ceramide Single molecule fluorescence clarifies the role of
- Slides: 38

ceramide Single molecule fluorescence clarifies the role of monosialoganglioside GM 1 and sphingomyelin in the in-membrane oligomerization of β-amyloid Sugar head Martin Hof http: //hof-fluorescence-group. weebly. com/

Lipid Topics • Size and character of ganglioside GM 1 nano-clusters • Sphingomyelin/cholesterol nanoheterogeneities; influence of GM 1 • In membrane oligomerisation of β-amyloid • • • Fluorescence Methods Monte Carlo simulations of Fluorescence Lifetime Imaging -Förster Resonance Energy Transfer Calibration free Fluorescence Correlation Spectroscopy: Z-scan FCS Fluorescence Correlation Cross Spectroscopy Šachl, Humpolíčková; Štefl, Johansson, Hof 2011, Biophys. J. , 101, L 60 -L 62. Amaro, Šachl, Jurkiewicz, Coutinho, Prieto, Hof Biophysical Journal. 107, 12 (2014), 2751 -2760.

Lipid Topics • Size and character of ganglioside GM 1 nano-clusters • Sphingomyelin/cholesterol nanoheterogeneities; influence of GM 1 • In membrane oligomerisation of β-amyloid • • • Fluorescence Methods Monte Carlo simulations of Fluorescence Lifetime Imaging -Förster Resonance Energy Transfer Calibration free Fluorescence Correlation Spectroscopy: Z-scan FCS Fluorescence Correlation Cross Spectroscopy Benda, Beneš, Mareček, Lhotský, Hermens, Hof Langmuir, 2003. 19(10) 7. Štefl, Kułakowska, Hof 2009, Biophysical Journal, 97(3): p. L 1 -L 3 for 2 -colour PIE Z-scan FCS

Lipid Topics • Size and character of ganglioside GM 1 nano-clusters • Sphingomyelin/cholesterol nanoheterogeneities; influence of GM 1 • In membrane oligomerisation of β-amyloid • • • Fluorescence Methods Monte Carlo simulations of Fluorescence Lifetime Imaging -Förster Resonance Energy Transfer Calibration free Fluorescence Correlation Spectroscopy: Z-scan FCS Fluorescence Correlation Cross Spectroscopy Schwille, Meyer-Almes, Rigler, Biophys J 72 (1997) 1878 -1886. Bacia, Kim, Schwille Nat Methods 3 (2006) 83 -89

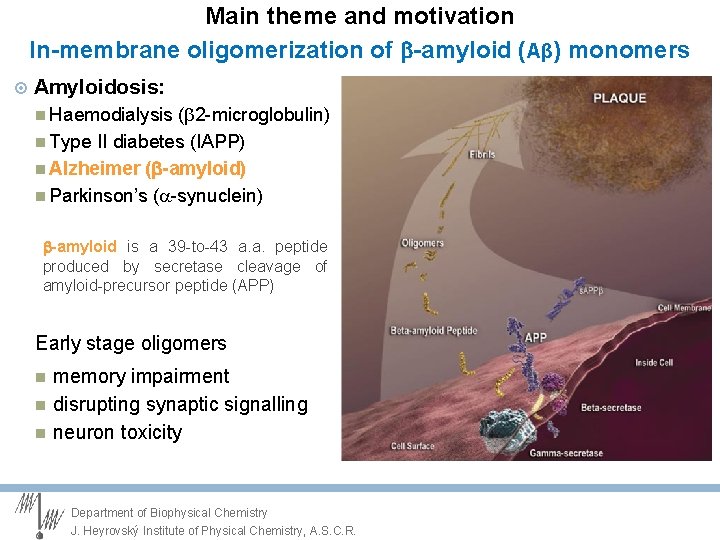
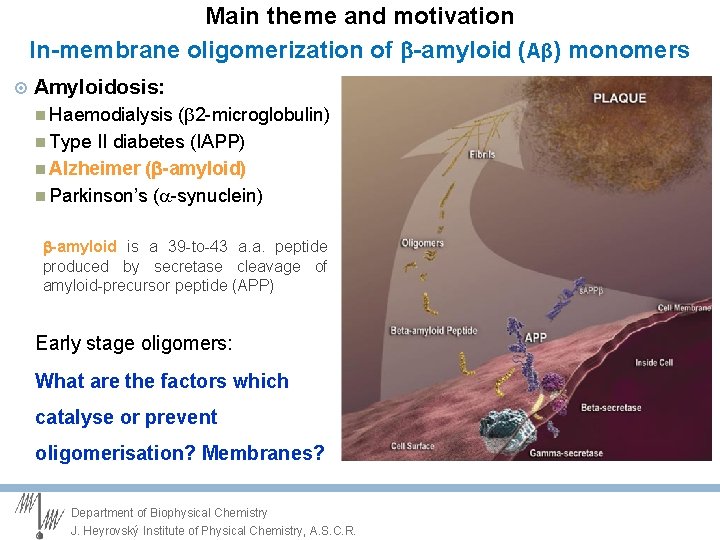
Main theme and motivation In-membrane oligomerization of -amyloid (Aβ) monomers Amyloidosis: Haemodialysis (b 2 -microglobulin) Type II diabetes (IAPP) Alzheimer ( -amyloid) Parkinson’s (a-synuclein) -amyloid is a 39 -to-43 a. a. peptide produced by secretase cleavage of amyloid-precursor peptide (APP) Early stage oligomers memory impairment disrupting synaptic signalling neuron toxicity Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

Main theme and motivation In-membrane oligomerization of -amyloid (Aβ) monomers Amyloidosis: Haemodialysis (b 2 -microglobulin) Type II diabetes (IAPP) Alzheimer ( -amyloid) Parkinson’s (a-synuclein) -amyloid is a 39 -to-43 a. a. peptide produced by secretase cleavage of amyloid-precursor peptide (APP) Early stage oligomers: What are the factors which catalyse or prevent oligomerisation? Membranes? Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

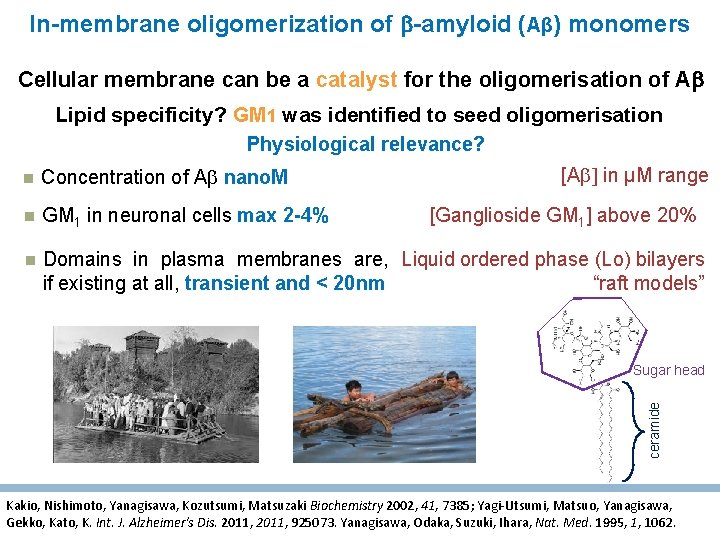
In-membrane oligomerization of -amyloid (Aβ) monomers Cellular membrane can be a catalyst for the oligomerisation of A Lipid specificity? GM 1 was identified to seed oligomerisation Physiological relevance? [Ab] in µM range Concentration of Ab nano. M GM 1 in neuronal cells max 2 -4% Domains in plasma membranes are, Liquid ordered phase (Lo) bilayers if existing at all, transient and < 20 nm “raft models” [Ganglioside GM 1] above 20% ceramide Sugar head Kakio, Nishimoto, Yanagisawa, Kozutsumi, Matsuzaki Biochemistry 2002, 41, 7385; Yagi-Utsumi, Matsuo, Yanagisawa, Gekko, Kato, K. Int. J. Alzheimer's Dis. 2011, 925073. Yanagisawa, Odaka, Suzuki, Ihara, Nat. Med. 1995, 1, 1062.

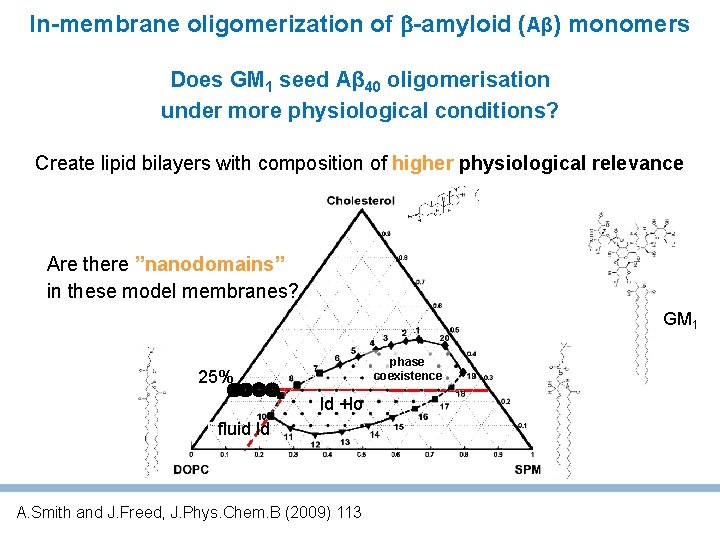
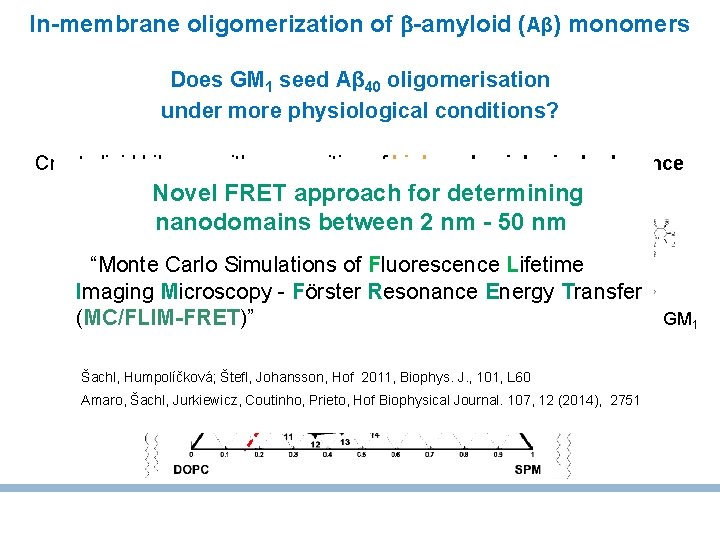
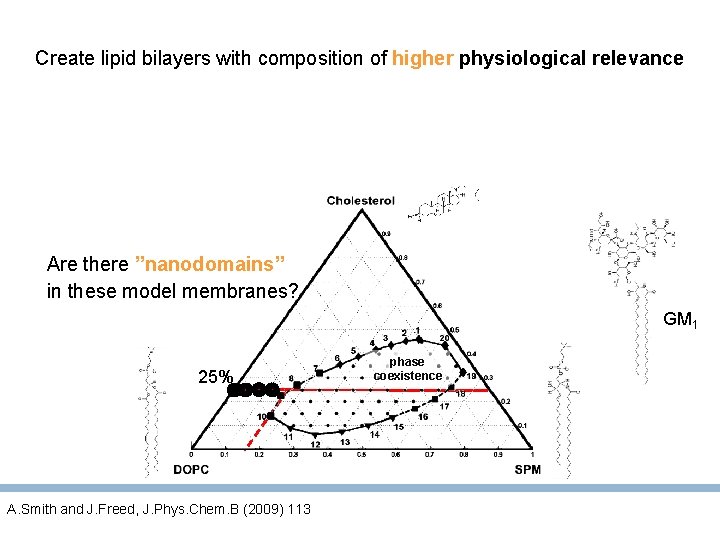
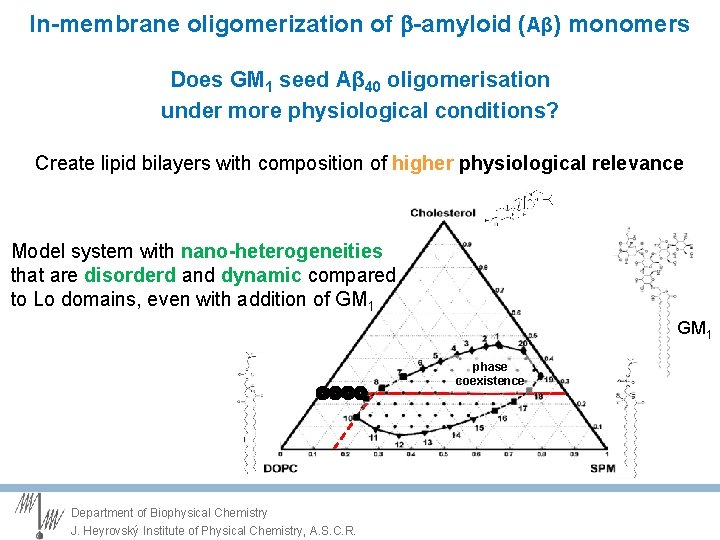
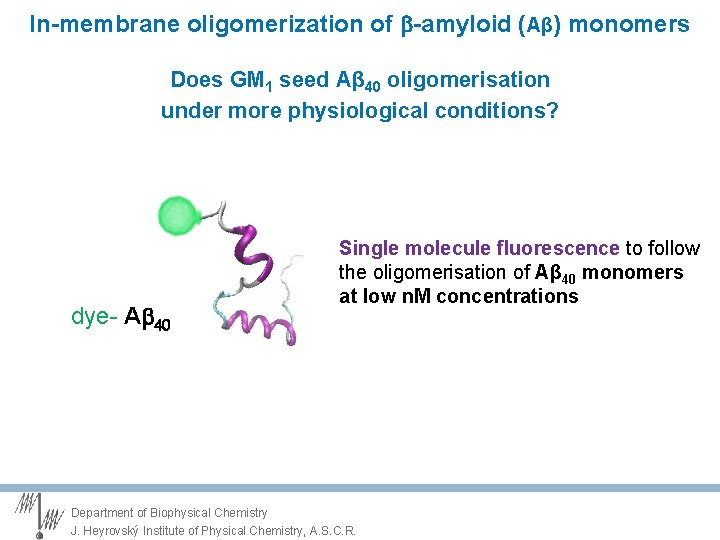
Central question Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Create lipid bilayers with composition of higher physiological relevance Lower GM 1 levels to 2 -4% avoid Liquid ordered (Lo) phase Lower Ab concentration to nano. M Single molecule fluorescence to follow the oligomerisation of Aβ monomers dye- A 40 Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Create lipid bilayers with composition of higher physiological relevance Are there ”nanodomains” in these model membranes? GM 1 phase coexistence 25% ld +lo Ffluid ld A. Smith and J. Freed, J. Phys. Chem. B (2009) 113

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Create lipid bilayers with composition of higher physiological relevance Novel FRET approach for determining nanodomains between 2 nm - 50 nm “Monte Carlo Simulations of Fluorescence Lifetime Imaging Microscopy - Förster Resonance Energy Transfer (MC/FLIM-FRET)” GM 1 phase coexistence Šachl, Humpolíčková; Štefl, Johansson, Hof 2011, Biophys. J. , 101, L 60 25% Amaro, Šachl, Jurkiewicz, Coutinho, Prieto, Hof Biophysical Journal. 107, 12 (2014), 2751

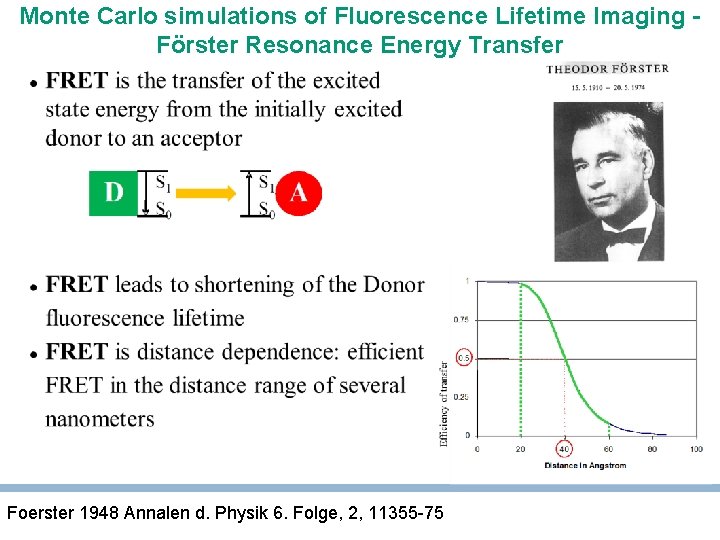
Monte Carlo simulations of Fluorescence Lifetime Imaging Förster Resonance Energy Transfer Foerster 1948 Annalen d. Physik 6. Folge, 2, 11355 -75

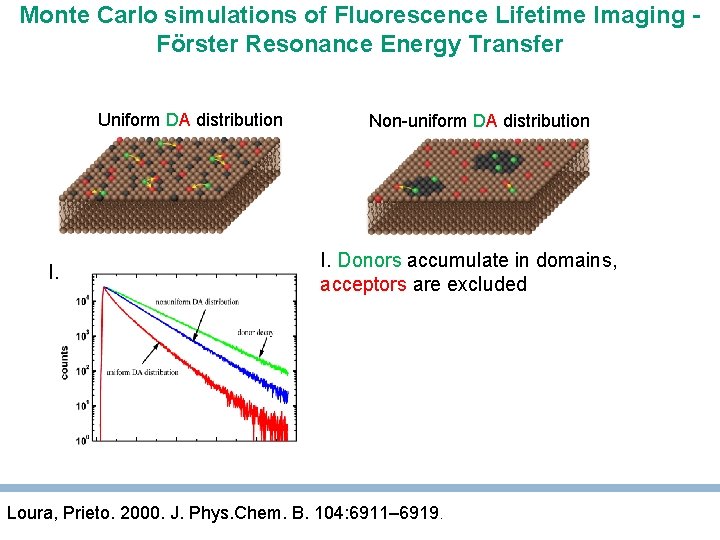
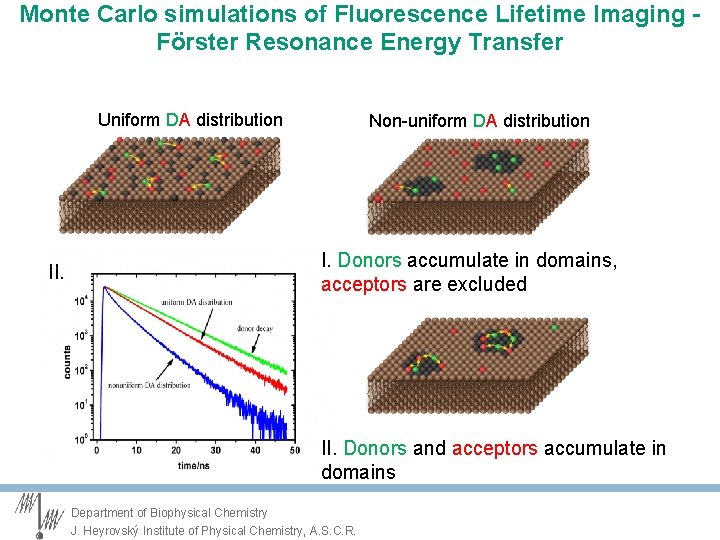
Monte Carlo simulations of Fluorescence Lifetime Imaging Förster Resonance Energy Transfer Uniform DA distribution I. Non-uniform DA distribution I. Donors accumulate in domains, acceptors are excluded Loura, Prieto. 2000. J. Phys. Chem. B. 104: 6911– 6919.

Monte Carlo simulations of Fluorescence Lifetime Imaging Förster Resonance Energy Transfer Uniform DA distribution II. Non-uniform DA distribution I. Donors accumulate in domains, acceptors are excluded II. Donors and acceptors accumulate in domains Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

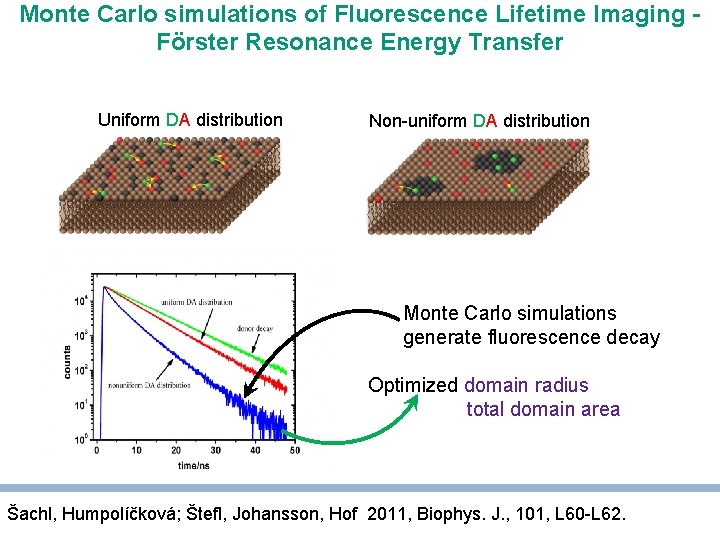
Monte Carlo simulations of Fluorescence Lifetime Imaging Förster Resonance Energy Transfer Uniform DA distribution Non-uniform DA distribution Monte Carlo simulations generate fluorescence decay Optimized domain radius total domain area Šachl, Humpolíčková; Štefl, Johansson, Hof 2011, Biophys. J. , 101, L 60 -L 62.

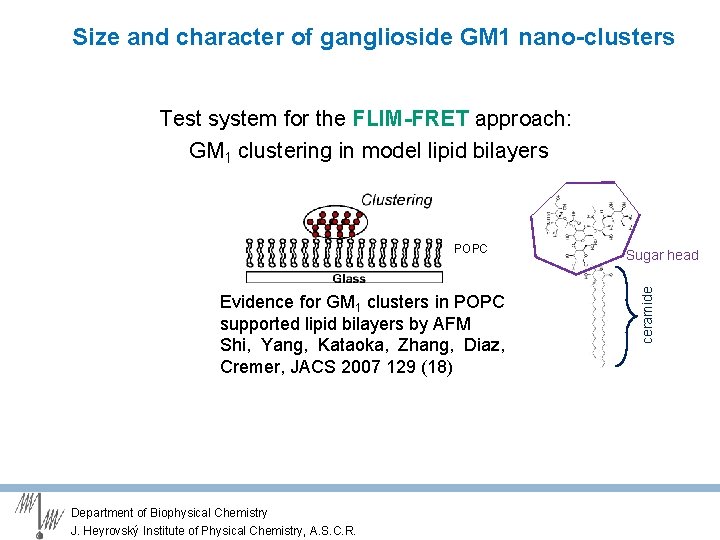
Size and character of ganglioside GM 1 nano-clusters Test system for the FLIM-FRET approach: GM 1 clustering in model lipid bilayers Evidence for GM 1 clusters in POPC supported lipid bilayers by AFM Shi, Yang, Kataoka, Zhang, Diaz, Cremer, JACS 2007 129 (18) Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R. Sugar head ceramide POPC

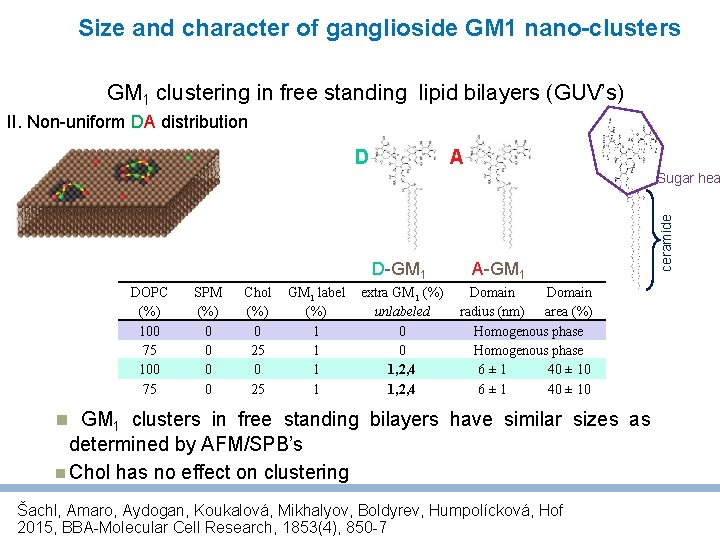
Size and character of ganglioside GM 1 nano-clusters GM 1 clustering in free standing lipid bilayers (GUV’s) II. Non-uniform DA distribution A D D-GM 1 DOPC (%) 100 75 SPM (%) 0 0 Chol (%) 0 25 GM 1 label (%) 1 1 extra GM 1 (%) unlabeled 0 0 1, 2, 4 A-GM 1 Domain radius (nm) area (%) Homogenous phase 6± 1 40 ± 10 GM 1 clusters in free standing bilayers have similar sizes as determined by AFM/SPB’s Chol has no effect on clustering Šachl, Amaro, Aydogan, Koukalová, Mikhalyov, Boldyrev, Humpolícková, Hof 2015, BBA-Molecular Cell Research, 1853(4), 850 -7 ceramide Sugar hea

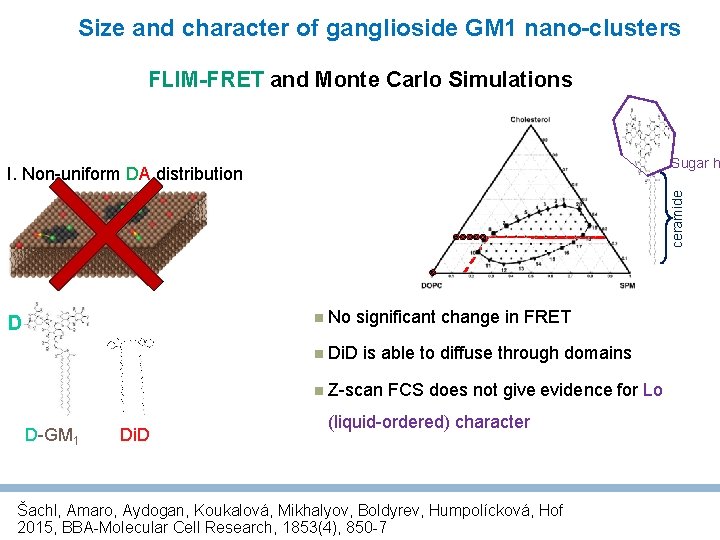
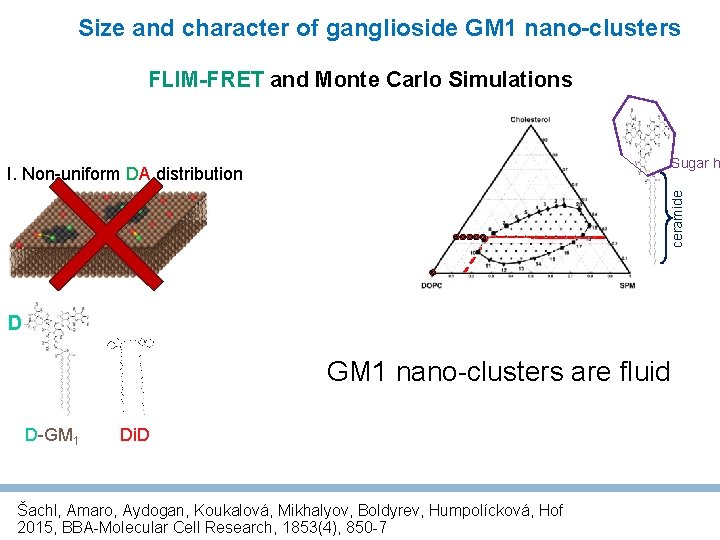
Size and character of ganglioside GM 1 nano-clusters FLIM-FRET and Monte Carlo Simulations Sugar h ceramide I. Non-uniform DA distribution No D significant change in FRET Di. D is able to diffuse through domains Z-scan D-GM 1 Di. D FCS does not give evidence for Lo (liquid-ordered) character Šachl, Amaro, Aydogan, Koukalová, Mikhalyov, Boldyrev, Humpolícková, Hof 2015, BBA-Molecular Cell Research, 1853(4), 850 -7

Size and character of ganglioside GM 1 nano-clusters FLIM-FRET and Monte Carlo Simulations Sugar h ceramide I. Non-uniform DA distribution D GM 1 nano-clusters are fluid D-GM 1 Di. D Šachl, Amaro, Aydogan, Koukalová, Mikhalyov, Boldyrev, Humpolícková, Hof 2015, BBA-Molecular Cell Research, 1853(4), 850 -7

Create lipid bilayers with composition of higher physiological relevance Are there ”nanodomains” in these model membranes? GM 1 25% A. Smith and J. Freed, J. Phys. Chem. B (2009) 113 phase coexistence

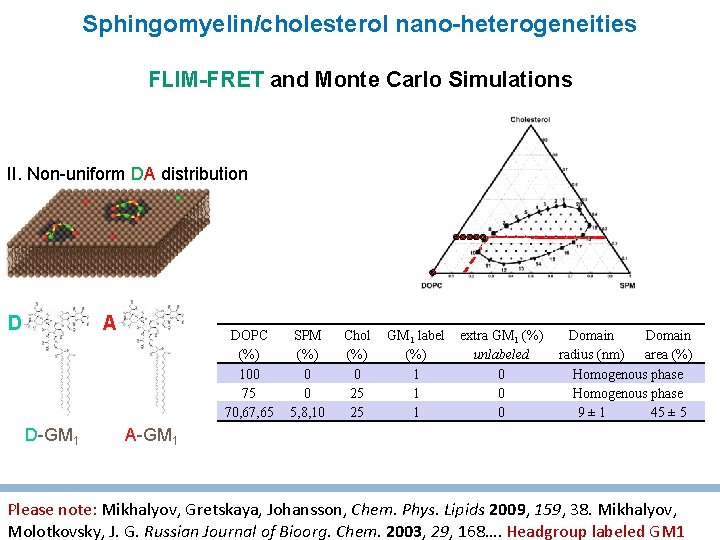
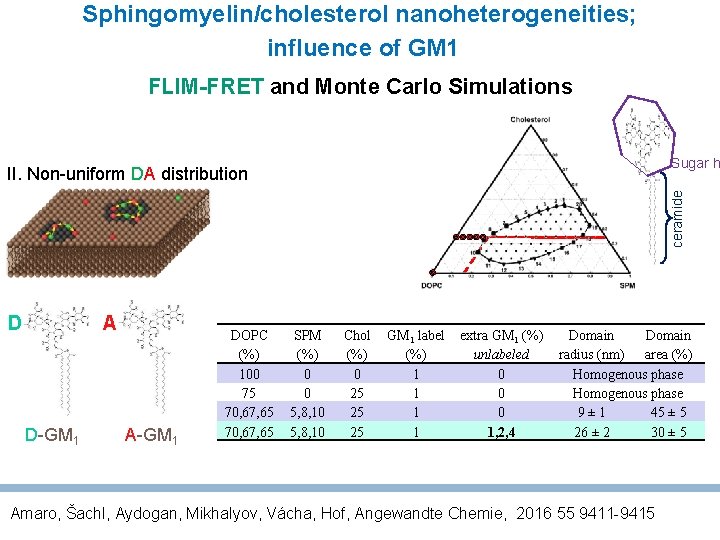
Sphingomyelin/cholesterol nano-heterogeneities FLIM-FRET and Monte Carlo Simulations II. Non-uniform DA distribution A D D-GM 1 DOPC (%) 100 75 70, 67, 65 SPM (%) 0 0 5, 8, 10 Chol (%) 0 25 25 GM 1 label (%) 1 1 1 extra GM 1 (%) unlabeled 0 0 0 Domain radius (nm) area (%) Homogenous phase 9± 1 45 ± 5 A-GM 1 Please note: Mikhalyov, Gretskaya, Johansson, Chem. Phys. Lipids 2009, 159, 38. Mikhalyov, Molotkovsky, J. G. Russian Journal of Bioorg. Chem. 2003, 29, 168…. Headgroup labeled GM 1

Sphingomyelin/cholesterol nanoheterogeneities; influence of GM 1 FLIM-FRET and Monte Carlo Simulations Sugar h ceramide II. Non-uniform DA distribution A D D-GM 1 A-GM 1 DOPC (%) 100 75 70, 67, 65 SPM (%) 0 0 5, 8, 10 Chol (%) 0 25 25 25 GM 1 label (%) 1 1 extra GM 1 (%) unlabeled 0 0 0 1, 2, 4 Domain radius (nm) area (%) Homogenous phase 9± 1 45 ± 5 26 ± 2 30 ± 5 Amaro, Šachl, Aydogan, Mikhalyov, Vácha, Hof, Angewandte Chemie, 2016 55 9411 -9415

Sphingomyelin/cholesterol nanoheterogeneities; influence of GM 1 • FRET: GM 1 accumulates in “domains”, Di. D diffuses through • 2 -colour Z-scan FCS using GM 1 and Di. D • Solid state wideline and high resolution magic angle spinning NMR Nano-heterogeneities are still fluid and disordered, even with addition of GM 1 Amaro, Šachl, Aydogan, Mikhalyov, Vácha, Hof, Angewandte Chemie, 2016 55 9411 -9415; paper with alternative labels and NMR is in preparation

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Create lipid bilayers with composition of higher physiological relevance Model system with nano-heterogeneities that are disorderd and dynamic compared to Lo domains, even with addition of GM 1 phase coexistence Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? dye- A 40 Single molecule fluorescence to follow the oligomerisation of Aβ 40 monomers at low n. M concentrations Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

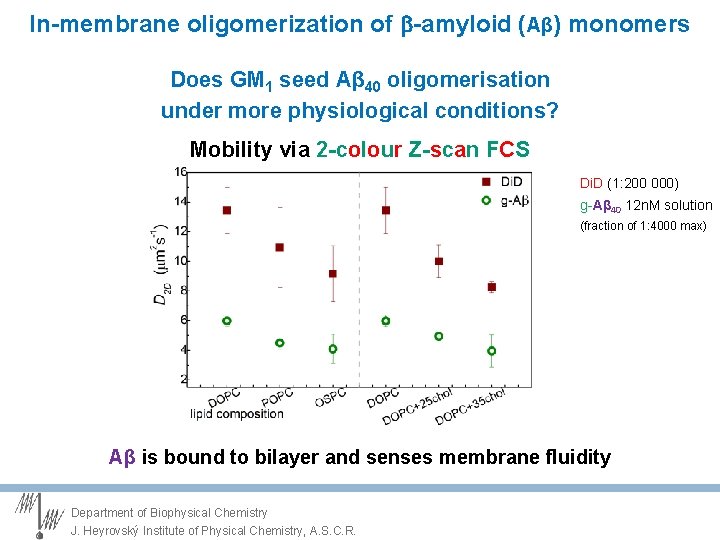
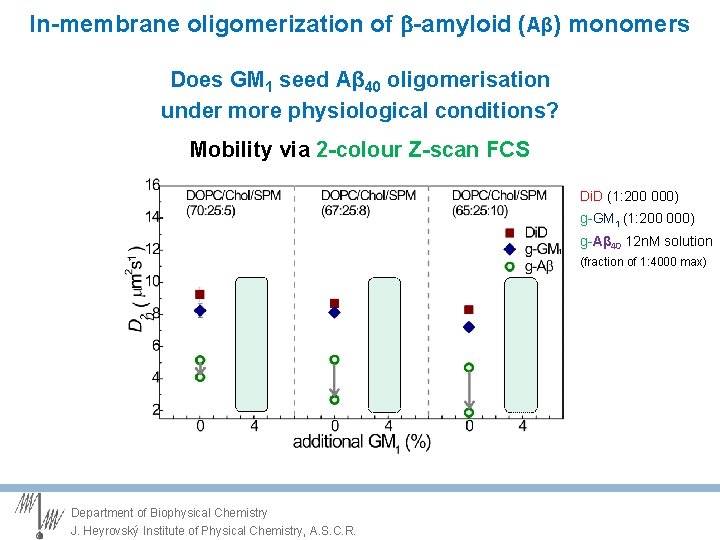
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Mobility via 2 -colour Z-scan FCS Di. D (1: 200 000) g-Aβ 40 12 n. M solution (fraction of 1: 4000 max) Aβ is bound to bilayer and senses membrane fluidity Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

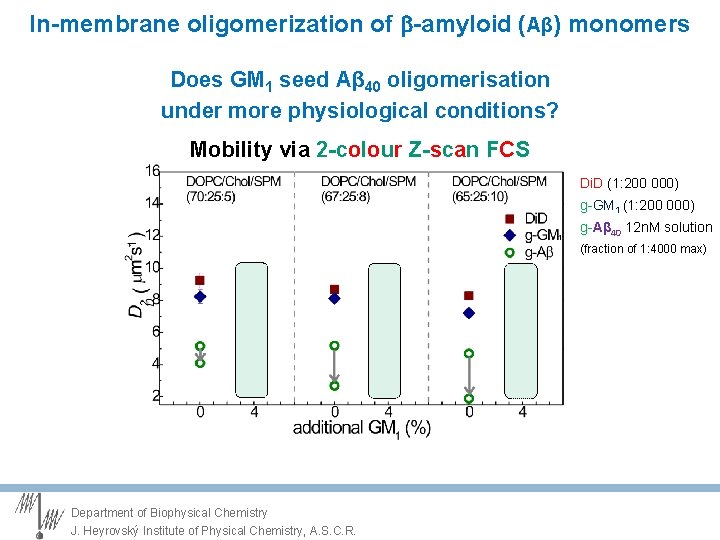
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Mobility via 2 -colour Z-scan FCS Di. D (1: 200 000) g-GM 1 (1: 200 000) g-Aβ 40 12 n. M solution (fraction of 1: 4000 max) Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

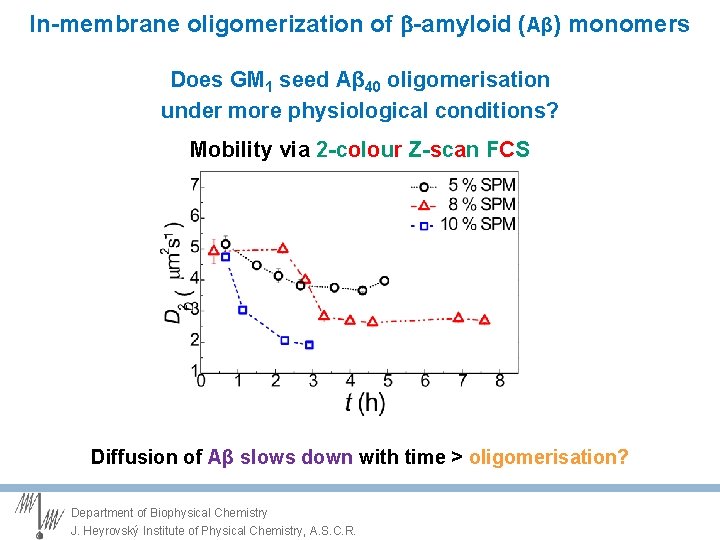
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Mobility via 2 -colour Z-scan FCS Diffusion of Aβ slows down with time > oligomerisation? Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

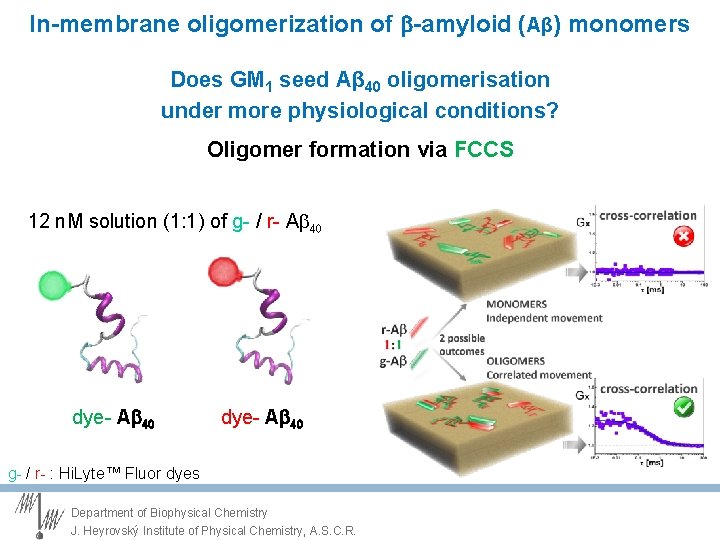
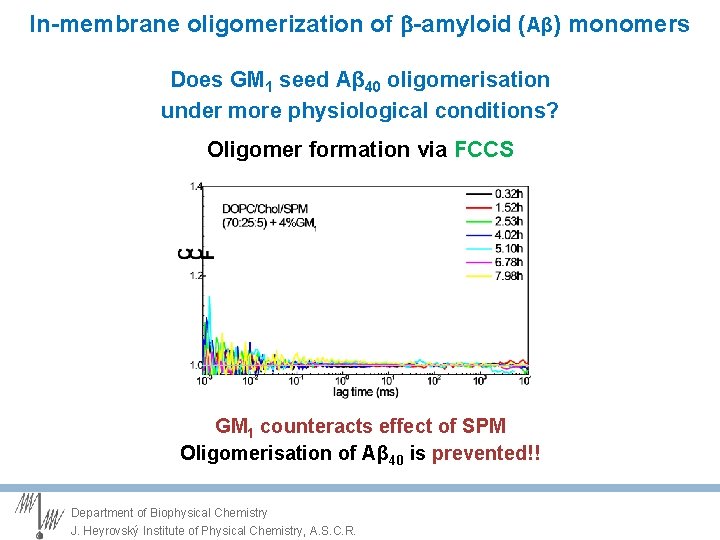
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Oligomer formation via FCCS 12 n. M solution (1: 1) of g- / r- Ab 40 dye- A 40 g- / r- : Hi. Lyte™ Fluor dyes Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

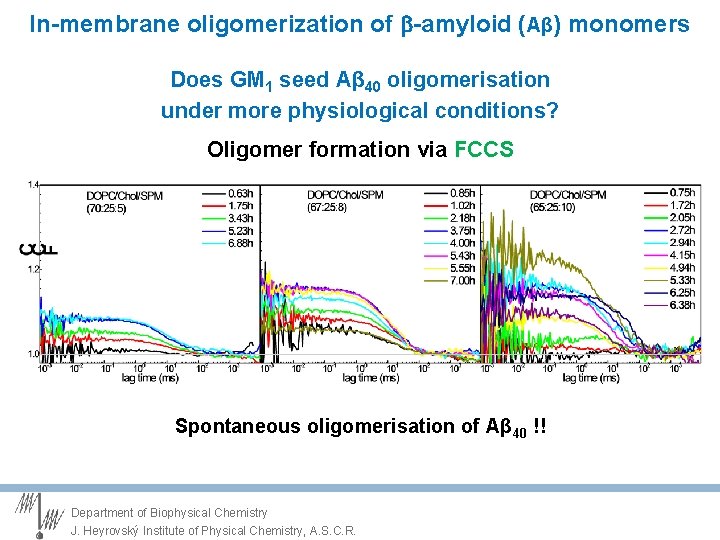
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Oligomer formation via FCCS Spontaneous oligomerisation of Aβ 40 !! Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

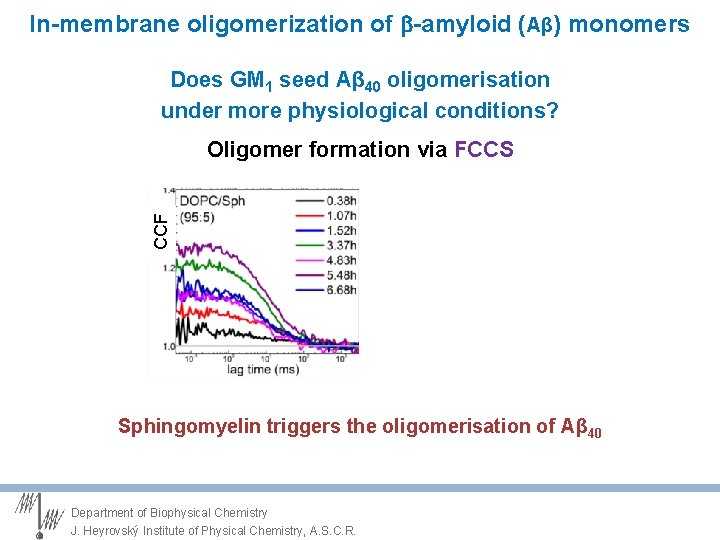
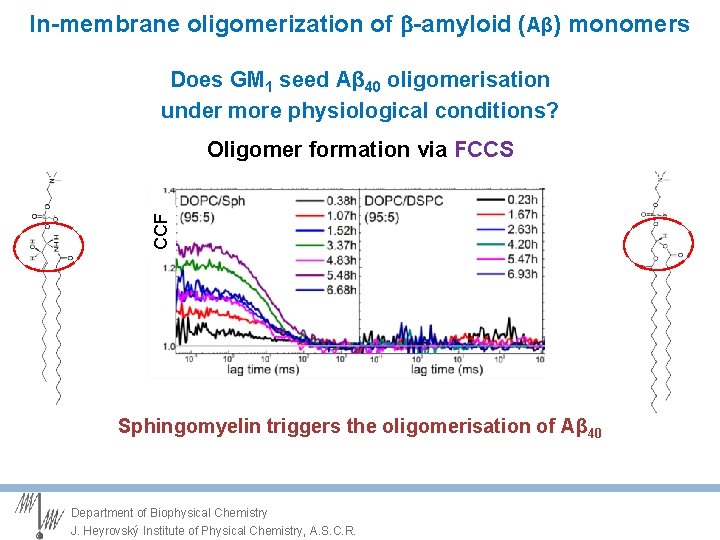
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? CCF Oligomer formation via FCCS Sphingomyelin triggers the oligomerisation of Aβ 40 Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? CCF Oligomer formation via FCCS Sphingomyelin triggers the oligomerisation of Aβ 40 Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Mobility via 2 -colour Z-scan FCS Di. D (1: 200 000) g-GM 1 (1: 200 000) g-Aβ 40 12 n. M solution (fraction of 1: 4000 max) Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Oligomer formation via FCCS GM 1 counteracts effect of SPM Oligomerisation of Aβ 40 is prevented!! Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

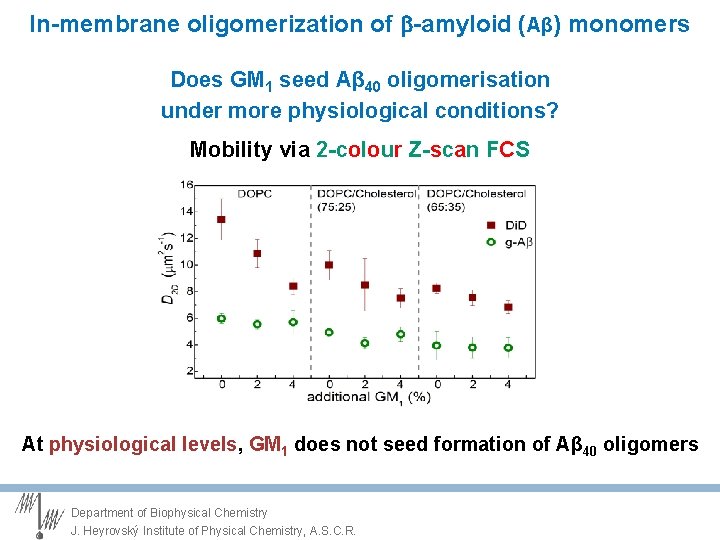
In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Mobility via 2 -colour Z-scan FCS At physiological levels, GM 1 does not seed formation of Aβ 40 oligomers Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

In-membrane oligomerization of -amyloid (Aβ) monomers Does GM 1 seed Aβ 40 oligomerisation under more physiological conditions? Physiological levels of GM 1 do not seed oligomer formation. Sphingomyelin is a key trigger of oligomerisation Molecular evidence for GM 1 as inhibitor of oligomer formation Amaro, Šachl, Aydogan, Mikhalyov, Vácha, Hof, Angewandte Chemie, 2016 55 9411 -9415

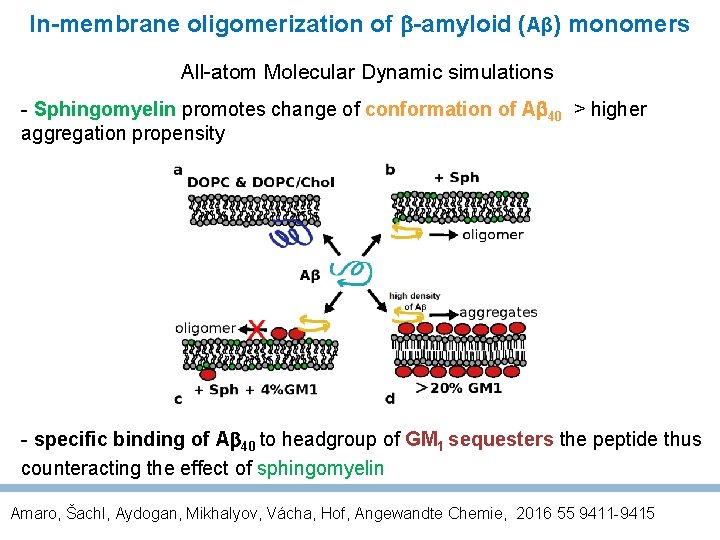
In-membrane oligomerization of -amyloid (Aβ) monomers All-atom Molecular Dynamic simulations - Sphingomyelin promotes change of conformation of A 40 > higher aggregation propensity - specific binding of A 40 to headgroup of GM 1 sequesters the peptide thus counteracting the effect of sphingomyelin Amaro, Šachl, Aydogan, Mikhalyov, Vácha, Hof, Angewandte Chemie, 2016 55 9411 -9415

Our molecular insights showing that a) Sphingomyelin triggers oligomerisation b) GM 1 inhibits oligomer formation • clarify the role of GM 1 in Aβ 40 oligomerisation • support experimental results on the protective role of GM 1 in cell cultures and rat models of Alzheimer’s disease • might be connected to decrease of GM 1 levels with aging a disease progression Department of Biophysical Chemistry J. Heyrovský Institute of Physical Chemistry, A. S. C. R.

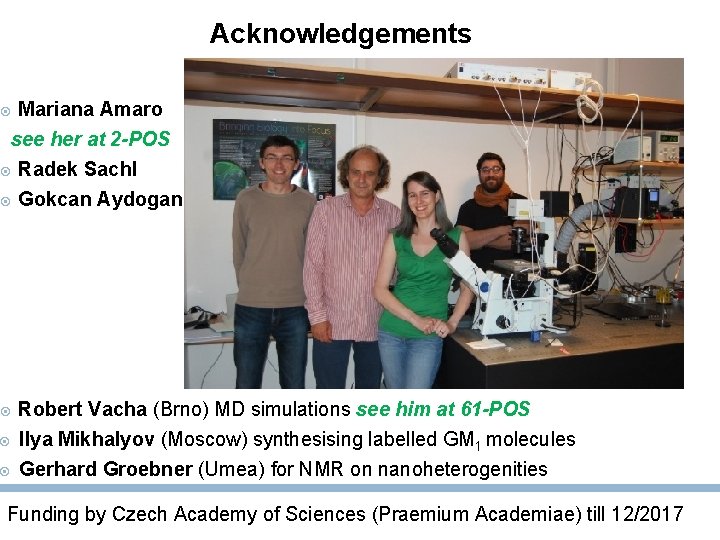
Acknowledgements Mariana Amaro see her at 2 -POS Radek Sachl Gokcan Aydogan Robert Vacha (Brno) MD simulations see him at 61 -POS Ilya Mikhalyov (Moscow) synthesising labelled GM 1 molecules Gerhard Groebner (Umea) for NMR on nanoheterogenities Funding by Czech Academy of Sciences (Praemium Academiae) till 12/2017