Chemoinformatics tools for lead discovery Virtual screening The







- Slides: 26

Chemoinformatics tools for lead discovery

Virtual screening �The huge numbers of molecules available in public and in-house databases means that there is a requirement for tools to rank compounds in order of decreasing probability of activity �Range of methods available, varying in the sophistication and the amount of information that is available �Use of structure-based methods when an X-ray structure for the biological target is available �If this is not the case then must make use of information about (potential) ligands

Ligand-Based Methods �Similarity searching �Use when just a single bioactive reference structure is available � 3 D pharmacophore searching �Use when it has been possible to carry out a pharmacophore mapping exercise �Machine learning �Use when a fair number of both actives and inactives have been identified

Similarity Searching: I �Use of a similarity measure to quantify the resemblance between an active target, or reference, structure and each database structure �The similar property principle means that highranked structures are likely to have similar activities to that of the target structure �Similarity searching hence provides an obvious way of following-up on an initial active

Similarity searching: II �Many ways in which the similarity between two molecules can be computed �A similarity measure has two components �A structure representation �A similarity coefficient to compare two representations �Most operational systems use similarity measures based on 2 D fingerprints and the Tanimoto coefficient

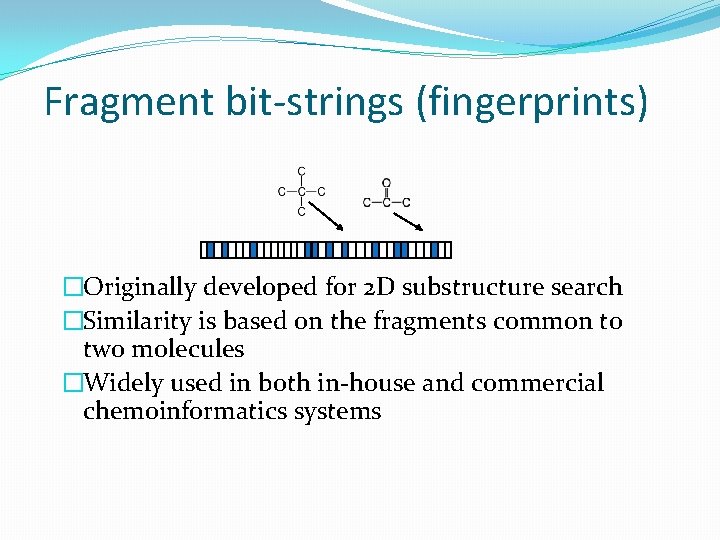
Fragment bit-strings (fingerprints) �Originally developed for 2 D substructure search �Similarity is based on the fragments common to two molecules �Widely used in both in-house and commercial chemoinformatics systems

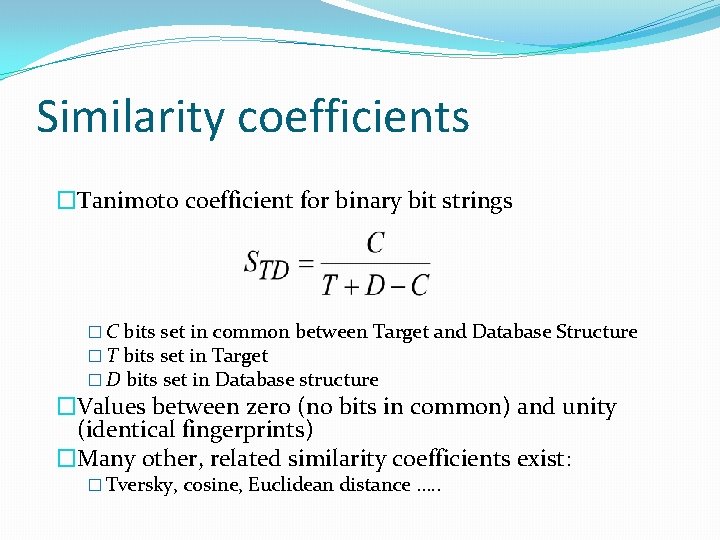
Similarity coefficients �Tanimoto coefficient for binary bit strings � C bits set in common between Target and Database Structure � T bits set in Target � D bits set in Database structure �Values between zero (no bits in common) and unity (identical fingerprints) �Many other, related similarity coefficients exist: � Tversky, cosine, Euclidean distance …. .

Combination of search techniques using data fusion: I �Tanimoto/fingerprint measures most common but many other types, e. g. , �Computed physicochemical properties � 3 D grid describing the molecular electrostatic potential �These reflect different molecular characteristics, so may enhance search performance by using more than one similarity measure �Data fusion or consensus scoring

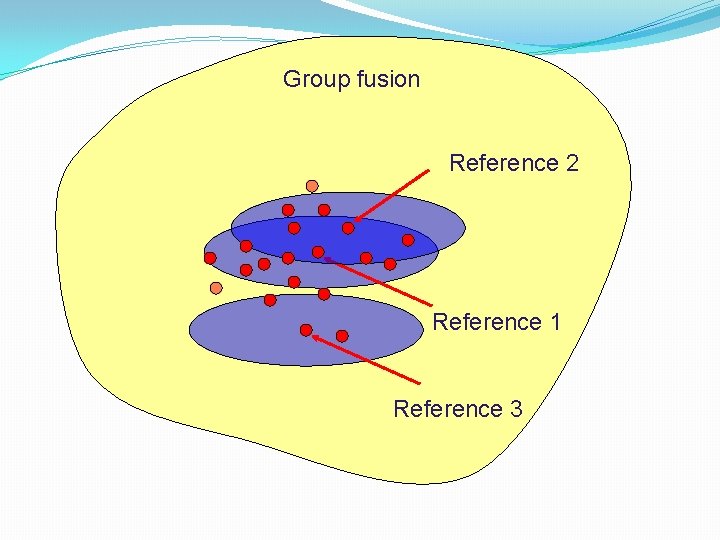
Combination of search techniques using data fusion: II �Combination of different rankings of the same sets of molecules �Two basic approaches • Generate rankings from the same molecule using different similarity measures (similarity fusion) • Generate rankings from different molecules using the same similarity measure but different molecules (group fusion)

Group fusion Reference 2 Reference 1 Reference 3

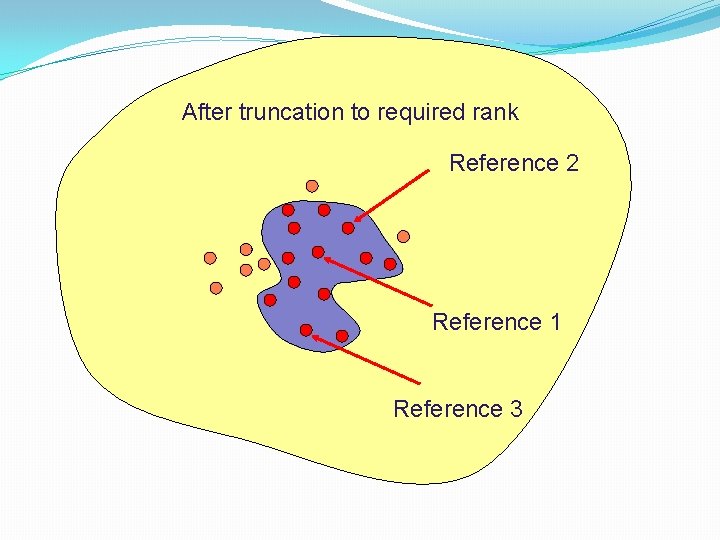
After truncation to required rank Reference 2 Reference 1 Reference 3

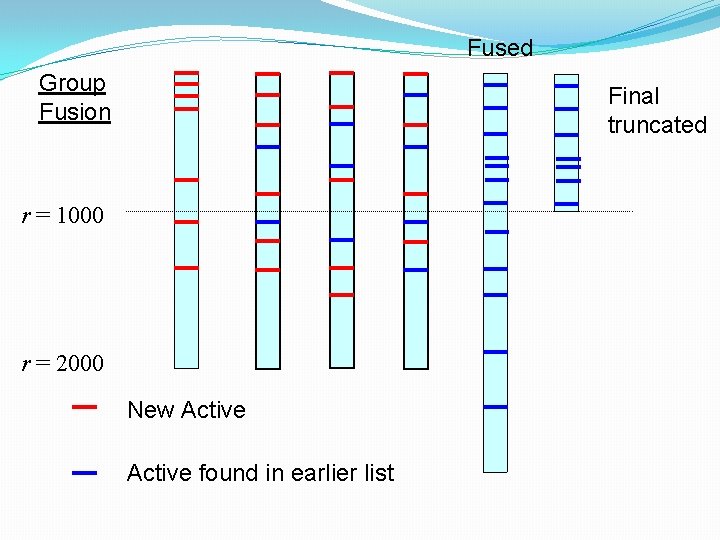
Fused Group Fusion Final truncated r = 1000 r = 2000 New Active found in earlier list

Group fusion rules �Useful performance increases, even with just 10 actives, as better coverage of structural space with multiple starting points �Improvement most obvious when searching for heterogeneous sets of active molecules �Best results obtained by • Fusing similarity coefficient values, rather than ranks • Re-ranking using the maximum of the similarity values associated with each molecule • Using the Tanimoto coefficient

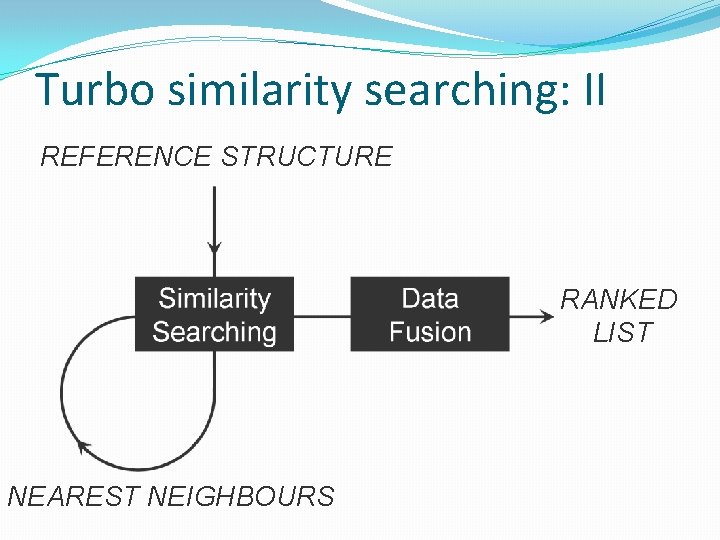
Turbo similarity searching: I �Similar property principle: nearest neighbours are likely to exhibit the same activity as the reference structure �Group fusion improves the identification of active compounds �Potential for further enhancements by group fusion of rankings from the reference structure and from its assumed active nearest neighbours

Turbo similarity searching: II REFERENCE STRUCTURE RANKED LIST NEAREST NEIGHBOURS

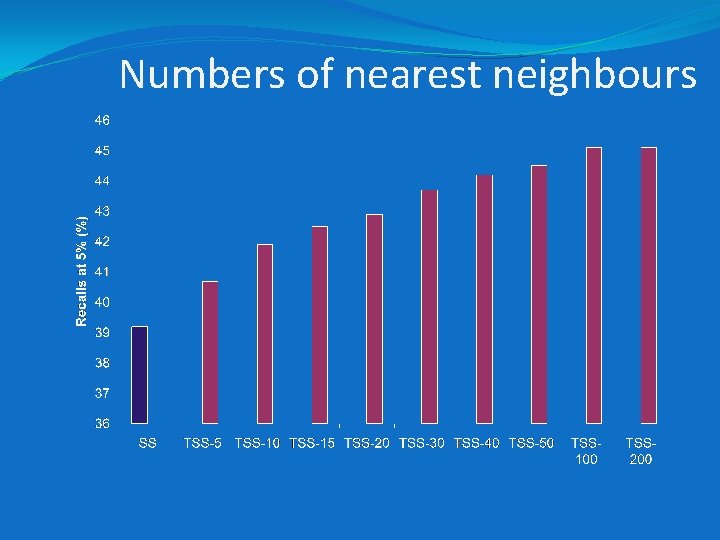
Experimental details �MDL Drug Data report (MDDR) dataset of 11 activity classes and 102 K structures • In all, 8294 actives in the 11 classes, with (turbo) similarity searches being carried out using each of these as the reference structure • ECFP_4 fingerprints/Tanimoto coefficient • MAX group fusion on similarity scores • Increasing numbers of nearest neighbours

Numbers of nearest neighbours

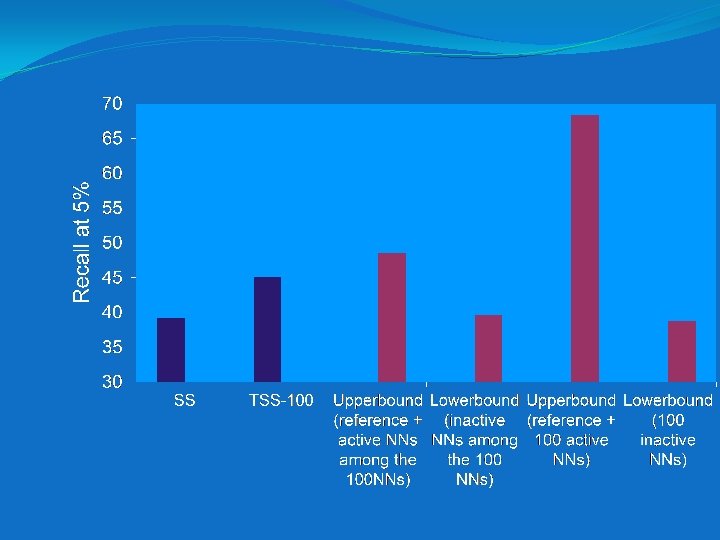
Upper and lower bound experiments

Rationale for upper bound results �The true actives in the set of assumed actives yield significant enhancements in performance �The true inactives in the set of assumed actives have little effect on performance �Taken together, the two groups of compounds yield the observed net enhancement

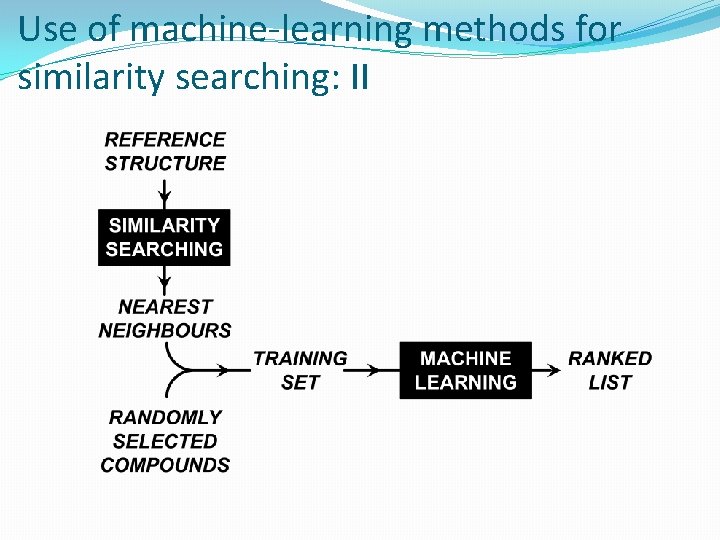
Use of machine-learning methods for similarity searching: I �Turbo similarity searching uses group fusion to enhance conventional similarity searching �Machine learning is a more powerful virtual screening tool than similarity searching �But requires a training-set containing known actives and inactives �Given an active reference structure, a training-set can be generated from �Using the k nearest neighbours of the reference structure as the actives �Using k randomly chosen, low-similarity compounds as the inactives

Use of machine-learning methods for similarity searching: II

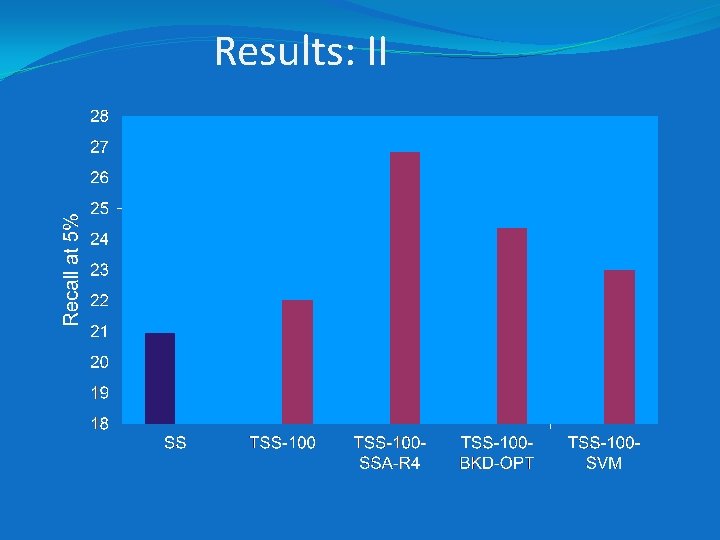
Results: I �Experiments with the MDDR dataset show that group fusion better than machine-learning methods when averaged over all of the classes �However, group fusion inferior for the most diverse datasets (as measured by the mean pairwise similarities) �Additional searches using 10 MDDR activity classes that are as structurally diverse as possible

Results: II

Conclusions: I �Fingerprint-based similarity searching using a known reference structure is long-established in chemoinformatics �When small numbers of actives are available, group fusion will enhance performance when the sought actives are structurally heterogeneous

Conclusions: II �Can also enhance conventional similarity search, even if there is just a single active, by assuming that the nearest neighbours are also active �Can be effected in two ways • Use of group fusion to combine similarity rankings (overall best approach) • Use of substructural analysis to compute fragment weights (best with highly heterogeneous sets of actives)

Soal untuk dipelajari �Tunjukkan peran khemoinformatik dalam QSAR �Data dan analisis dari khemoinformatik yang banyak digunakan dalam docking molekul �Indeks kemiripan (similarity index) banyak digunakan untuk mendapatkan informasi tentang senyawa baru yang memiliki aktivitas biologis tinggi. Jelaskan secara singkat sistem kerjanya �Dalam penemuan obat baru yang lebih potensial dari yang sudah dikenal, banyak memanfaatkan khemoinformatiks. Jelaskan dengan beberapa contoh. �Apa perbedaan penggunaan Khemoinformatiks dalam QSAR, molecular docking dan similarity searching?