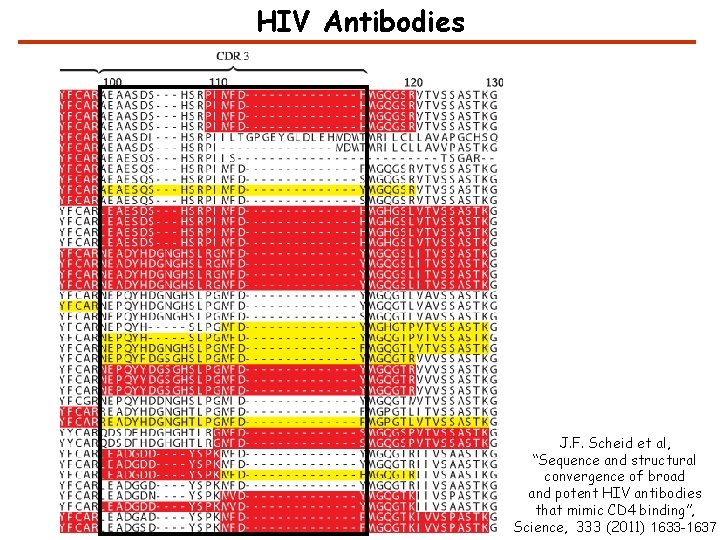
HIV Antibodies J. F. Scheid et al, “Sequence and structural convergence of broad and potent HIV antibodies that mimic CD 4 binding”, Science, 333 (2011) 1633 -1637

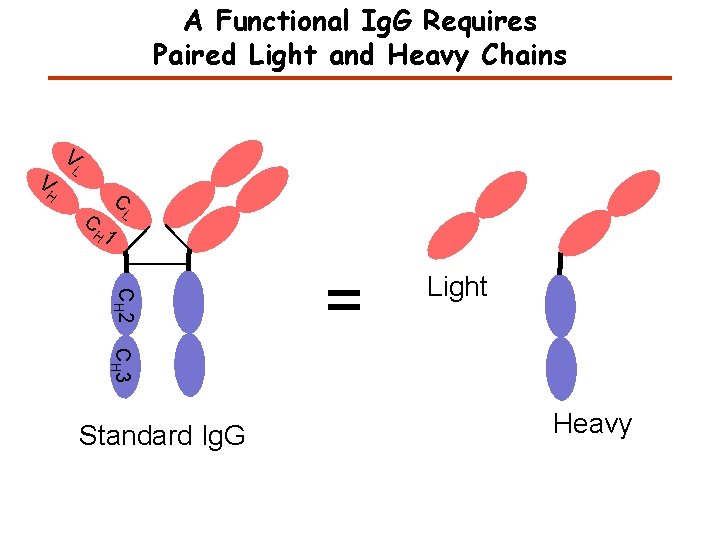
A Functional Ig. G Requires Paired Light and Heavy Chains V V L C H C L H 1 CH 2 = Light CH 3 Standard Ig. G Heavy

Cloning Single-Chain Llama Antibodies

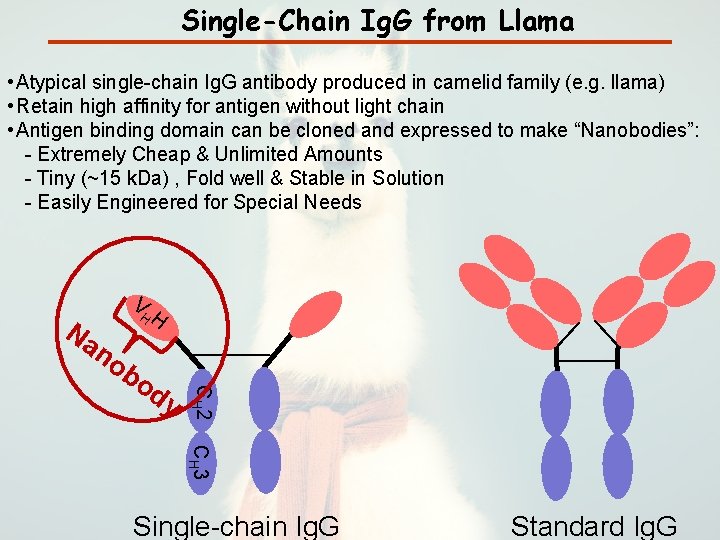
Single-Chain Ig. G from Llama • Atypical single-chain Ig. G antibody produced in camelid family (e. g. llama) • Retain high affinity for antigen without light chain • Antigen binding domain can be cloned and expressed to make “Nanobodies”: - Extremely Cheap & Unlimited Amounts - Tiny (~15 k. Da) , Fold well & Stable in Solution - Easily Engineered for Special Needs V Na HH no b CH 2 od y CH 3 Single-chain Ig. G Standard Ig. G

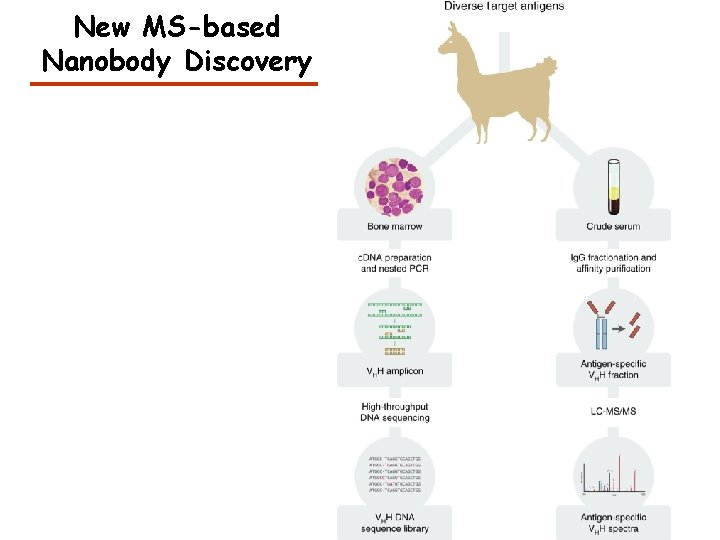
New MS-based Nanobody Discovery

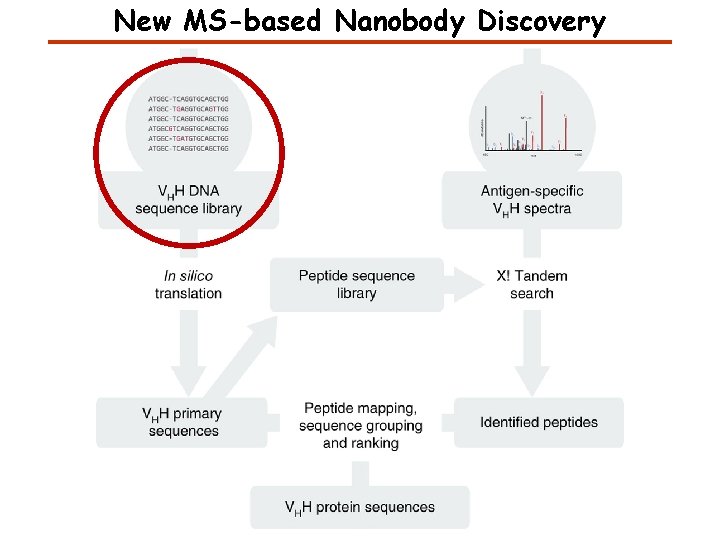
New MS-based Nanobody Discovery

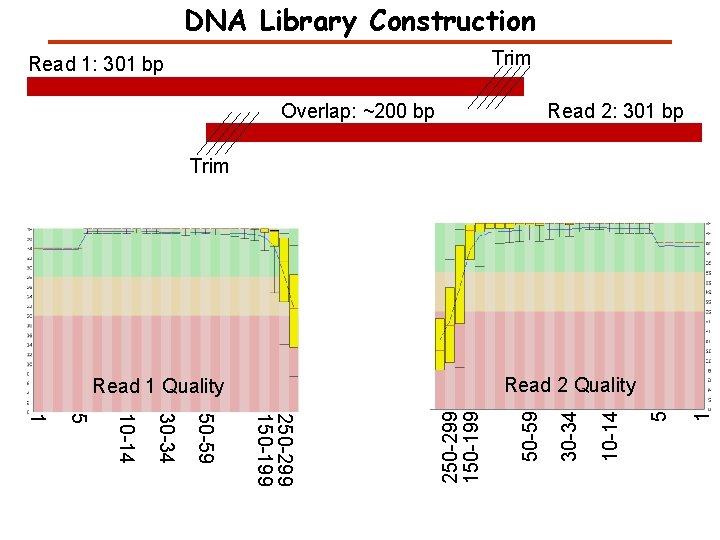
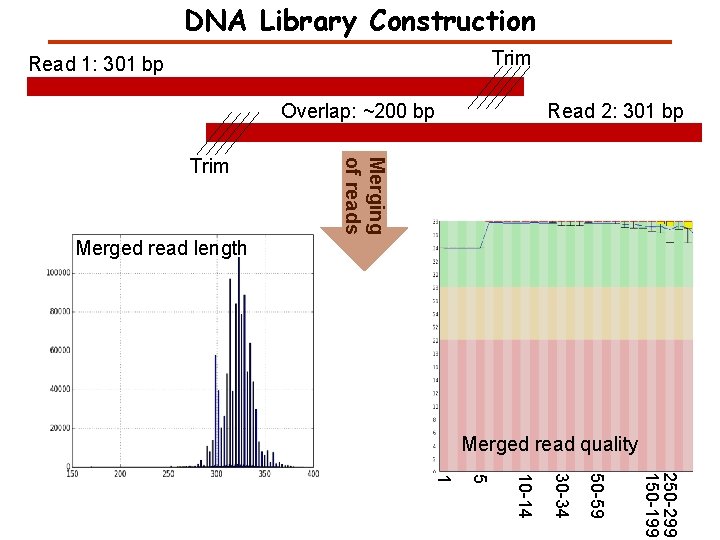
DNA Library Construction Trim Read 1: 301 bp Overlap: ~200 bp Read 2: 301 bp Trim 1 5 10 -14 30 -34 250 -299 150 -199 50 -59 30 -34 10 -14 5 1 50 -59 Read 2 Quality Read 1 Quality

DNA Library Construction Trim Read 1: 301 bp Overlap: ~200 bp Merging of reads Trim Read 2: 301 bp Merged read length Merged read quality 250 -299 150 -199 50 -59 30 -34 10 -14 5 1

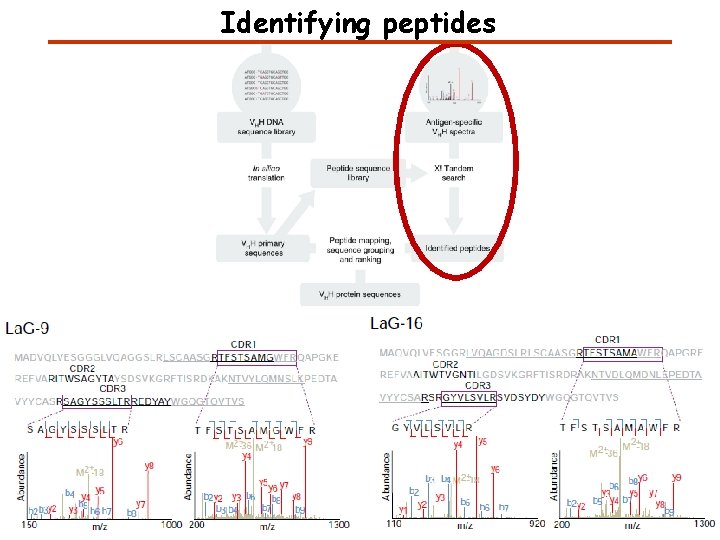
Identifying peptides

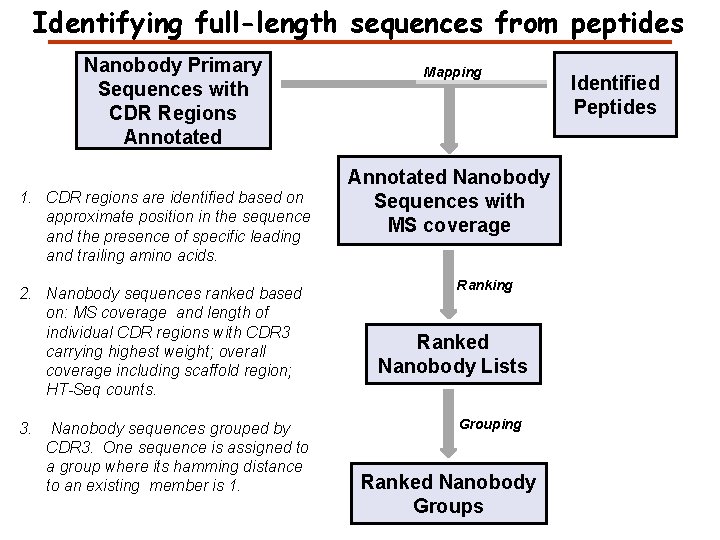
Identifying full-length sequences from peptides Nanobody Primary Sequences with CDR Regions Annotated 1. CDR regions are identified based on approximate position in the sequence and the presence of specific leading and trailing amino acids. 2. Nanobody sequences ranked based on: MS coverage and length of individual CDR regions with CDR 3 carrying highest weight; overall coverage including scaffold region; HT-Seq counts. 3. Nanobody sequences grouped by CDR 3. One sequence is assigned to a group where its hamming distance to an existing member is 1. Mapping Annotated Nanobody Sequences with MS coverage Ranking Ranked Nanobody Lists Grouping Ranked Nanobody Groups Identified Peptides

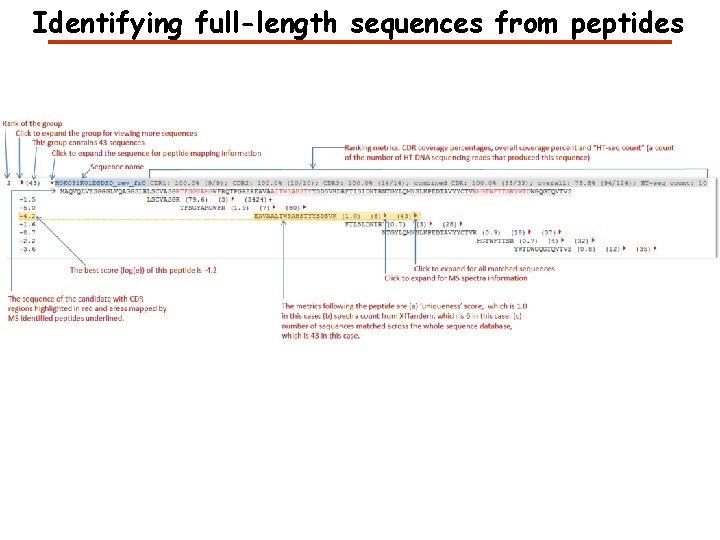
Identifying full-length sequences from peptides

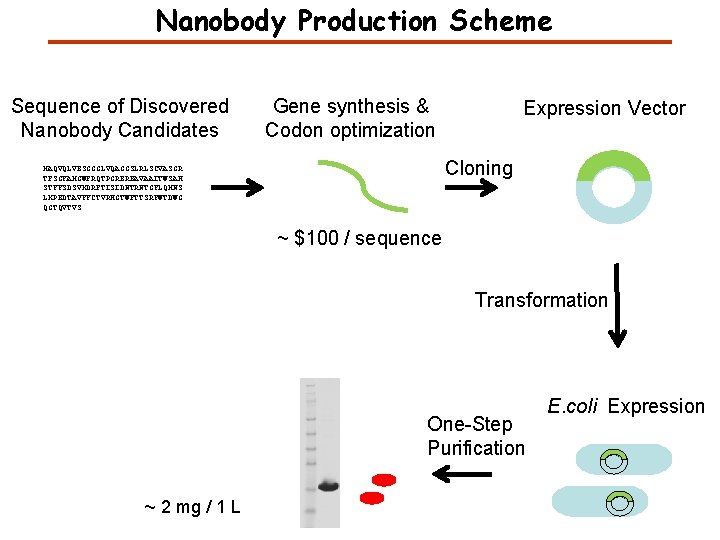
Nanobody Production Scheme Sequence of Discovered Nanobody Candidates Gene synthesis & Codon optimization Expression Vector Cloning MAQVQLVESGGGLVQAGGSLRLSCVASGR TFSGYAMGWFRQTPGREREAVAAITWSAH STYYSDSVKDRFTISIDNTRNTGYLQMNS LKPEDTAVYYCTVRHGTWFTTSRYWTDWG QGTQVTVS ~ $100 / sequence Transformation One-Step Purification ~ 2 mg / 1 L E. coli Expression

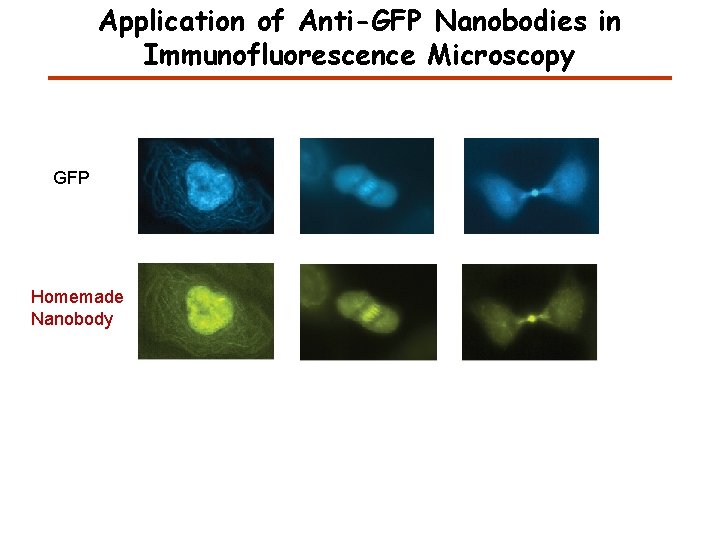
Application of Anti-GFP Nanobodies in Immunofluorescence Microscopy GFP Homemade Nanobody

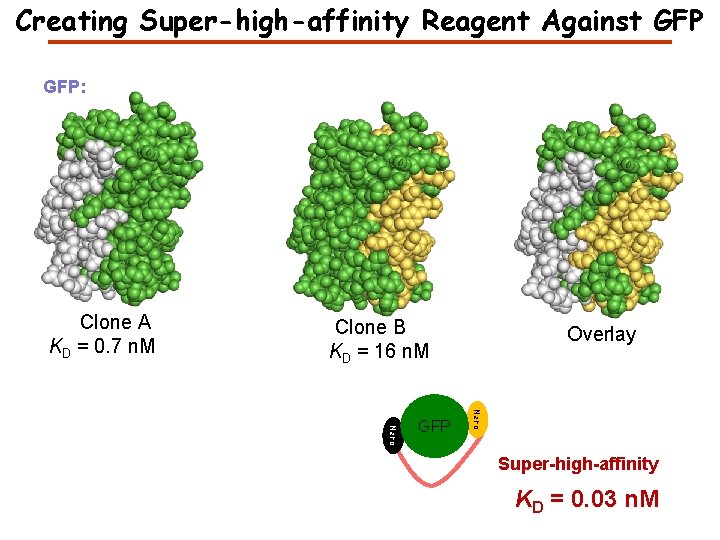
Creating Super-high-affinity Reagent Against GFP: Clone A KD = 0. 7 n. M Clone B KD = 16 n. M Nano GFP Overlay Super-high-affinity KD = 0. 03 n. M

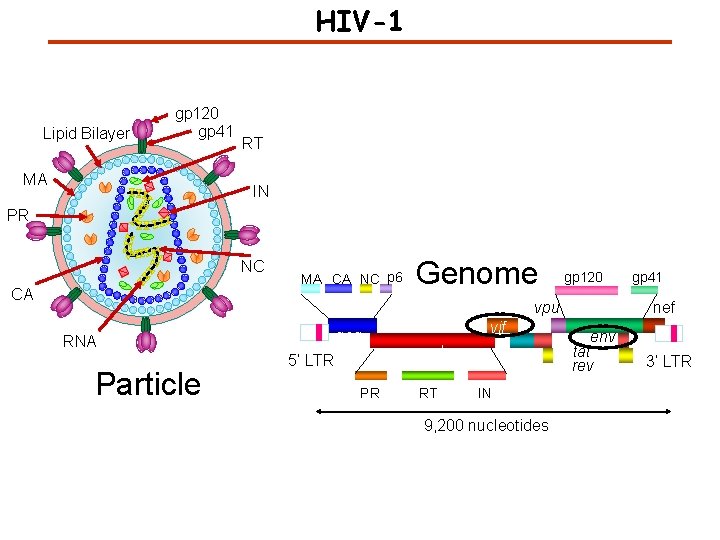
HIV-1 Lipid Bilayer gp 120 gp 41 MA RT IN PR NC CA MA CA NC p 6 Genome gp 120 vpu gag RNA Particle 5’ LTR PR vif pol RT vpr IN 9, 200 nucleotides gp 41 nef env tat rev 3’ LTR

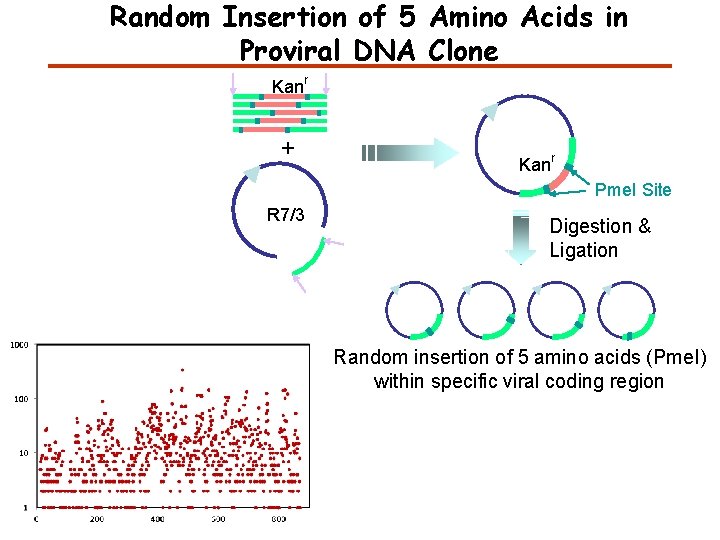
Random Insertion of 5 Amino Acids in Proviral DNA Clone Kan r + Kan r Pme. I Site R 7/3 Digestion & Ligation Random insertion of 5 amino acids (Pme. I) within specific viral coding region

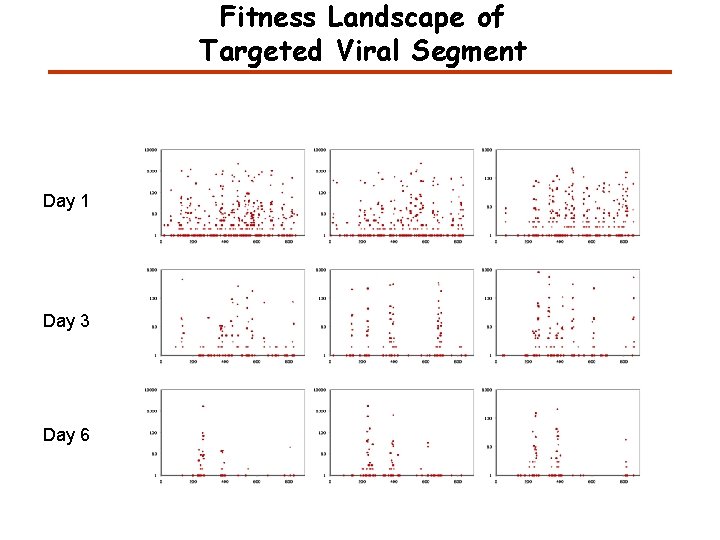
Fitness Landscape of Targeted Viral Segment Day 1 Day 3 Day 6

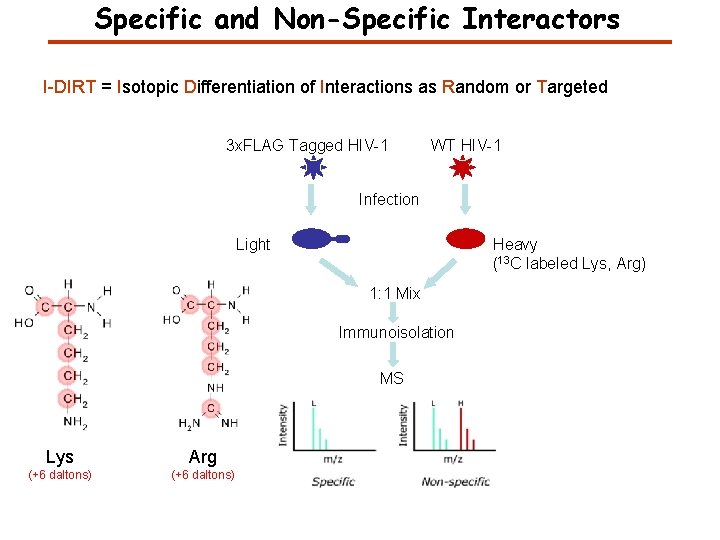
Specific and Non-Specific Interactors I-DIRT = Isotopic Differentiation of Interactions as Random or Targeted 3 x. FLAG Tagged HIV-1 WT HIV-1 Infection Light Heavy (13 C labeled Lys, Arg) 1: 1 Mix Immunoisolation MS Lys Arg (+6 daltons) Modified from Tackett AJ et al. , J Proteome Res. (2005) 4, 1752 -6.

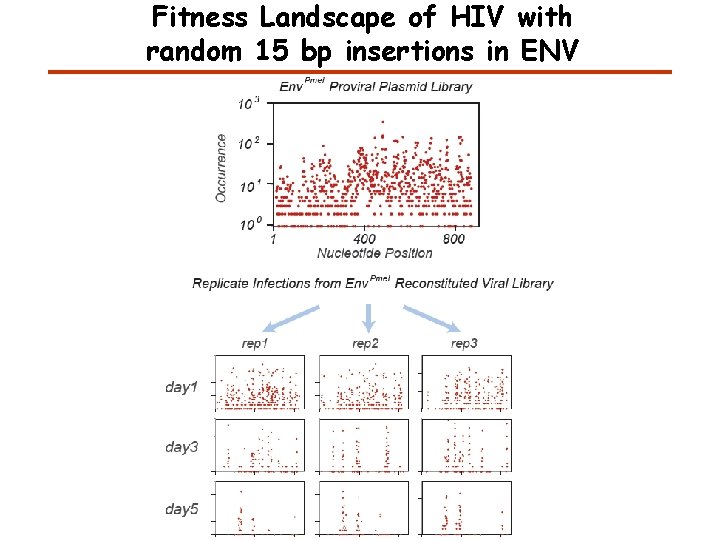
Fitness Landscape of HIV with random 15 bp insertions in ENV

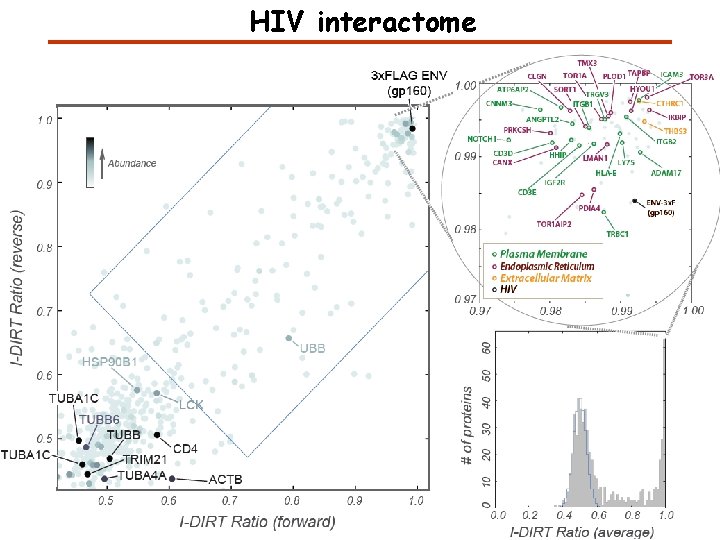
HIV interactome

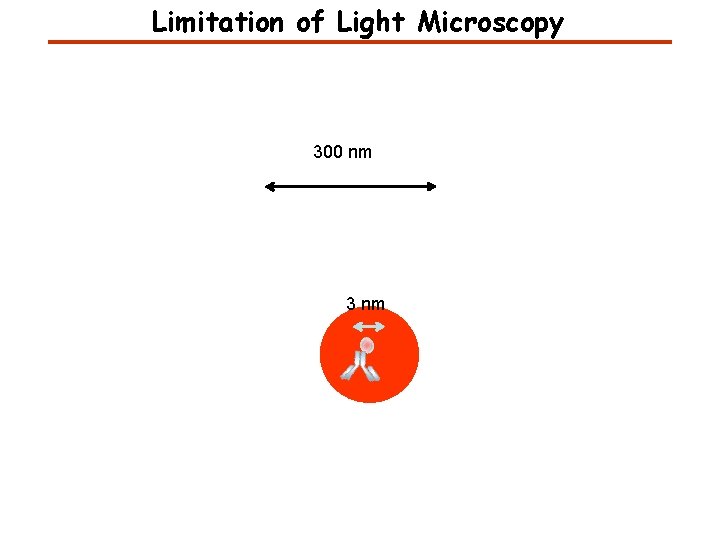
Limitation of Light Microscopy 300 nm 3 nm

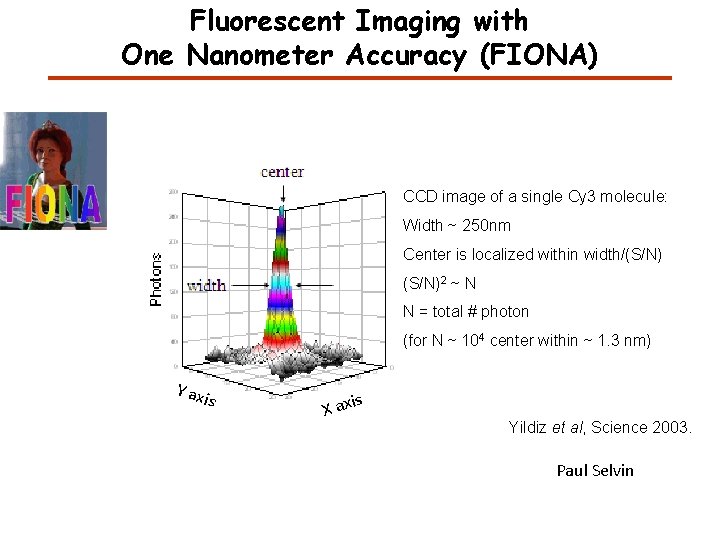
Fluorescent Imaging with One Nanometer Accuracy (FIONA) CCD image of a single Cy 3 molecule: Width ~ 250 nm Center is localized within width/(S/N)2 ~ N N = total # photon (for N ~ 104 center within ~ 1. 3 nm) Y ax is s X axi Yildiz et al, Science 2003. Paul Selvin

Limitation of Light Microscopy 3 nm 3 nm 3 nm

Limitation of Light Microscopy 3 nm 3 nm 3 nm

Limitation of Light Microscopy 3 nm 3 nm 3 nm

Limitation of Light Microscopy 3 nm 3 nm 3 nm

Limitation of Light Microscopy 20 nm 20 nm 20 nm

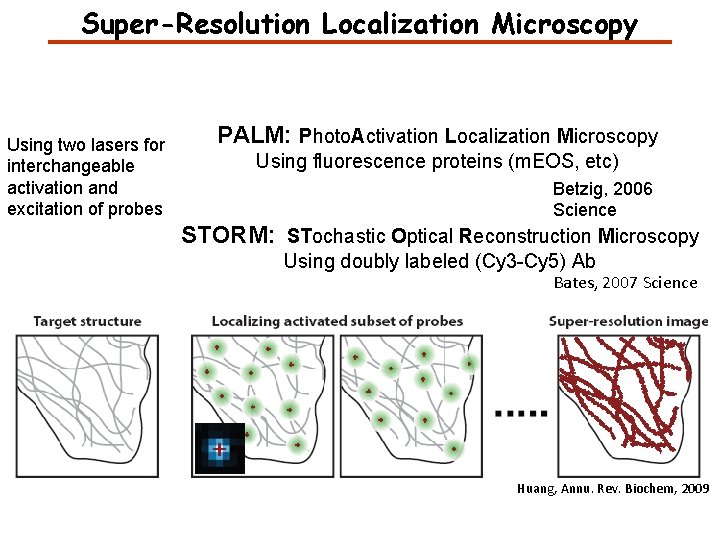
Super-Resolution Localization Microscopy Using two lasers for interchangeable activation and excitation of probes PALM: Photo. Activation Localization Microscopy Using fluorescence proteins (m. EOS, etc) Betzig, 2006 Science STORM: STochastic Optical Reconstruction Microscopy Using doubly labeled (Cy 3 -Cy 5) Ab Bates, 2007 Science Huang, Annu. Rev. Biochem, 2009

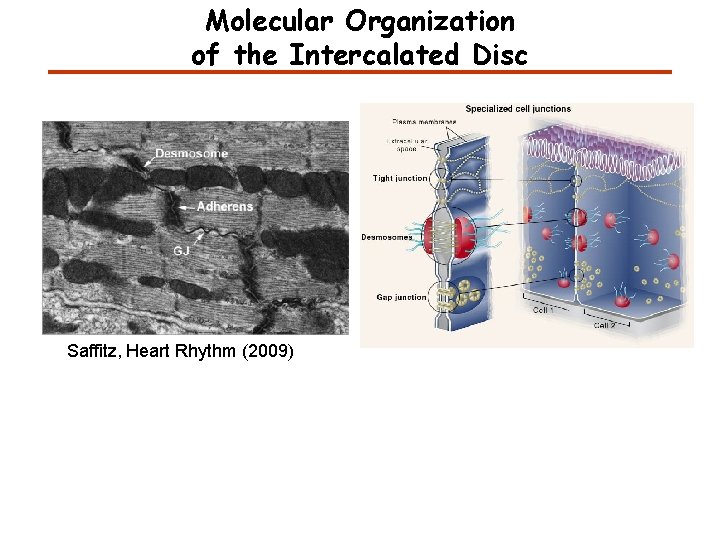
Molecular Organization of the Intercalated Disc Saffitz, Heart Rhythm (2009)

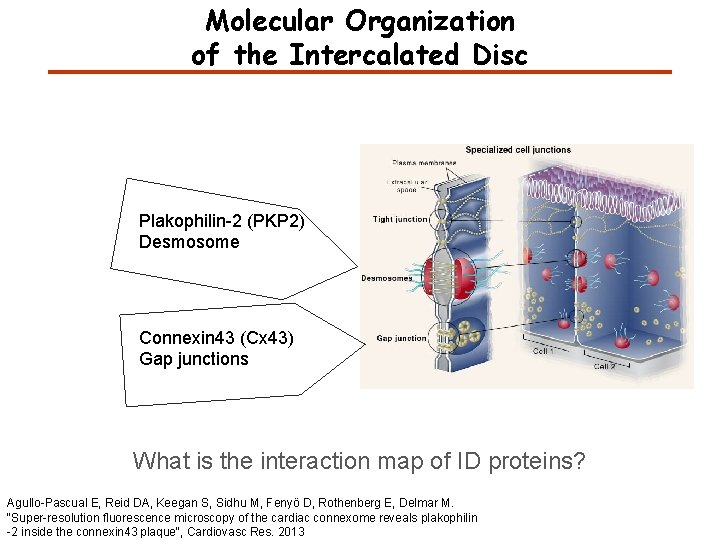
Molecular Organization of the Intercalated Disc Plakophilin-2 (PKP 2) Desmosome Connexin 43 (Cx 43) Gap junctions What is the interaction map of ID proteins? Agullo-Pascual E, Reid DA, Keegan S, Sidhu M, Fenyö D, Rothenberg E, Delmar M. "Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin -2 inside the connexin 43 plaque“, Cardiovasc Res. 2013

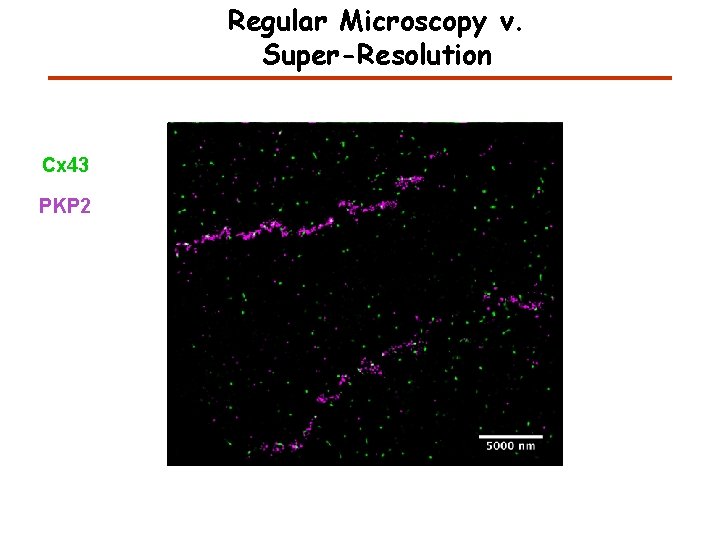
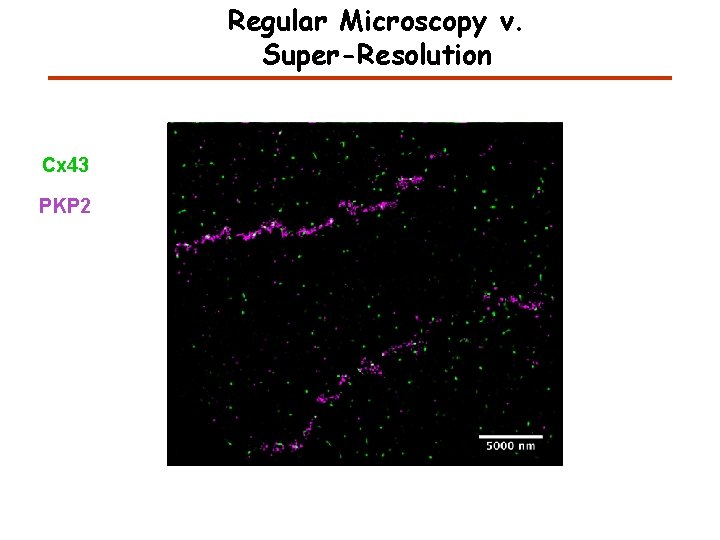
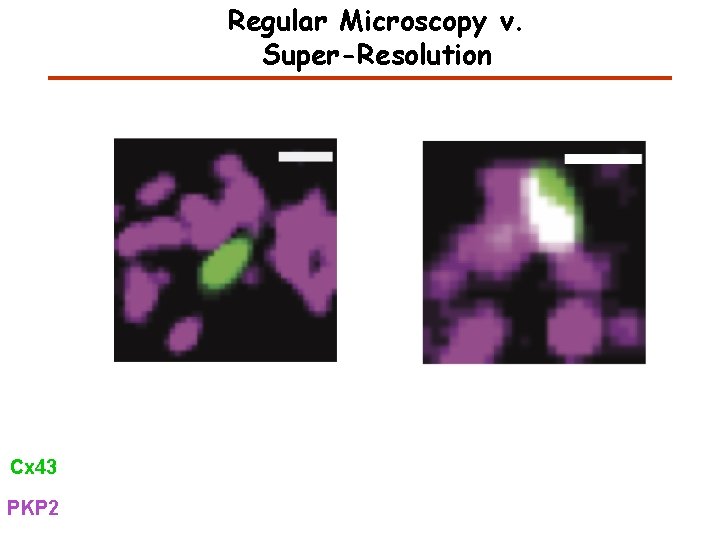
Regular Microscopy v. Super-Resolution Cx 43 PKP 2

Regular Microscopy v. Super-Resolution Cx 43 PKP 2

Regular Microscopy v. Super-Resolution Cx 43 PKP 2

What Do We Mean by Colocalization?

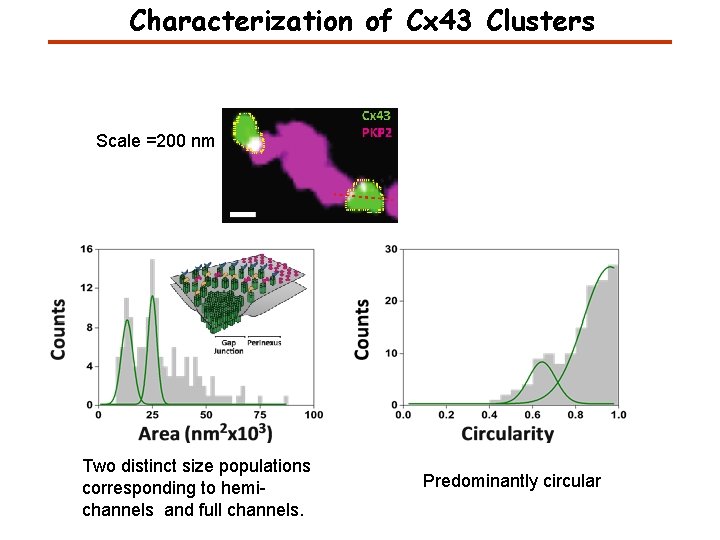
Characterization of Cx 43 Clusters Scale =200 nm Two distinct size populations corresponding to hemichannels and full channels. Predominantly circular

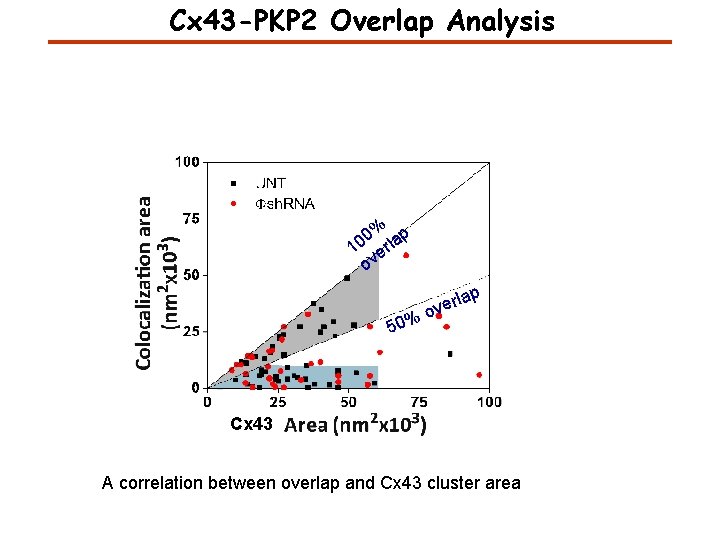
Cx 43 -PKP 2 Overlap Analysis % p 0 10 erla ov 50% lap r e ov Cx 43 A correlation between overlap and Cx 43 cluster area

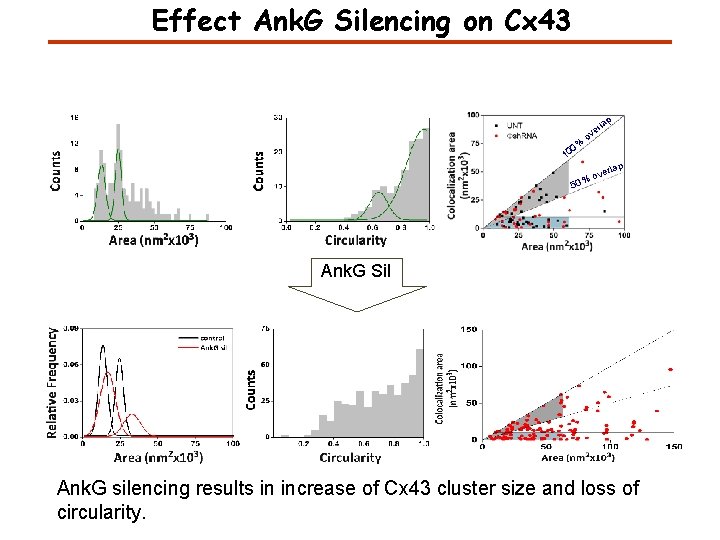
Effect Ank. G Silencing on Cx 43 lap % 0 10 r ve o rlap ve %o 50 Ank. G Sil Ank. G silencing results in increase of Cx 43 cluster size and loss of circularity.

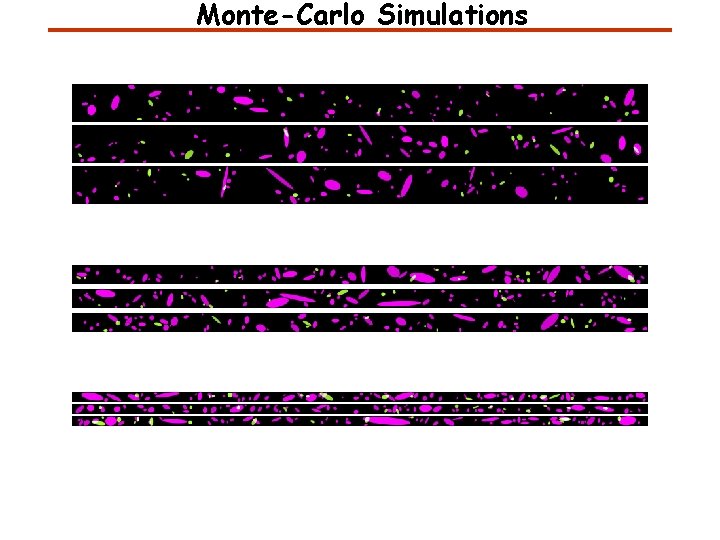
Monte-Carlo Simulations

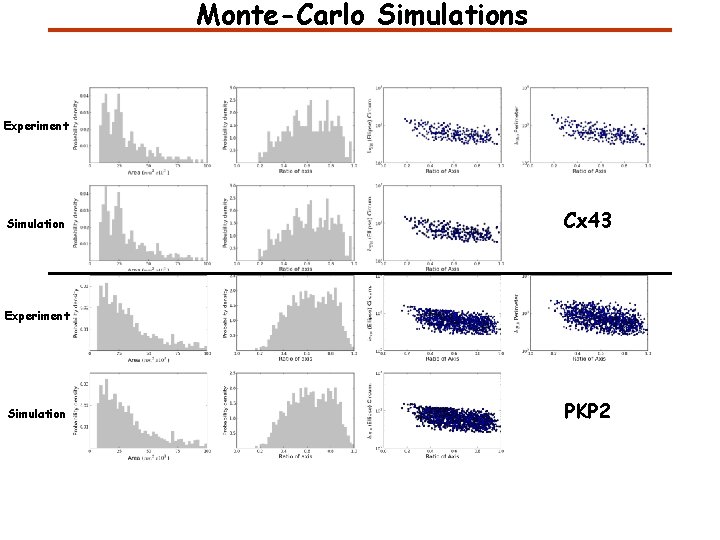
Monte-Carlo Simulations Experiment Simulation Cx 43 Experiment Simulation PKP 2

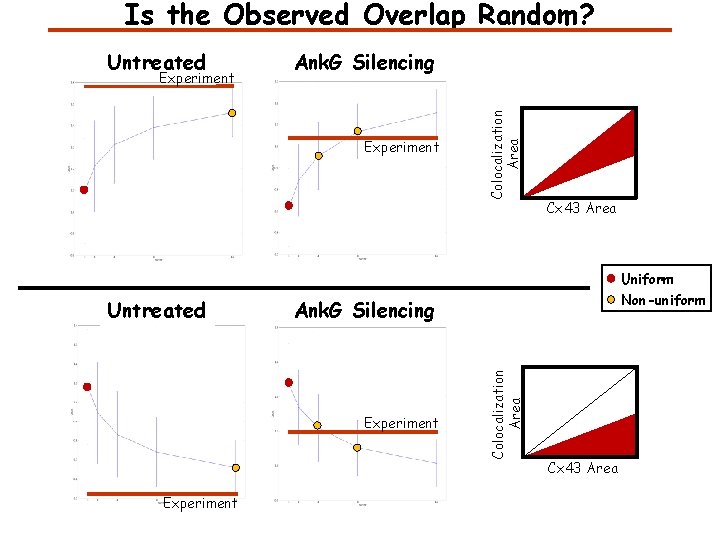
Is the Observed Overlap Random? Experiment Ank. G Silencing Experiment Colocalization Area Untreated Cx 43 Area Uniform Experiment Non-uniform Ank. G Silencing Colocalization Area Untreated Cx 43 Area

Proteomics Informatics – Protein Characterization II: Protein Interactions (Week 12)