46 complete Sargasso Sea 1600 Methaneoxidizing archaea from



































- Slides: 55



46 метагеномов • “complete” – – – – – Sargasso Sea 1600 Мб Methane-oxidizing archaea from deep sea sediments 111 Мб Minnesota Soil 100 Мб Acid mine drainage (AMD) 75 Мб Woolly mammoth 28 Мб Whale falls 25 Мб Antarctic Marine Bacterioplankton 12 Мб Pleistocene Cave Bears 1 Мб DOE Hanford Site • “incomplete”, но с оценкой размера или опубликованые – – – – Hawaii Ocean Times Series Station (HOT) 2000 Мб Rice endophyte community 100 Мб Poplar endophytic bacteria 37 Мб Alaskan Soil 8 Мб Pacific Beach Sand 3 Мб Calyptogena magnifica symbiont 1. 2 Мб Biofilms in drinking-water networks

Метагеномы в другом смысле • • Escherichia 5+32 + Shigella 6+5 Salmonella 5+18 Staphylococcus 17+9 (aureus 9+5) Streptococcus 17+35 – S. pyogenes 7+7 – S. pneumoniae ~7+7 • Bacillus anthracis+cereus+thuringiensis 7+22 • Mycoplasma 12+16 • Chlamydia 4+5 + Chlamydophyla 6+2





базы данных результатов экспериментов по анализу экспрессии • Array. Express http: //www. ebi. ac. uk/microarray/Array. Express/arrayxpress. html • Stanford Microarray Database http: //genome-www 5. stanford. edu/Micro. Array/NDEV/index. shtml • GEO (Gene Expression Omnibus) http: //www. ncbi. nlm. nih. gov/geo/ • Стандартизованная форма данных об эксперименте (MIAME: Minimal Iinformation About a Microarray Experiment)




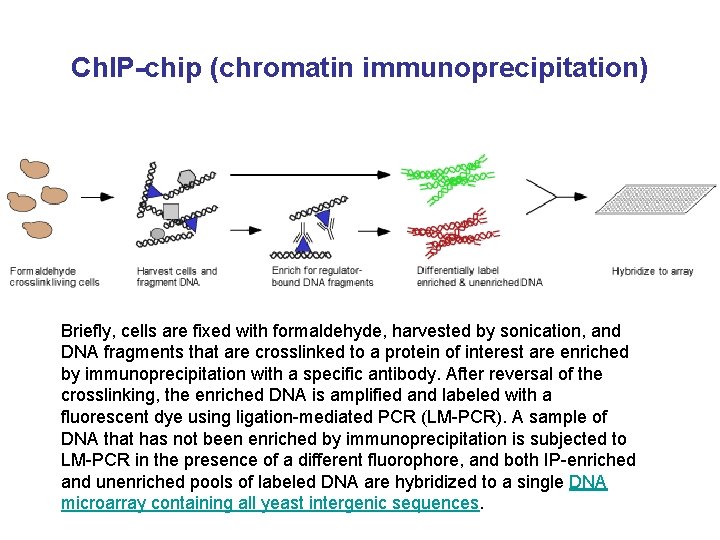
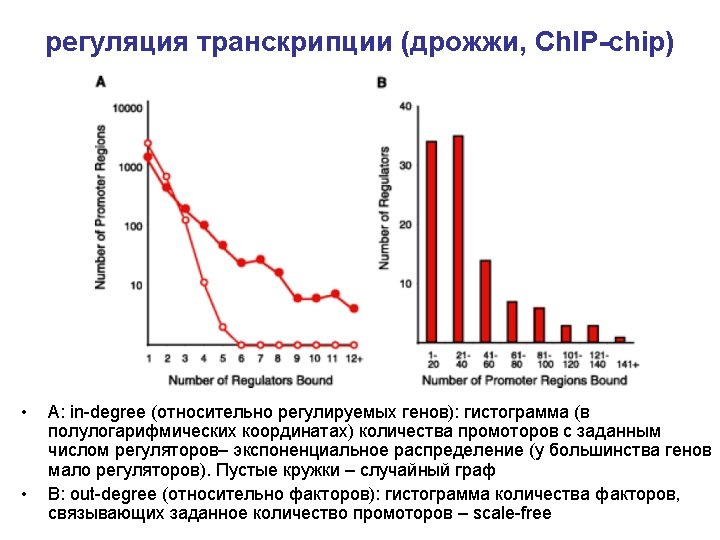
Ch. IP-chip (chromatin immunoprecipitation) Briefly, cells are fixed with formaldehyde, harvested by sonication, and DNA fragments that are crosslinked to a protein of interest are enriched by immunoprecipitation with a specific antibody. After reversal of the crosslinking, the enriched DNA is amplified and labeled with a fluorescent dye using ligation-mediated PCR (LM-PCR). A sample of DNA that has not been enriched by immunoprecipitation is subjected to LM-PCR in the presence of a different fluorophore, and both IP-enriched and unenriched pools of labeled DNA are hybridized to a single DNA microarray containing all yeast intergenic sequences.



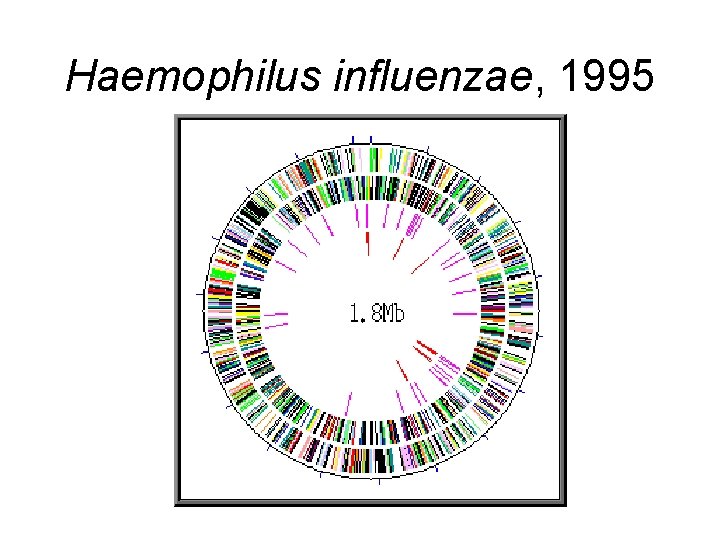
Haemophilus influenzae, 1995

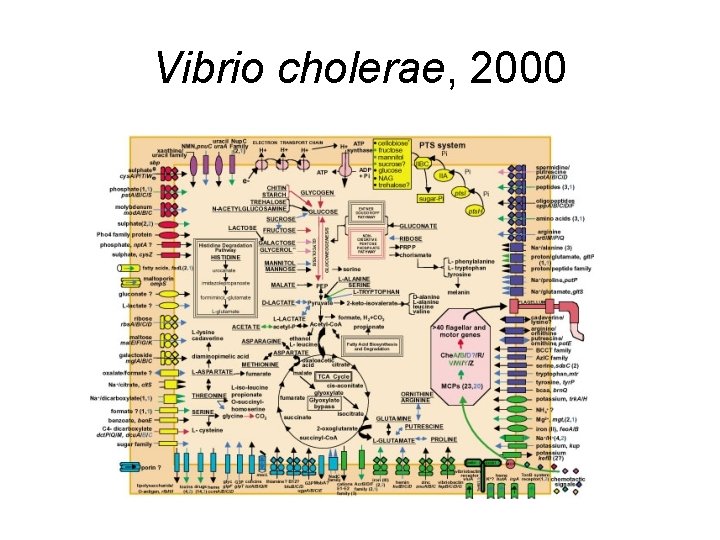
Vibrio cholerae, 2000




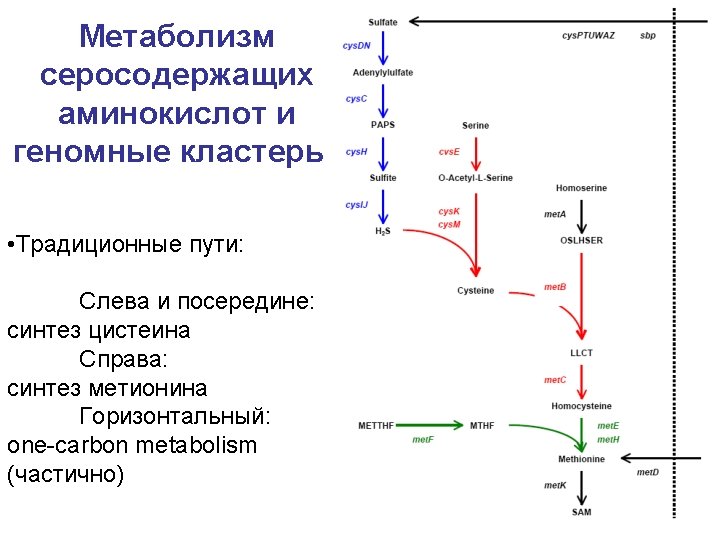
Metabolic pathways

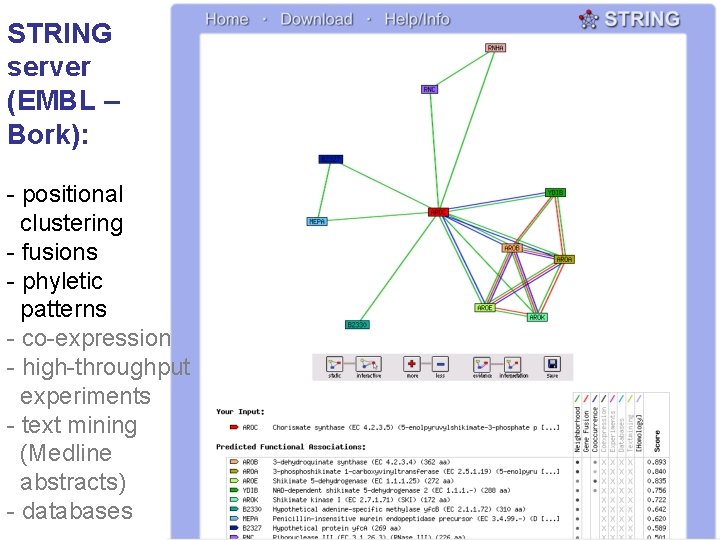
STRING server (EMBL – Bork): - positional clustering - fusions - phyletic patterns - co-expression - high-throughput experiments - text mining (Medline abstracts) - databases

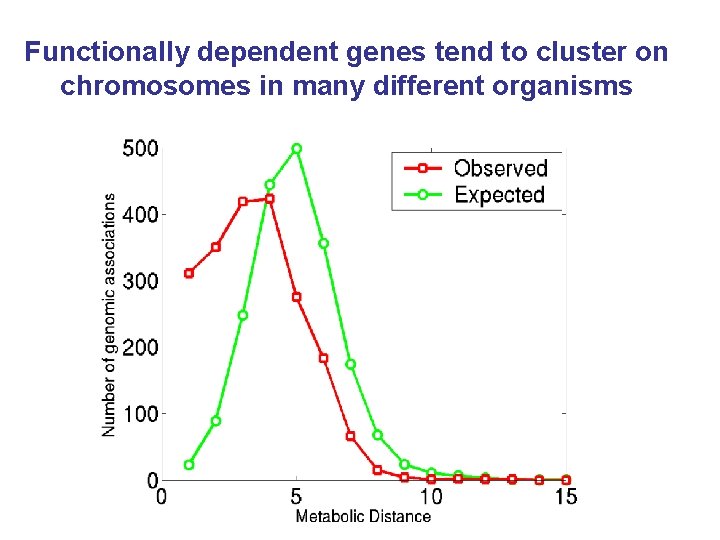
Functionally dependent genes tend to cluster on chromosomes in many different organisms

More genomes (stronger links) => highly significant clustering










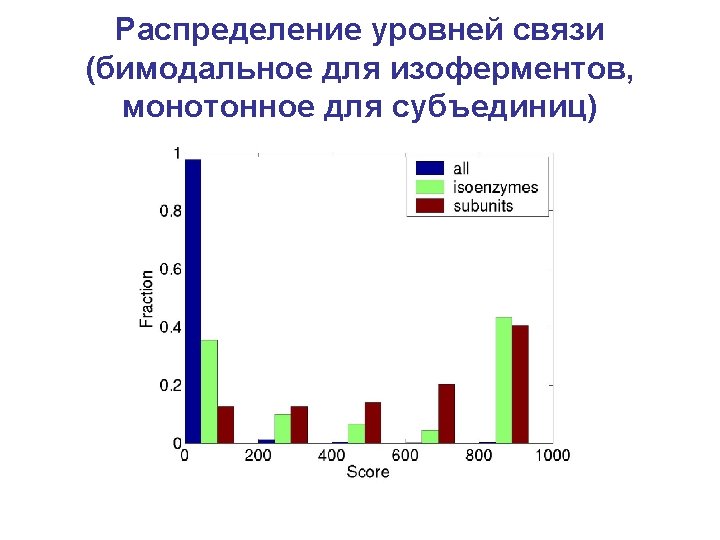
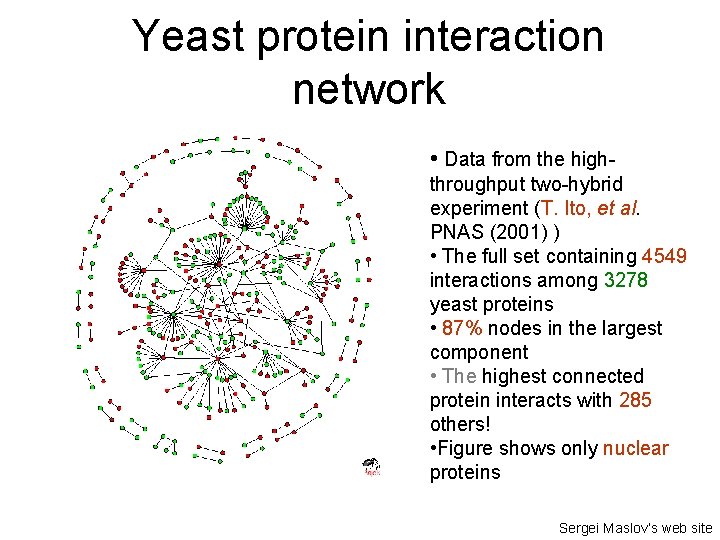
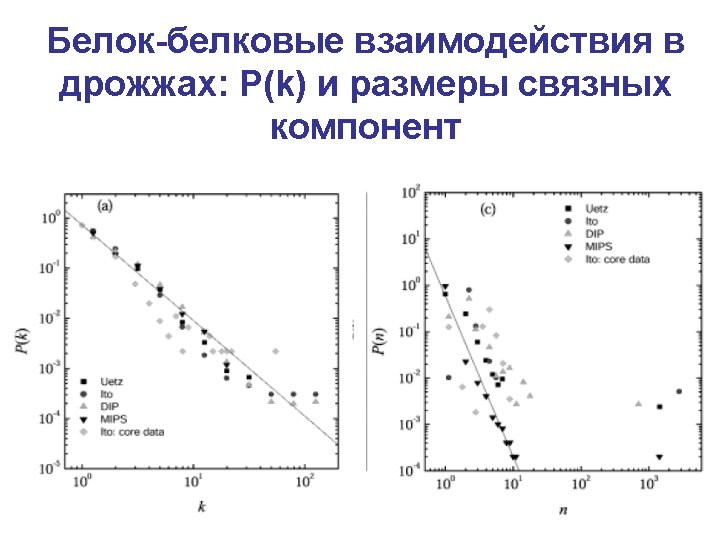
Yeast protein interaction network • Data from the highthroughput two-hybrid experiment (T. Ito, et al. PNAS (2001) ) • The full set containing 4549 interactions among 3278 yeast proteins • 87% nodes in the largest component • The highest connected protein interacts with 285 others! • Figure shows only nuclear proteins Sergei Maslov’s web site




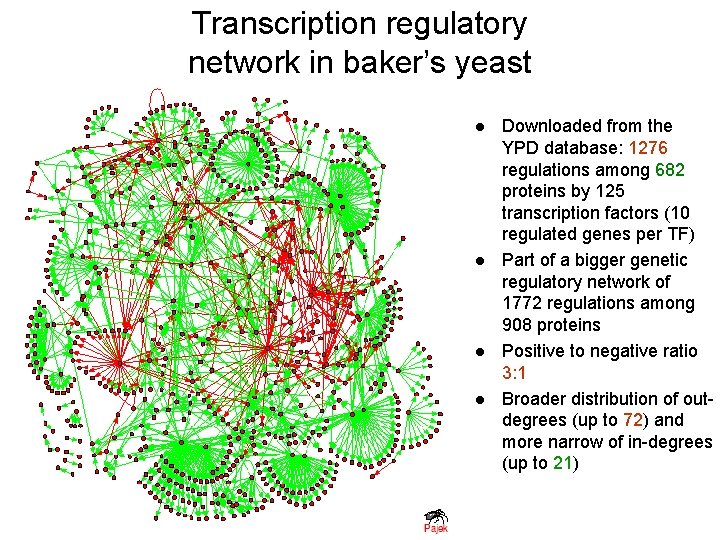
Transcription regulatory network in baker’s yeast l l Downloaded from the YPD database: 1276 regulations among 682 proteins by 125 transcription factors (10 regulated genes per TF) Part of a bigger genetic regulatory network of 1772 regulations among 908 proteins Positive to negative ratio 3: 1 Broader distribution of outdegrees (up to 72) and more narrow of in-degrees (up to 21)


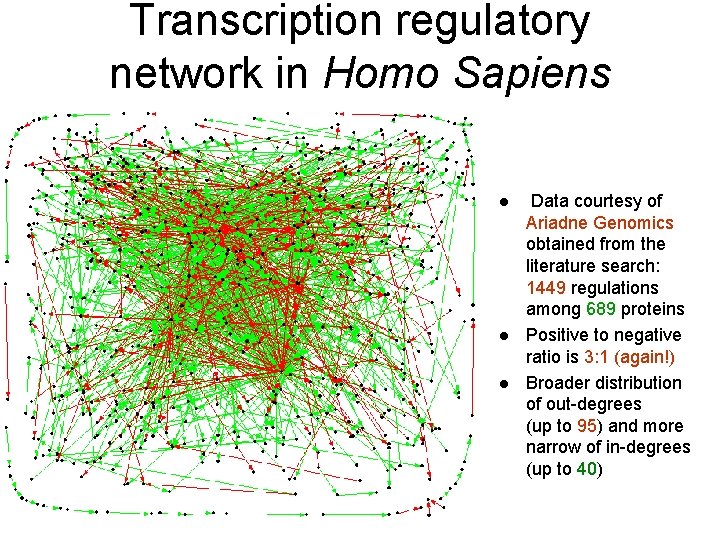
Transcription regulatory network in Homo Sapiens l l l Data courtesy of Ariadne Genomics obtained from the literature search: 1449 regulations among 689 proteins Positive to negative ratio is 3: 1 (again!) Broader distribution of out-degrees (up to 95) and more narrow of in-degrees (up to 40)

Transcription regulatory network in E. coli l l l Data (courtesy of Uri Alon) was curated from the Regulon database: 606 interactions between 424 operons (by 116 TFs) Positive to negative ratio is 3: 2 (different from eukaryots!) Broader distribution of out-degrees (up to 85) and more narrow of in-degrees (only up to 6 !)













