Using Hap Map Org A Tutorial Lincoln Stein



![8: Adjust Tagger (cont) Select population Select tagging algorithm and parameters. [optional] upload list 8: Adjust Tagger (cont) Select population Select tagging algorithm and parameters. [optional] upload list](https://slidetodoc.com/presentation_image_h/2348fd5ca46413e7ae466c462cfe6267/image-14.jpg)





- Slides: 30

Using Hap. Map. Org A Tutorial Lincoln Stein, Cold Spring Harbor Laboratory

Goal of This Tutorial • This tutorial will show you how to: – Find Hap. Map SNPs near a gene or region of interest (ROI). – View patterns of LD in the ROI. – Select tag SNPs in the ROI. – Download information on the SNPs in ROI for use in Haploview. – Generate customized extracts of the entire data set. – Download the entire data set in bulk.

Finding Hap. Map SNPs in a Region of Interest • Find the region of the genome containing the DTNBP 1 gene. • Identify the characterized SNPs in the region. • View the patterns of LD in the region. • Pick tag SNPs. • Download the region in Haploview format.

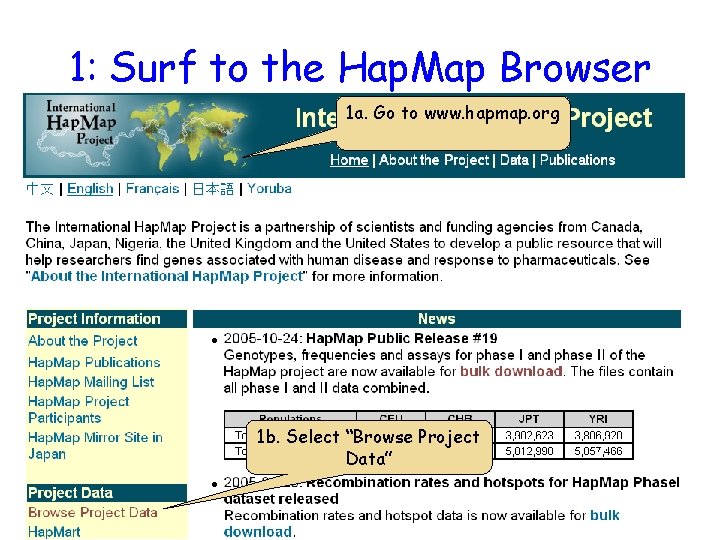
1: Surf to the Hap. Map Browser 1 a. Go to www. hapmap. org 1 b. Select “Browse Project Data”

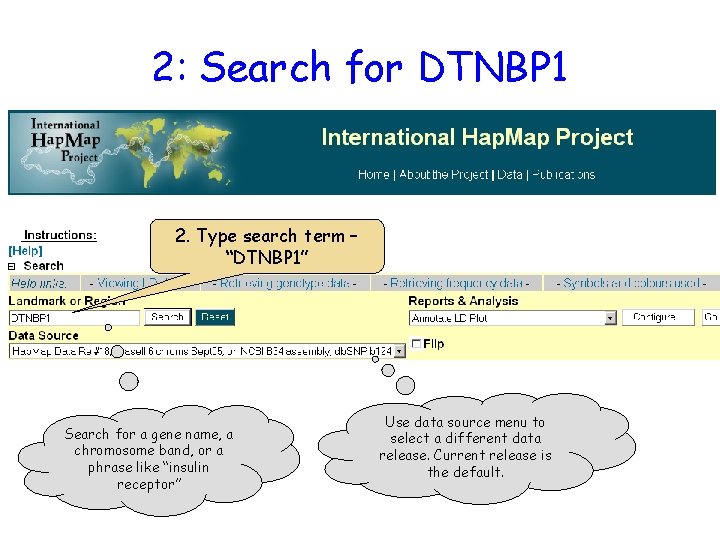
2: Search for DTNBP 1 2. Type search term – “DTNBP 1” Search for a gene name, a chromosome band, or a phrase like “insulin receptor” Use data source menu to select a different data release. Current release is the default.

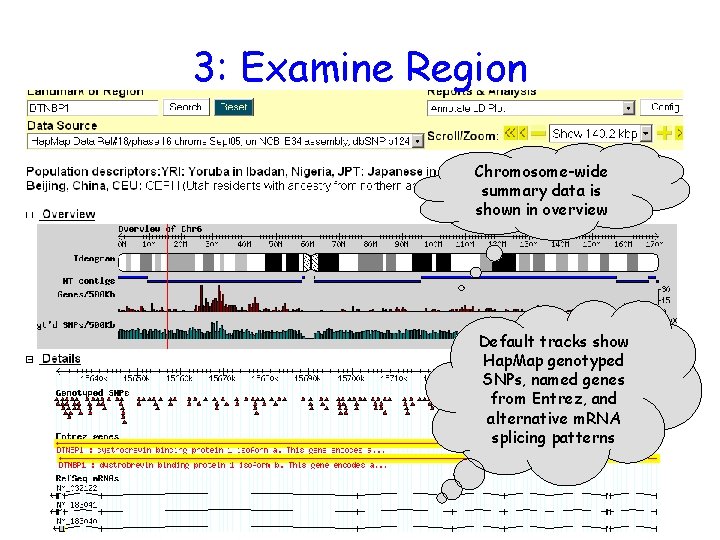
3: Examine Region Chromosome-wide summary data is shown in overview Default tracks show Hap. Map genotyped SNPs, named genes from Entrez, and alternative m. RNA splicing patterns

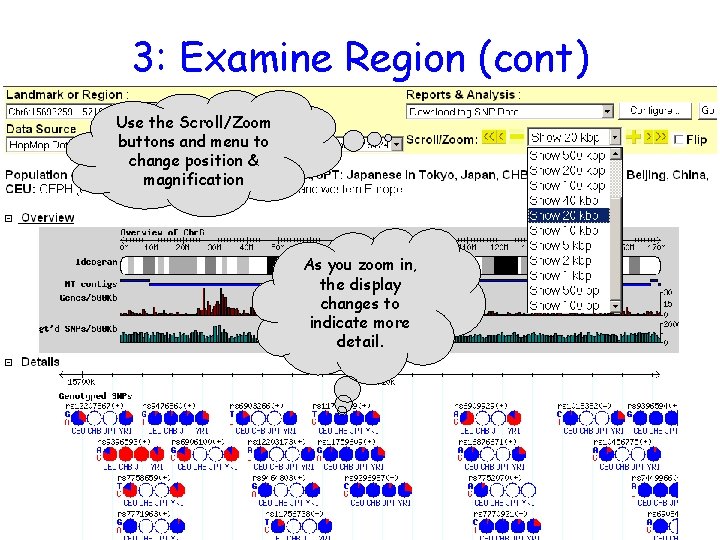
3: Examine Region (cont) Use the Scroll/Zoom buttons and menu to change position & magnification As you zoom in, the display changes to indicate more detail.

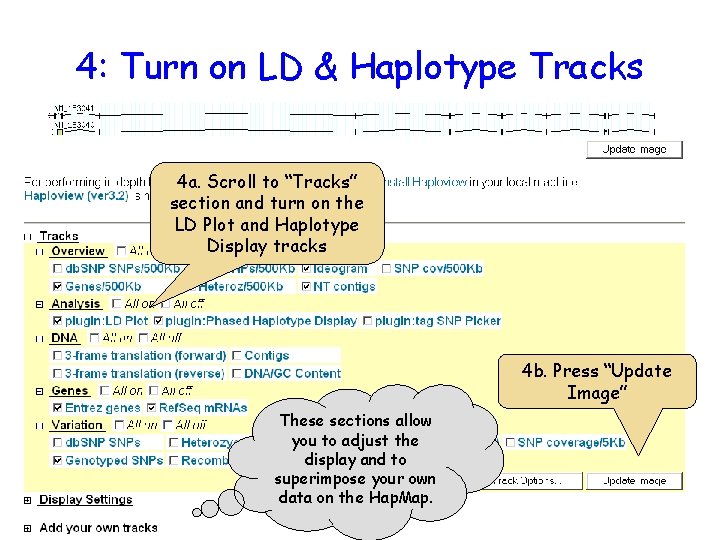
4: Turn on LD & Haplotype Tracks 4 a. Scroll to “Tracks” section and turn on the LD Plot and Haplotype Display tracks 4 b. Press “Update Image” These sections allow you to adjust the display and to superimpose your own data on the Hap. Map.

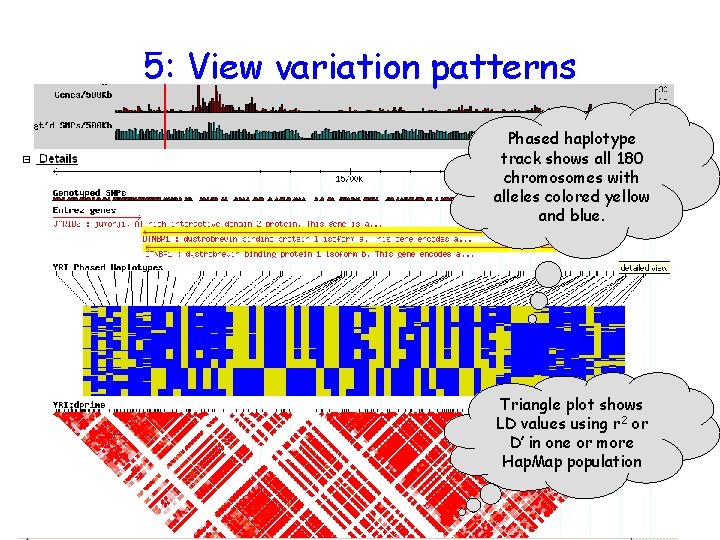
5: View variation patterns Phased haplotype track shows all 180 chromosomes with alleles colored yellow and blue. Triangle plot shows LD values using r 2 or D’ in one or more Hap. Map population

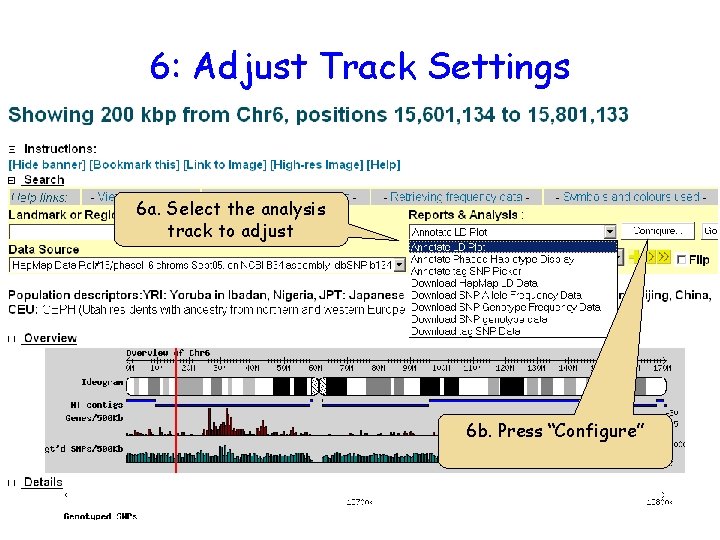
6: Adjust Track Settings 6 a. Select the analysis track to adjust 6 b. Press “Configure”

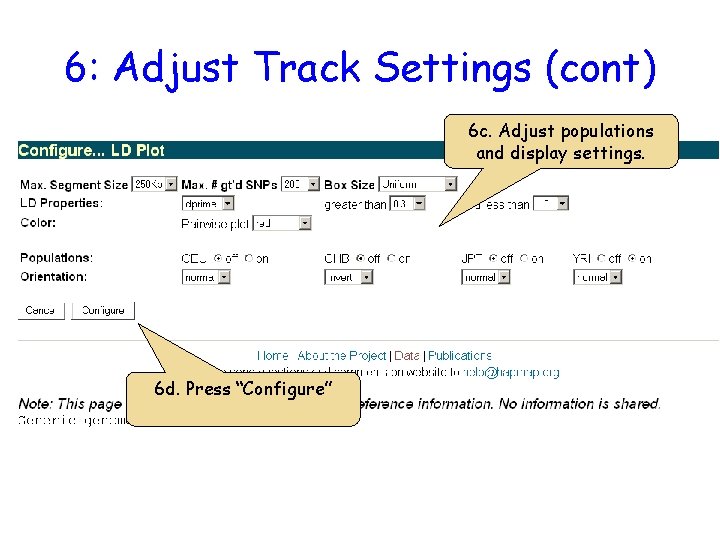
6: Adjust Track Settings (cont) 6 c. Adjust populations and display settings. 6 d. Press “Configure”

7: Turn on Tag SNP Track 7 a. Activate the “tag SNP Picker” and press “Update Image”

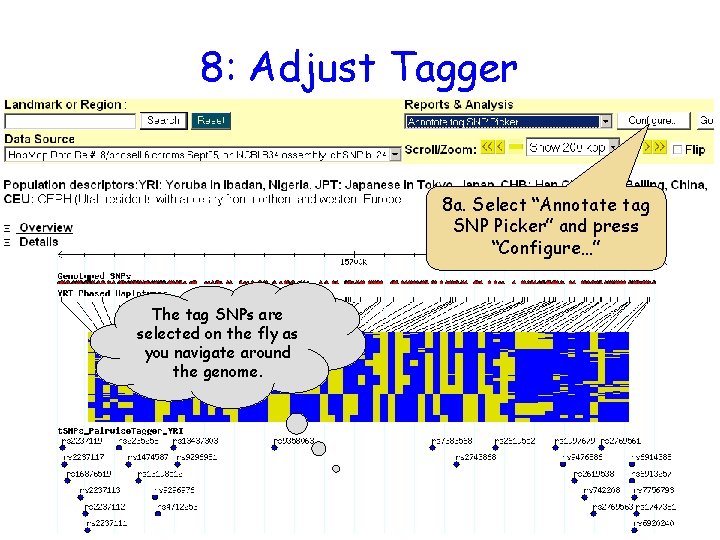
8: Adjust Tagger 8 a. Select “Annotate tag SNP Picker” and press “Configure…” The tag SNPs are selected on the fly as you navigate around the genome.
![8 Adjust Tagger cont Select population Select tagging algorithm and parameters optional upload list 8: Adjust Tagger (cont) Select population Select tagging algorithm and parameters. [optional] upload list](https://slidetodoc.com/presentation_image_h/2348fd5ca46413e7ae466c462cfe6267/image-14.jpg)
8: Adjust Tagger (cont) Select population Select tagging algorithm and parameters. [optional] upload list of SNPs to be included, excluded, or design scores. 8 b. Press “Configure” to save changes.

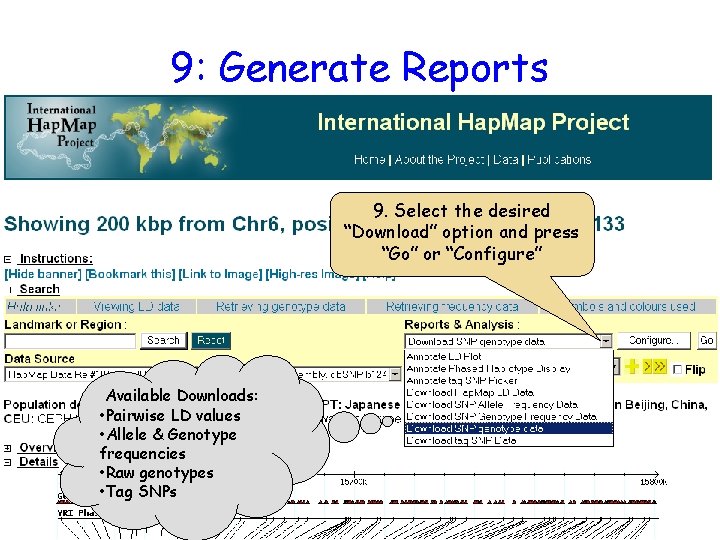
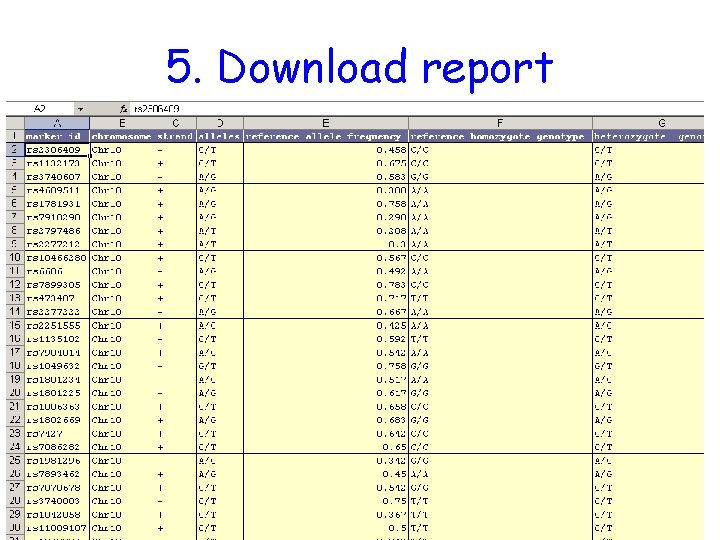
9: Generate Reports 9. Select the desired “Download” option and press “Go” or “Configure” Available Downloads: • Pairwise LD values • Allele & Genotype frequencies • Raw genotypes • Tag SNPs

9: Generate Reports (cont) The Genotype download format can be saved to disk and loaded into Haploview.

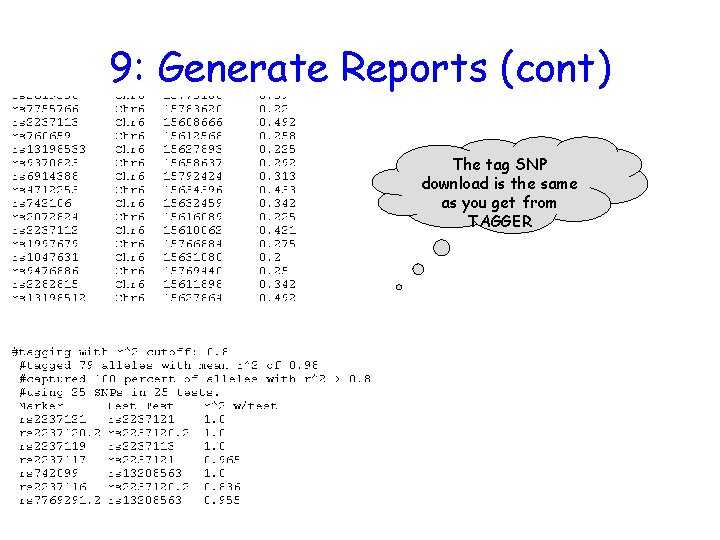
9: Generate Reports (cont) The tag SNP download is the same as you get from TAGGER

Generating Extracts of the Hap. Map Dataset • Find all Hap. Map characterized SNPs that: – 1. Have a MAF > 0. 20. – 2. Cause a nonsynonymous amino acid change.

1. Surf to hapmart. hapmap. org 1. From www. hapmap. org, click on “Hap. Mart”

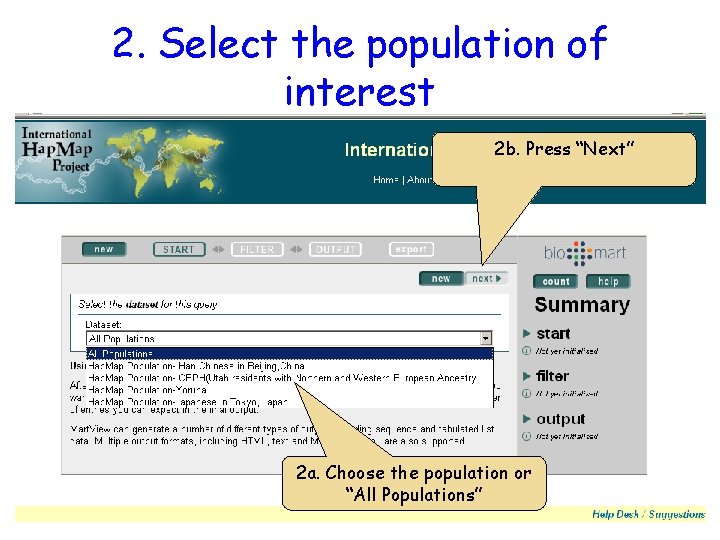
2. Select the population of interest 2 b. Press “Next” 2 a. Choose the population or “All Populations”

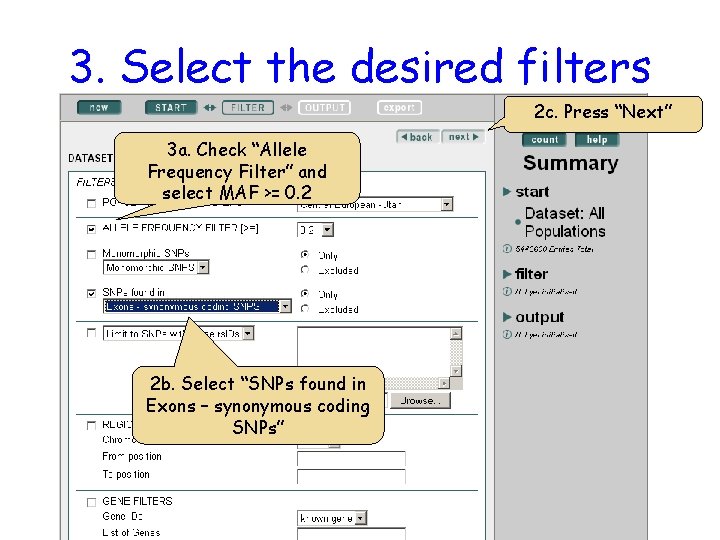
3. Select the desired filters 2 c. Press “Next” 3 a. Check “Allele Frequency Filter” and select MAF >= 0. 2 2 b. Select “SNPs found in Exons – synonymous coding SNPs”

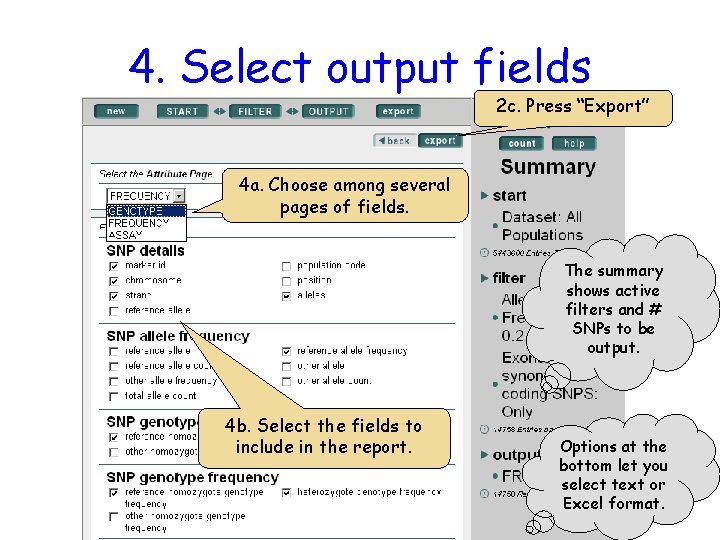
4. Select output fields 2 c. Press “Export” 4 a. Choose among several pages of fields. The summary shows active filters and # SNPs to be output. 4 b. Select the fields to include in the report. Options at the bottom let you select text or Excel format.

5. Download report

Download the Complete Data • Download the entire Hap. Map data set to your own computer

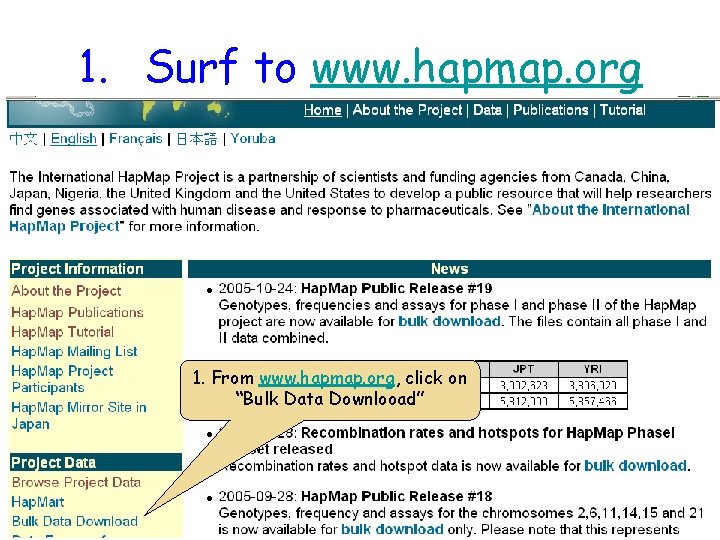
1. Surf to www. hapmap. org 1. From www. hapmap. org, click on “Bulk Data Downlooad”

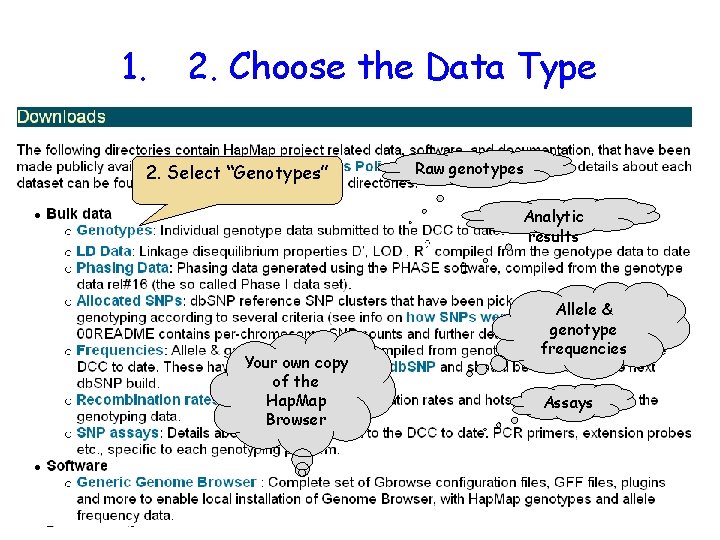
1. 2. Choose the Data Type 2. Select “Genotypes” Raw genotypes Analytic results Your own copy of the Hap. Map Browser Allele & genotype frequencies Assays

1. 3. Choose the Desired Release 3. Select “latest”

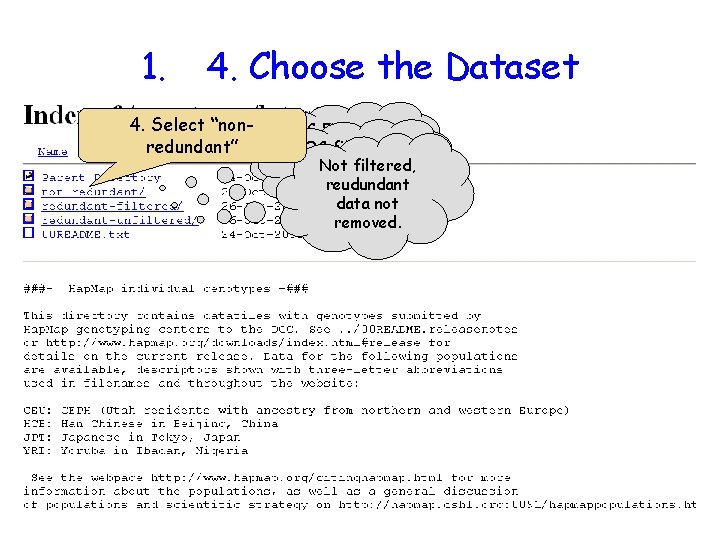
1. 4. Choose the Dataset 4. Select “nonredundant” QC Filtered QCduplicate filtered, with Not filtered, duplicate data removed. notreudundant removed. data not removed.

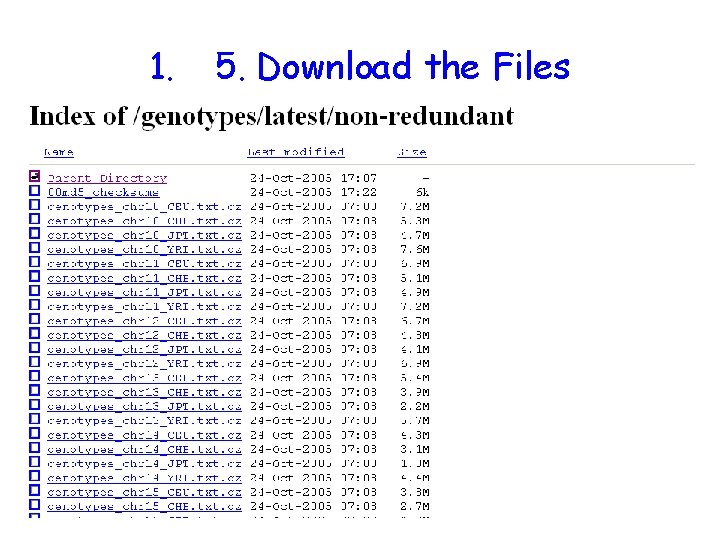
1. 5. Download the Files

For Further Information • Users guide to Hap. Map. org – http: //hapmap. cshl. org/downloads/presentations /users_guide_to_hapmap. pdf • Hap. Map Publications & Guidelines – http: //hapmap. cshl. org/publications. html. en