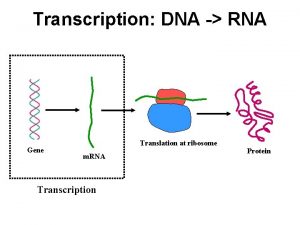
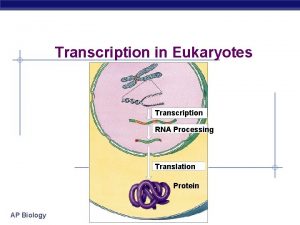
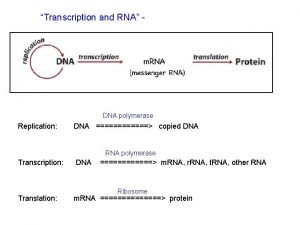
Transcription and Translation 1201 120320 RNA polymerase II














- Slides: 65

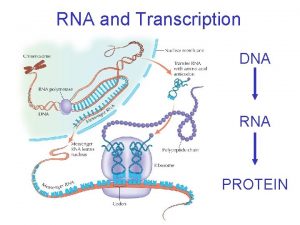
Transcription and Translation 12/01 – 12/03/20


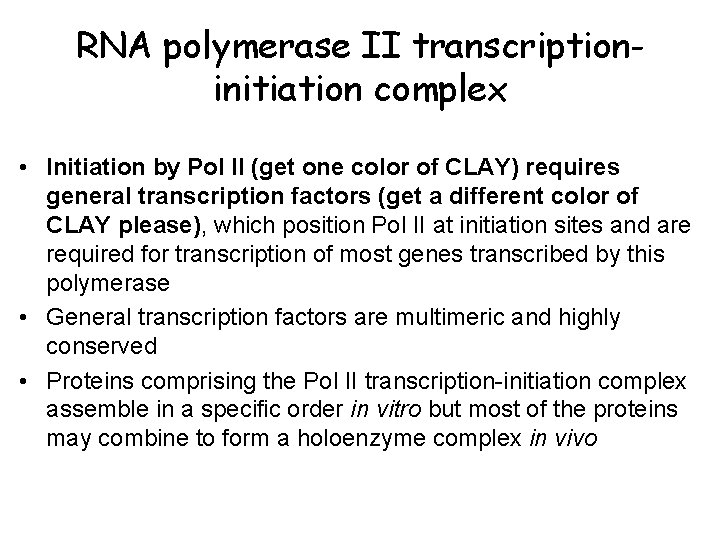
RNA polymerase II transcriptioninitiation complex • Initiation by Pol II (get one color of CLAY) requires general transcription factors (get a different color of CLAY please), which position Pol II at initiation sites and are required for transcription of most genes transcribed by this polymerase • General transcription factors are multimeric and highly conserved • Proteins comprising the Pol II transcription-initiation complex assemble in a specific order in vitro but most of the proteins may combine to form a holoenzyme complex in vivo

Get two new colors of Clay • Color 1 = 5’ to 3’ of DNA • Color 2 = 3’ to 5’ direction of DNA • Go ahead and make your helix!

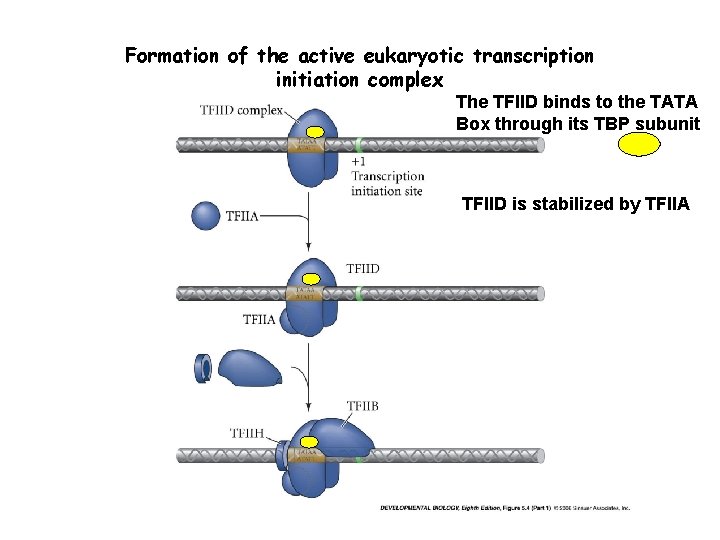
Formation of the active eukaryotic transcription initiation complex The TFIID binds to the TATA Box through its TBP subunit TFIID is stabilized by TFIIA

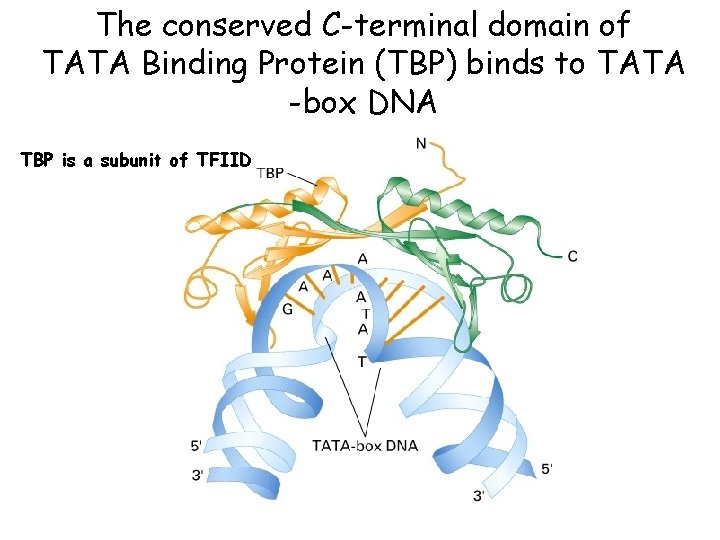
The conserved C-terminal domain of TATA Binding Protein (TBP) binds to TATA -box DNA TBP is a subunit of TFIID

TFIIH Te Cal in rm TATA ng i nd Bi RNA Polymerase II n ai m Do

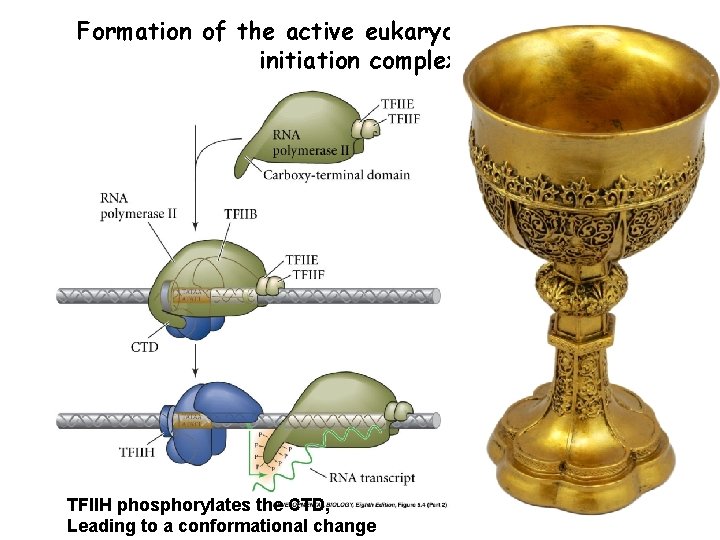
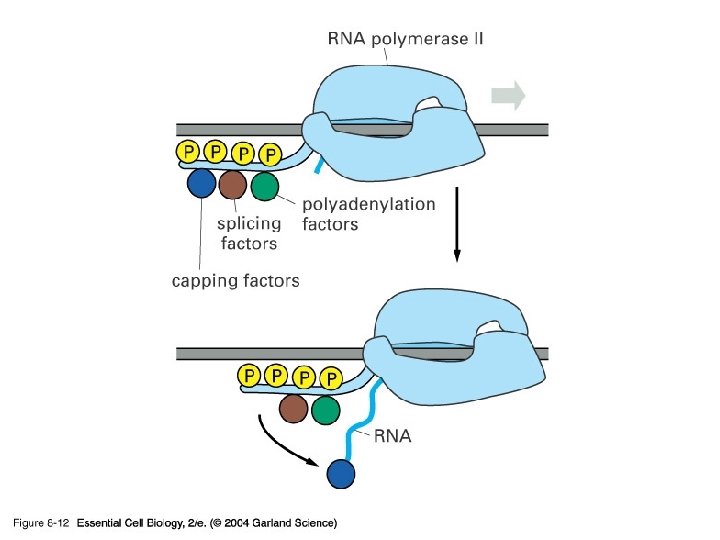
Formation of the active eukaryotic transcription initiation complex TFIIH phosphorylates the CTD, Leading to a conformational change

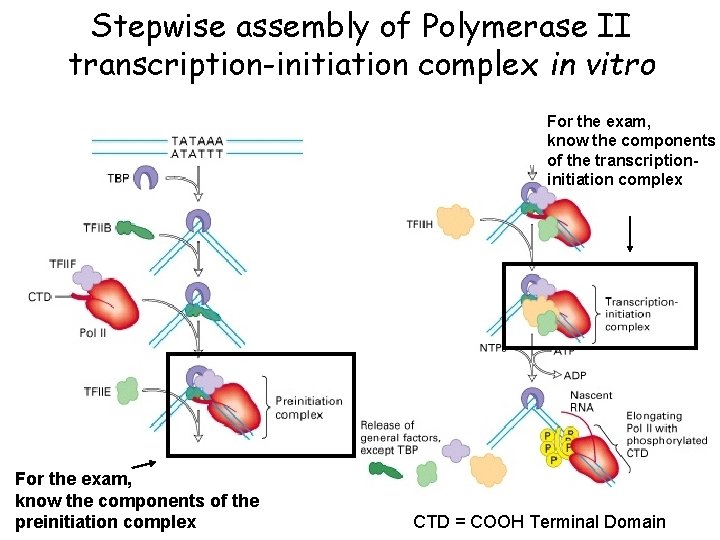
Stepwise assembly of Polymerase II transcription-initiation complex in vitro For the exam, know the components of the transcriptioninitiation complex For the exam, know the components of the preinitiation complex CTD = COOH Terminal Domain

08_13_gene. activation. jpg

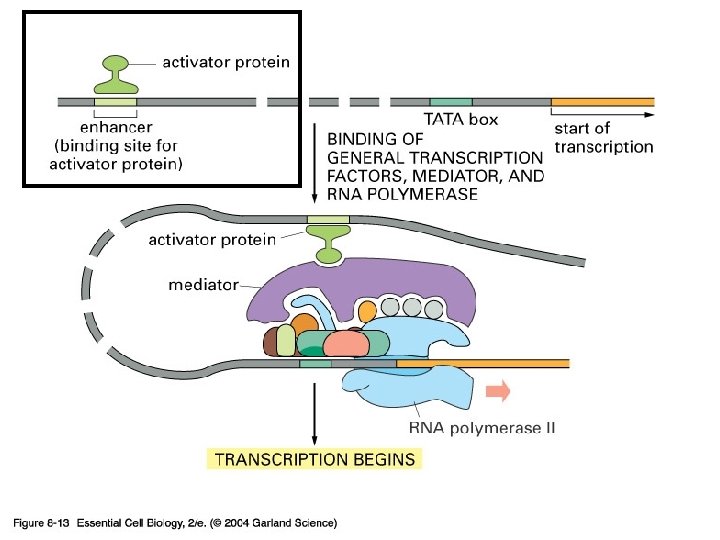
Don’t need to know this for the exam, just appreciate that: Multiprotein complexes form on enhancers

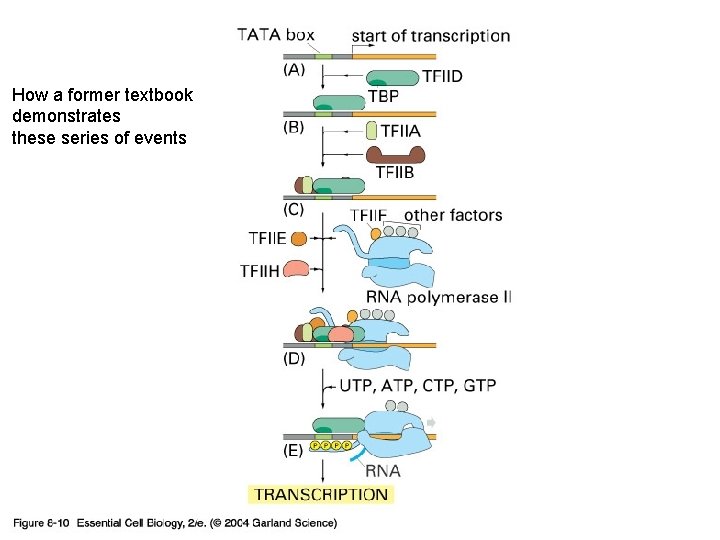
08_10_transcr. factors. jpg How a former textbook demonstrates these series of events

08_12_Phosphoryltn. jpg

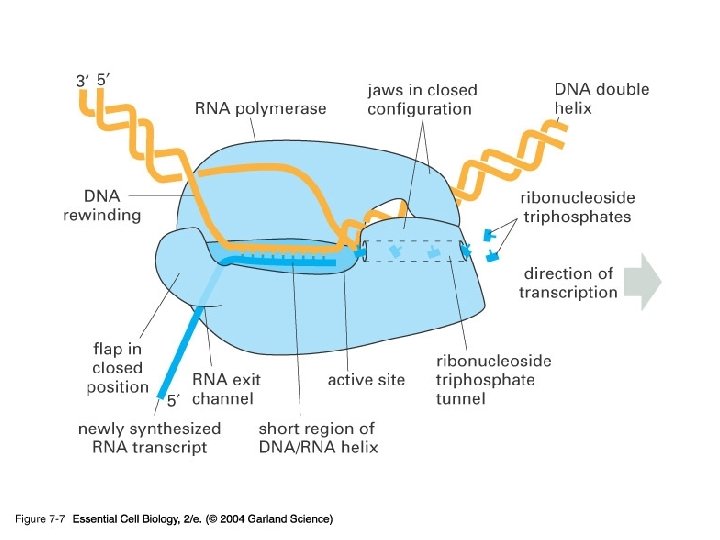
07_07_RNApolymer. jpg

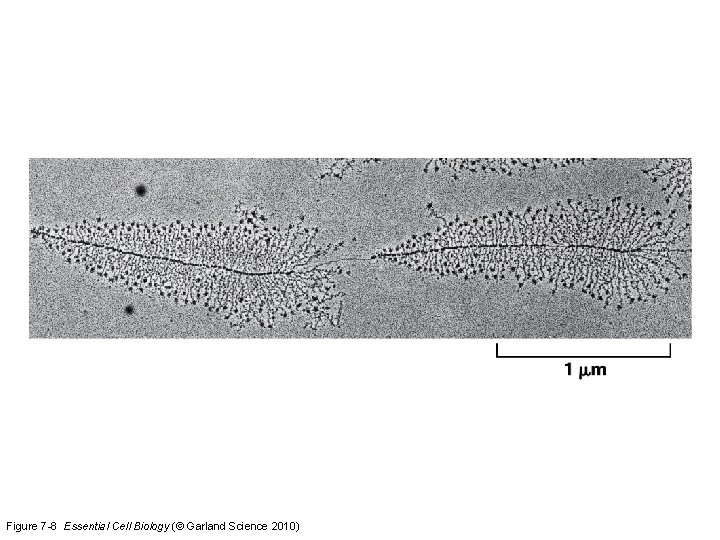
Figure 7 -8 Essential Cell Biology (© Garland Science 2010)

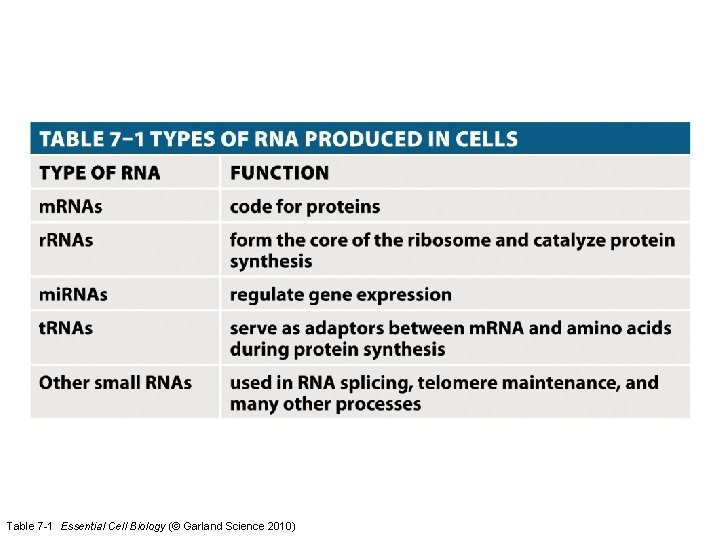
Table 7 -1 Essential Cell Biology (© Garland Science 2010)


Movie Time……Transcription Website: http: //www. youtube. com/watch? v=Wsof. H 466 lqk&feature=related


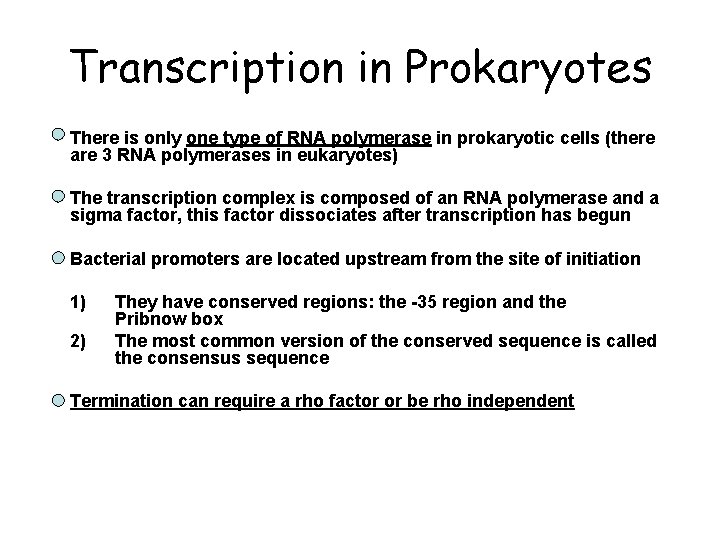
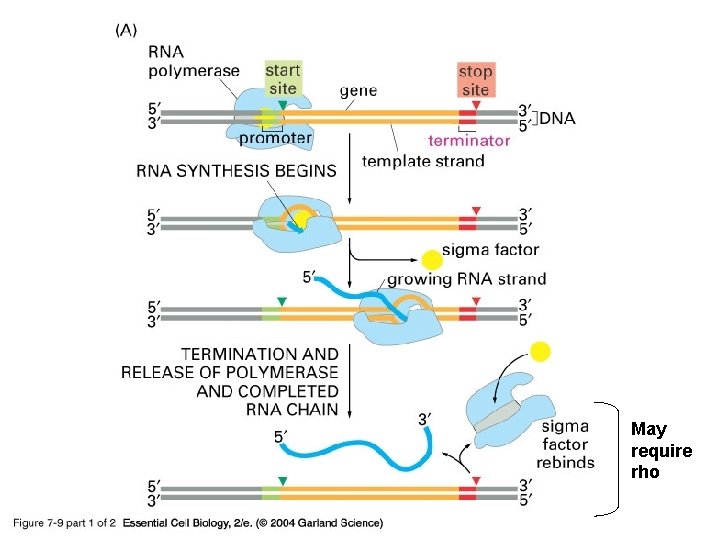
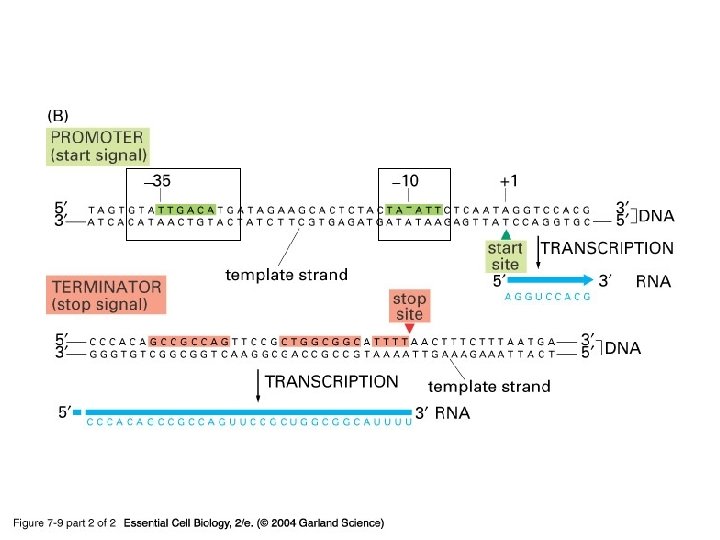
Transcription in Prokaryotes • There is only one type of RNA polymerase in prokaryotic cells (there are 3 RNA polymerases in eukaryotes) • The transcription complex is composed of an RNA polymerase and a sigma factor, this factor dissociates after transcription has begun • Bacterial promoters are located upstream from the site of initiation 1) 2) They have conserved regions: the -35 region and the Pribnow box The most common version of the conserved sequence is called the consensus sequence Termination can require a rho factor or be rho independent

07_09_1_bacterial gene. jpg May require rho

07_09_2_bacterial gene. jpg

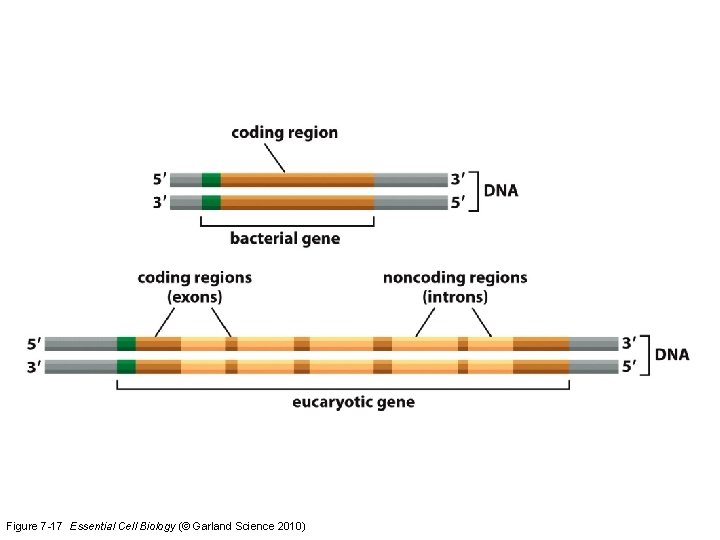
Figure 7 -17 Essential Cell Biology (© Garland Science 2010)

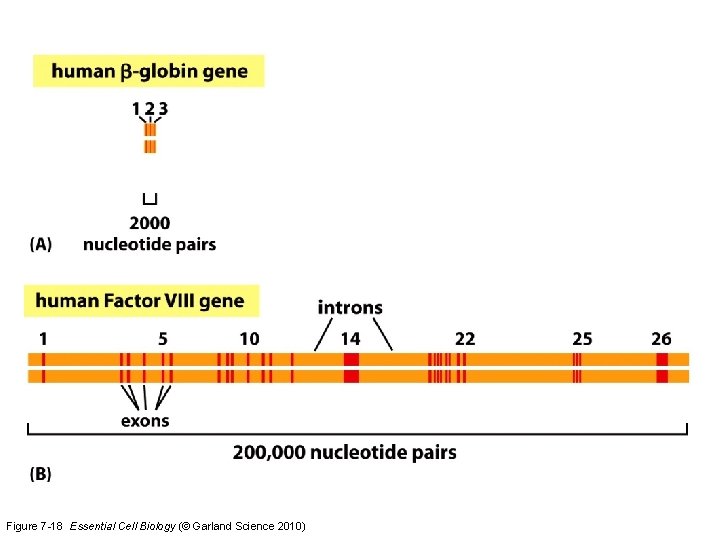
Figure 7 -18 Essential Cell Biology (© Garland Science 2010)

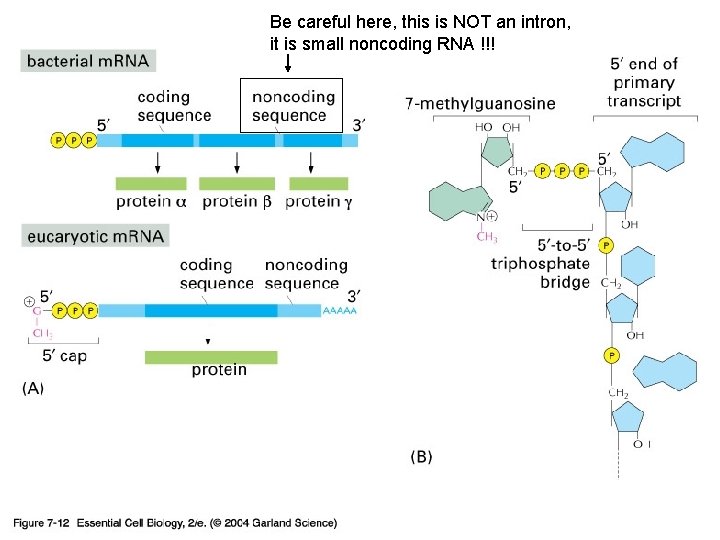
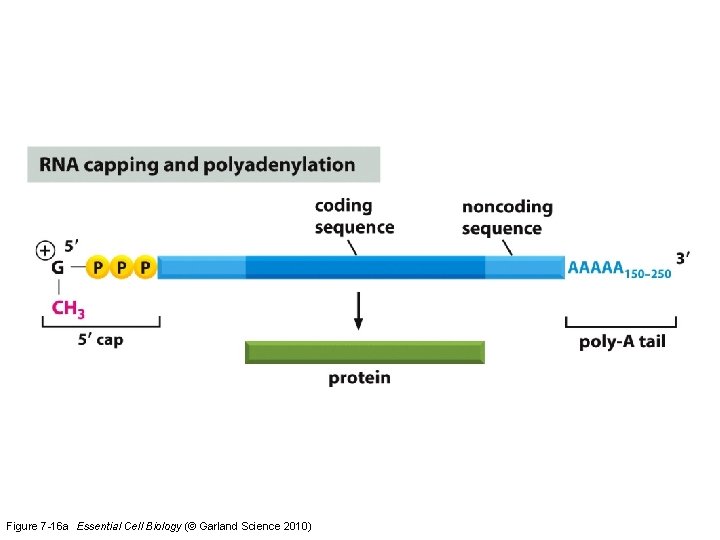
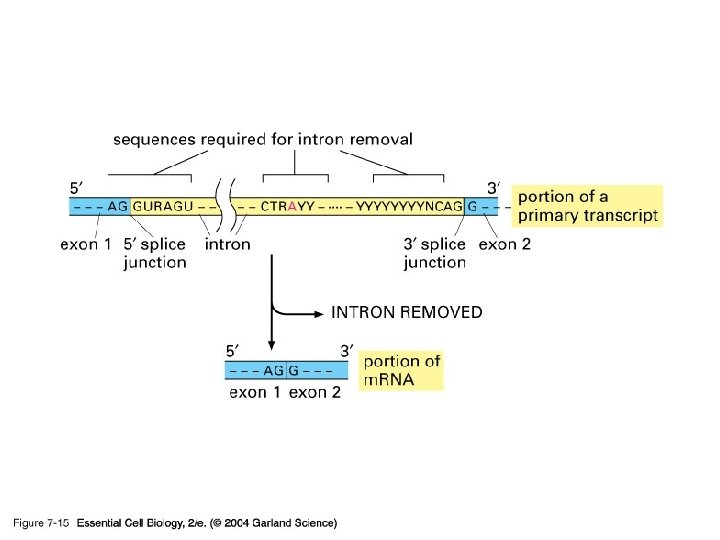
Eukaryotic transcripts need further processing to leave the nucleus…

Be careful here, this is NOT an intron, it is small noncoding RNA !!! 07_12_capping. jpg

Figure 7 -16 a Essential Cell Biology (© Garland Science 2010)

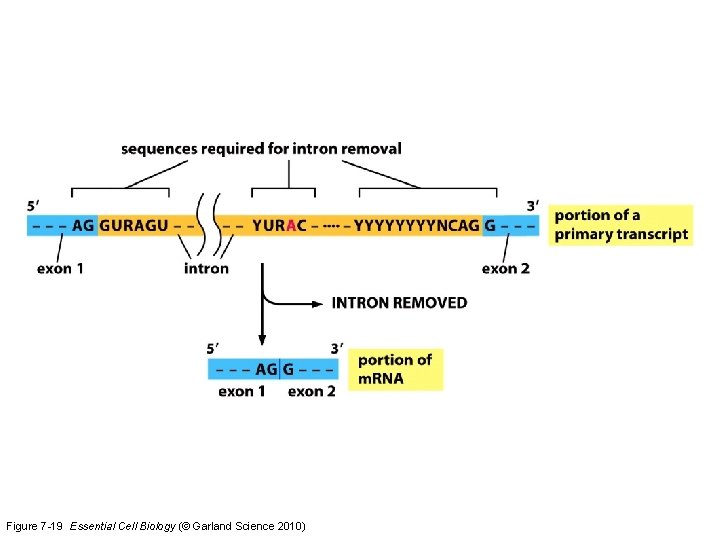
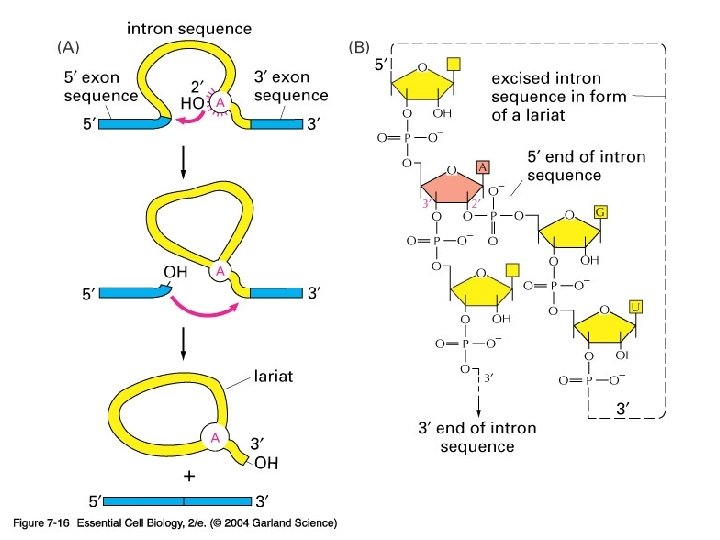
Okay, eukaryotic m. RNA needs to be processed further…. Splicing time to remove introns

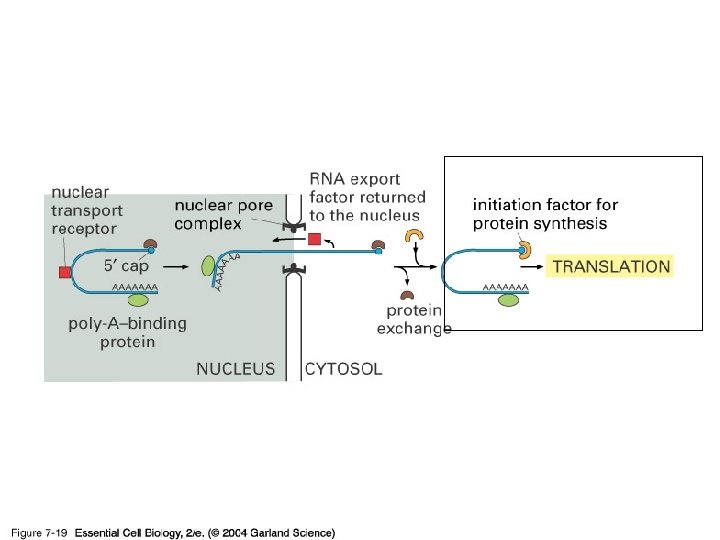
Figure 7 -19 Essential Cell Biology (© Garland Science 2010)

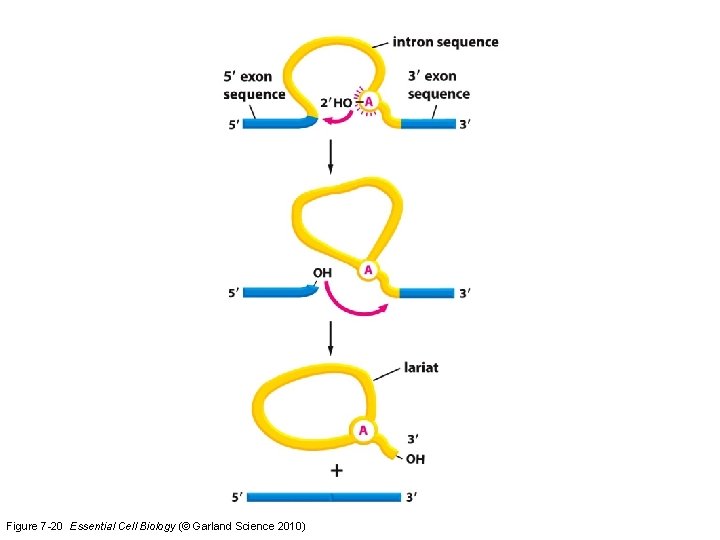
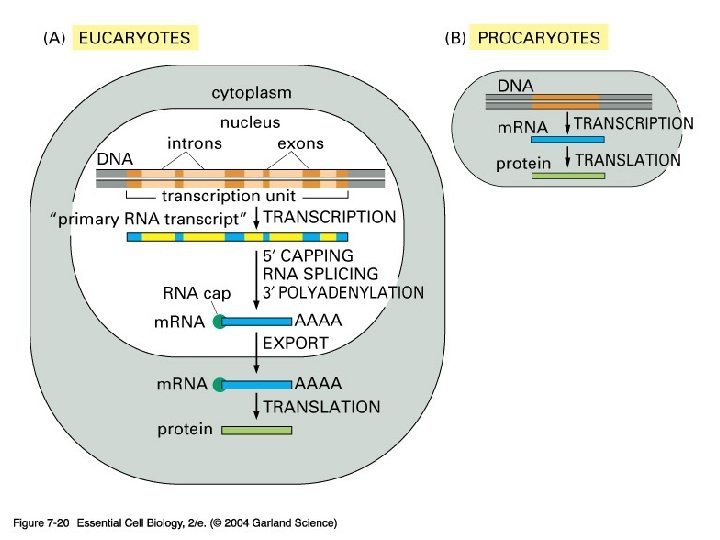
Figure 7 -20 Essential Cell Biology (© Garland Science 2010)

07_15_end_intron. jpg

07_16_RNA_chain. jpg

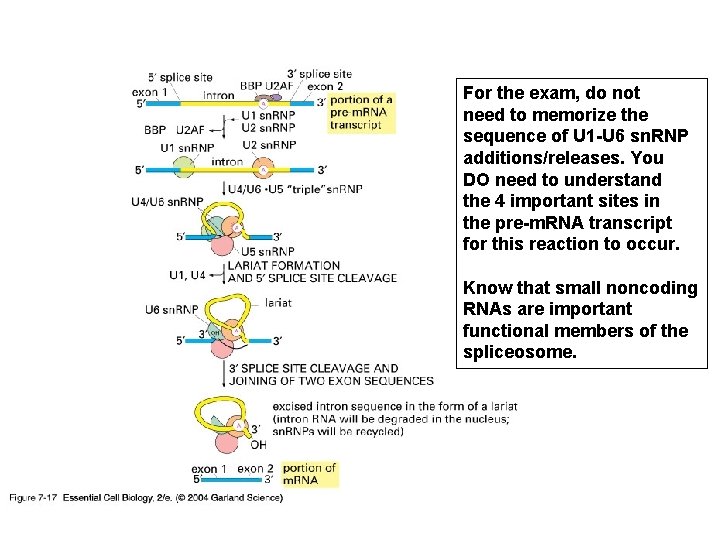
For the exam, do not need to memorize the sequence of U 1 -U 6 sn. RNP additions/releases. You DO need to understand the 4 important sites in the pre-m. RNA transcript for this reaction to occur. Know that small noncoding RNAs are important functional members of the spliceosome.

Spliceosome

Movie Time: The Spliceosome http: //www. youtube. com/watch? v=FVu. Aw. BGw_p. Q And Garland Science Video to show function of sn. RNPs


For more videos please see http: //vcell. ndsu. edu/animations This is a great site showcasing many videos of the virtual cell Enjoy!!!

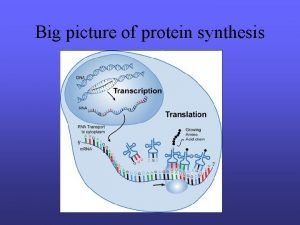
Translation

07_19_export_cytop. jpg

07_20_Pro_v_Eucar. jpg

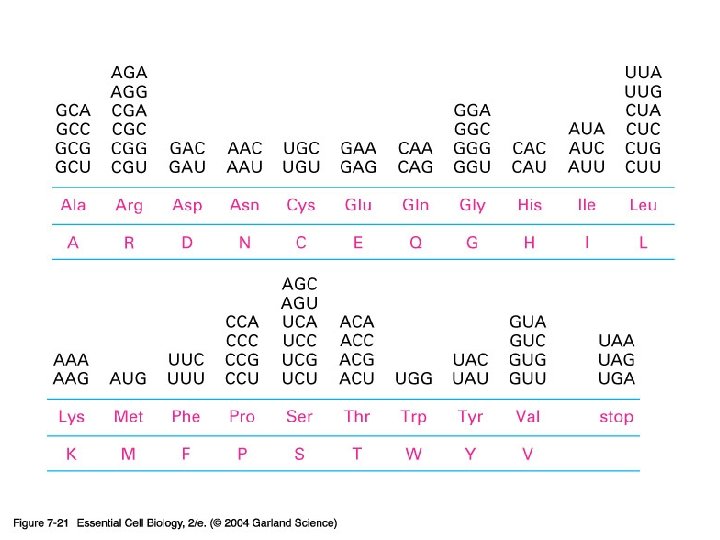
07_21_nucl_sequence. jpg

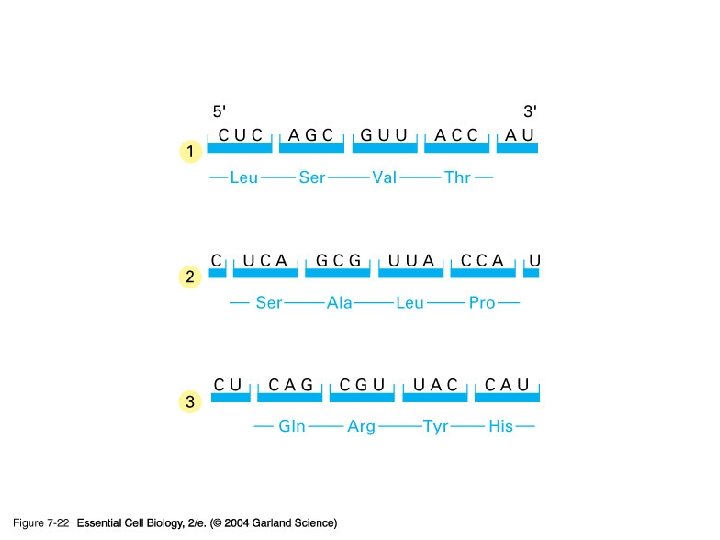
07_22_readingframes. jpg

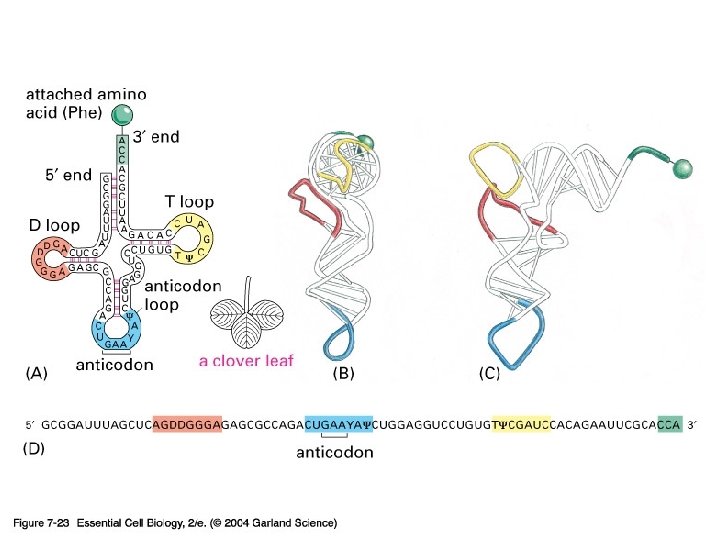
07_23_t. RNA. jpg

What molecules are needed for translation? m. RNA Transfer RNA (t. RNA) Ribosomes A good attitude! You always need this!

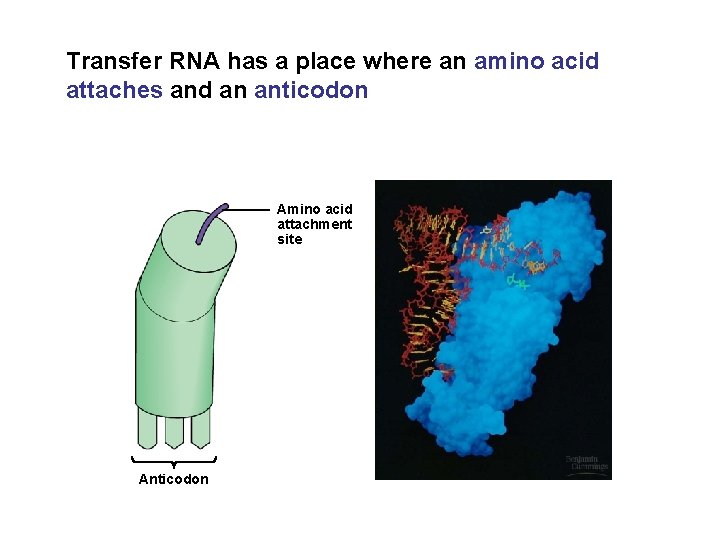
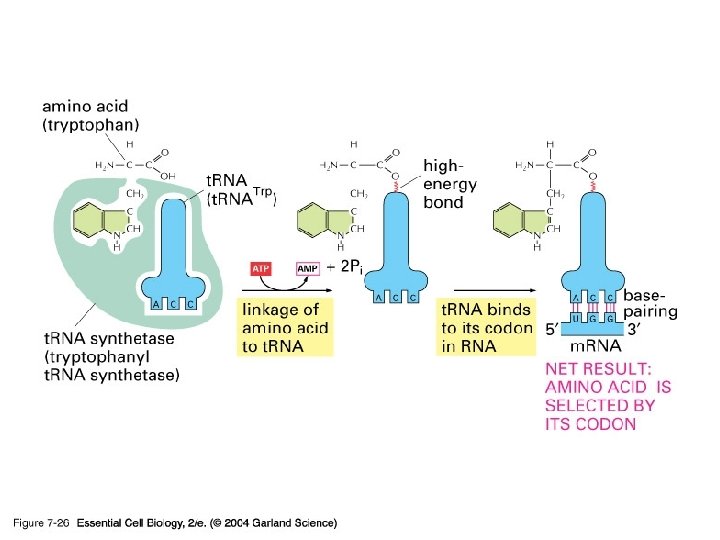
Transfer RNA has a place where an amino acid attaches and an anticodon Amino acid attachment site Anticodon

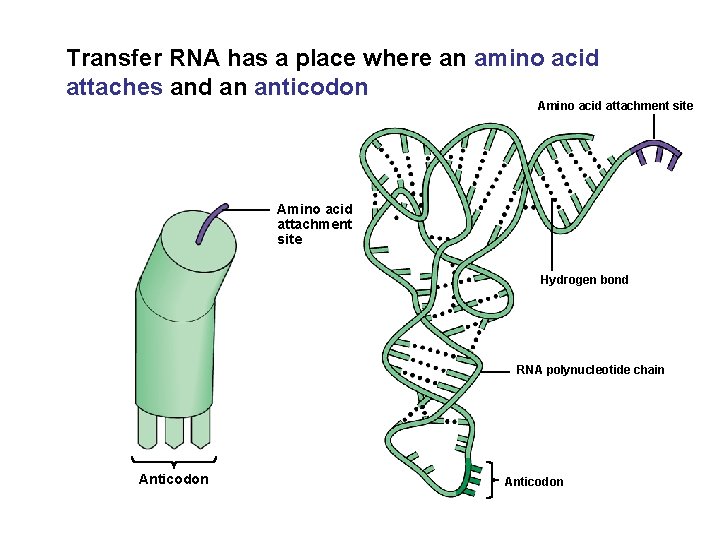
Transfer RNA has a place where an amino acid attaches and an anticodon Amino acid attachment site Hydrogen bond RNA polynucleotide chain Anticodon

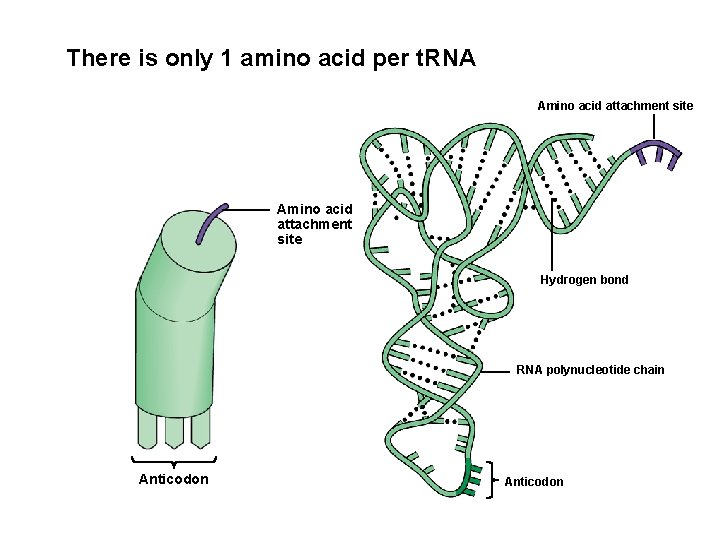
There is only 1 amino acid per t. RNA Amino acid attachment site Hydrogen bond RNA polynucleotide chain Anticodon

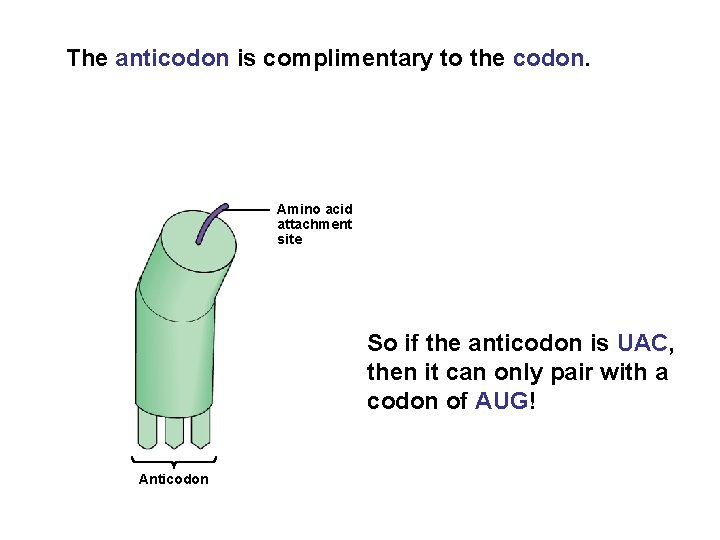
The anticodon is complimentary to the codon. Amino acid attachment site So if the anticodon is UAC, then it can only pair with a codon of AUG! Anticodon


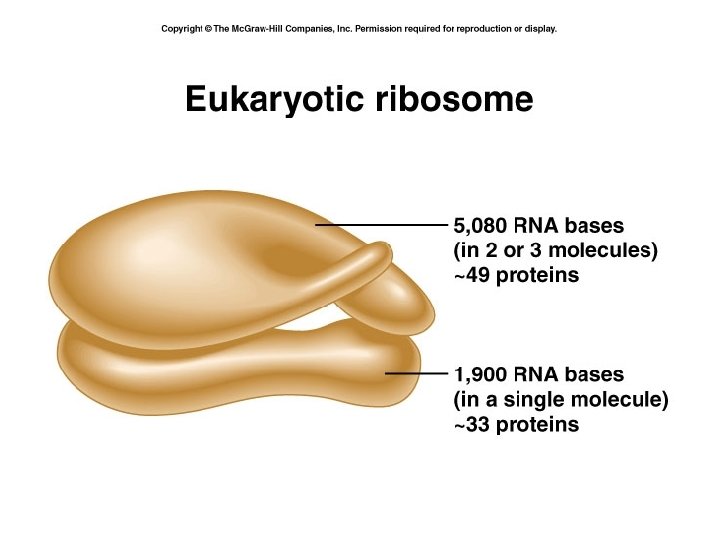
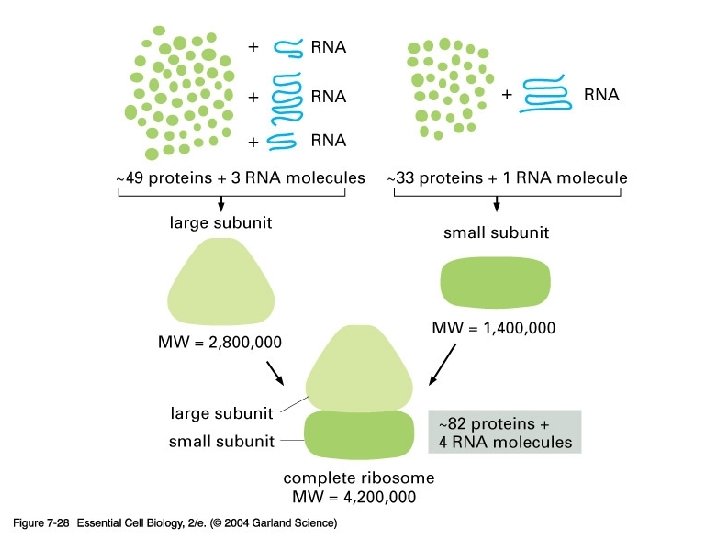
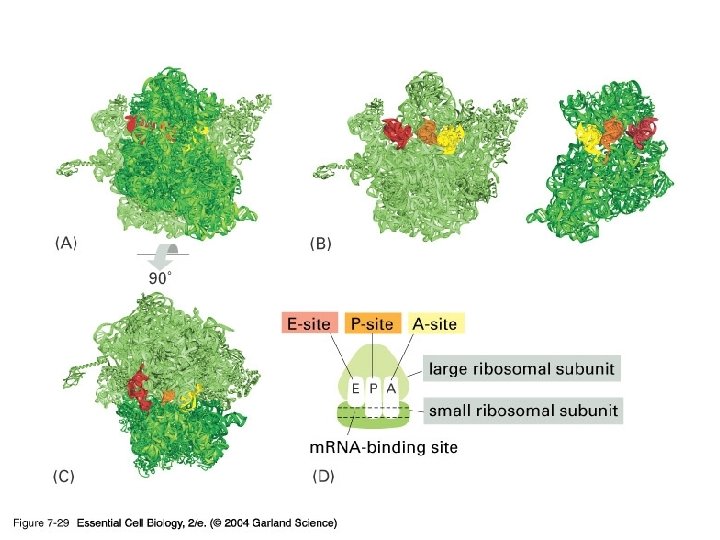
07_28_ribosome. jpg

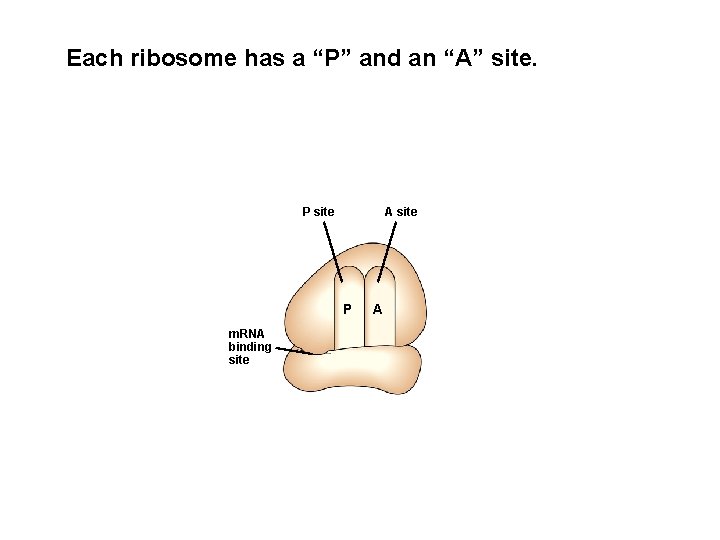
Each ribosome has a “P” and an “A” site. P site A site P m. RNA binding site A

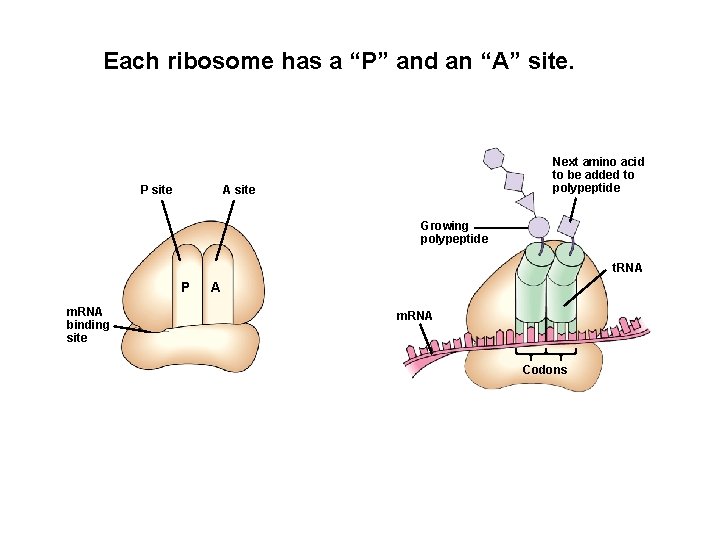
Each ribosome has a “P” and an “A” site. P site Next amino acid to be added to polypeptide A site Growing polypeptide t. RNA P m. RNA binding site A m. RNA Codons

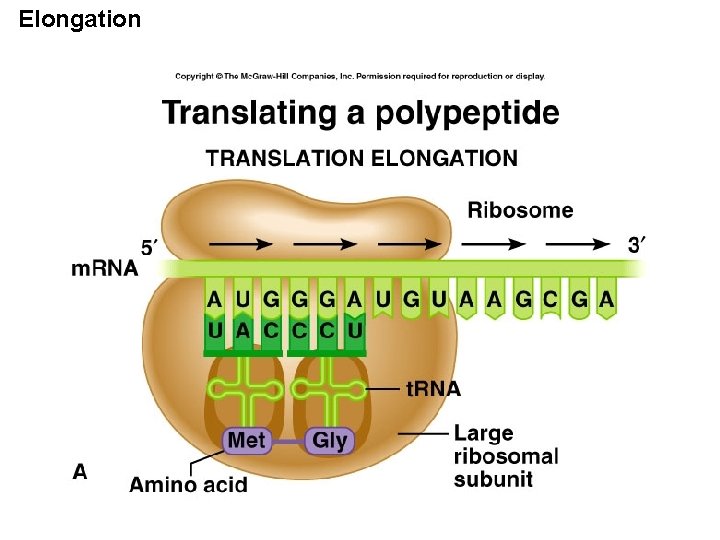
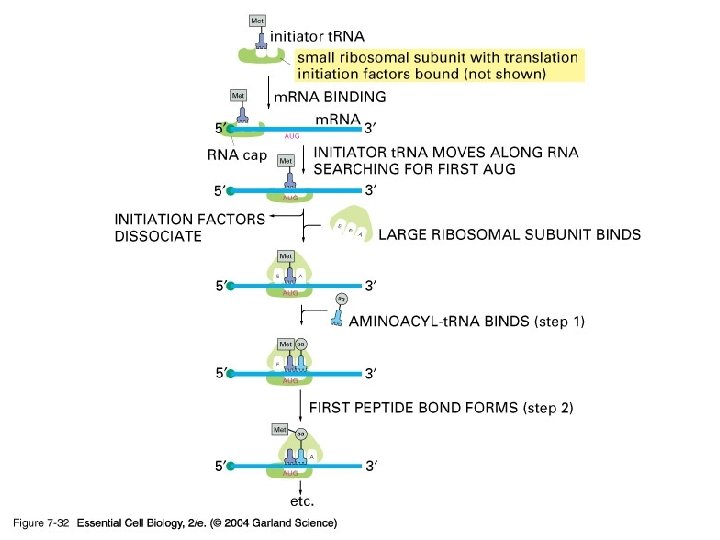
Translation consists of three steps Initiation Elongation Termination

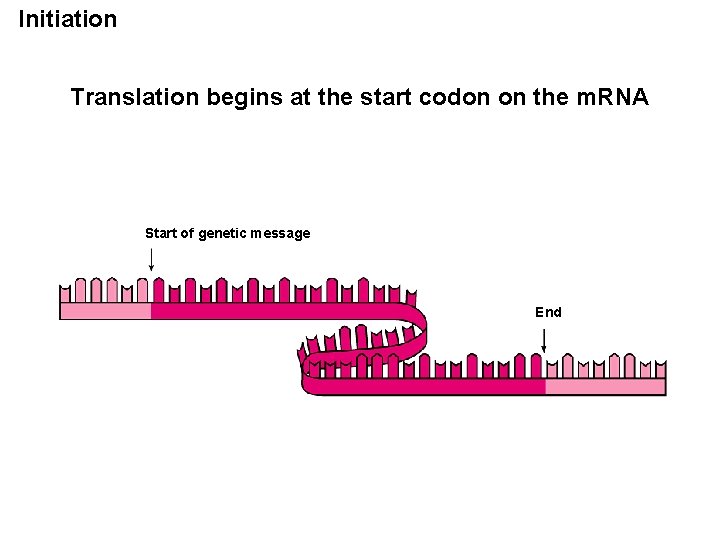
Initiation Translation begins at the start codon on the m. RNA Start of genetic message End

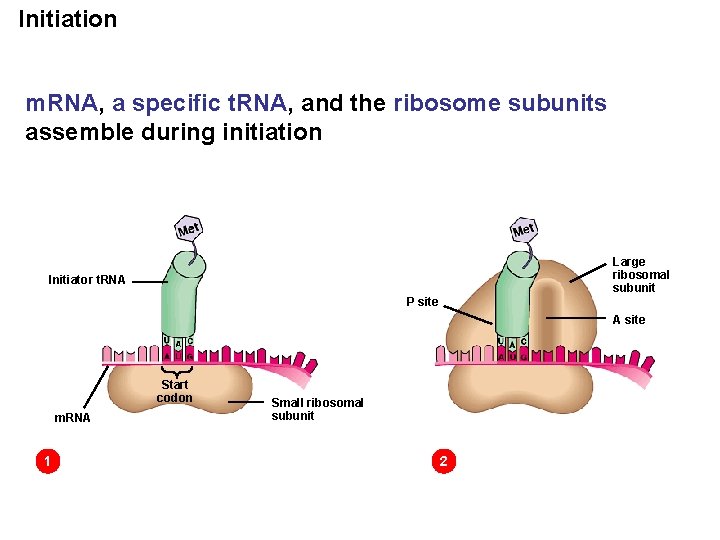
Initiation m. RNA, a specific t. RNA, and the ribosome subunits assemble during initiation Large ribosomal subunit Initiator t. RNA P site A site Start codon m. RNA 1 Small ribosomal subunit 2

07_29_binding. site. jpg

07_26_2_adaptors. jpg

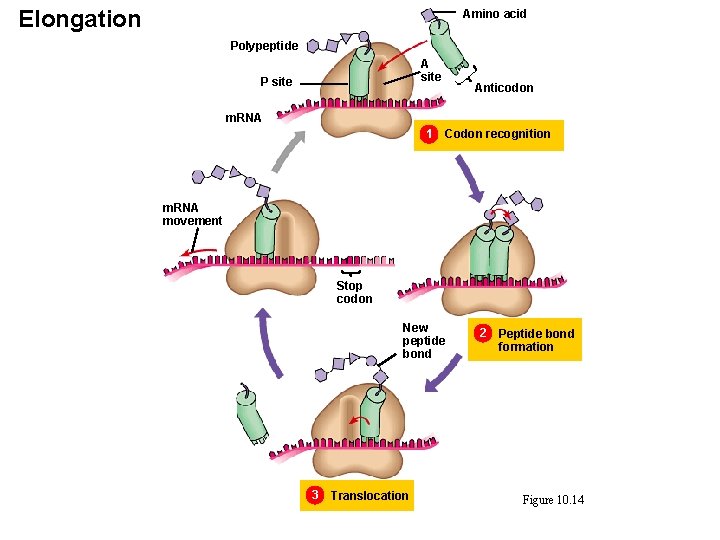
Elongation

Elongation Amino acid Polypeptide A site P site Anticodon m. RNA 1 Codon recognition m. RNA movement Stop codon New peptide bond 3 Translocation 2 Peptide bond formation Figure 10. 14

07_32_initiation. jpg

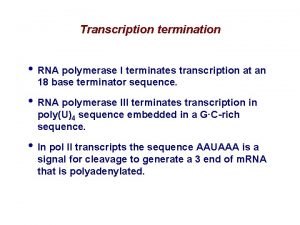
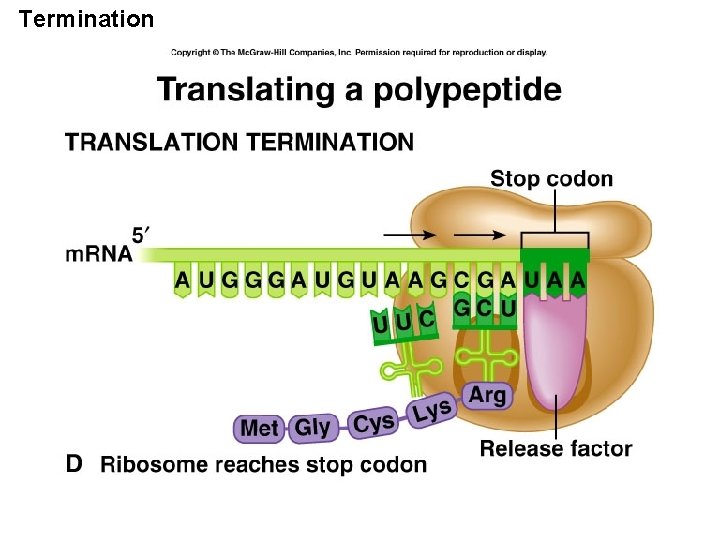
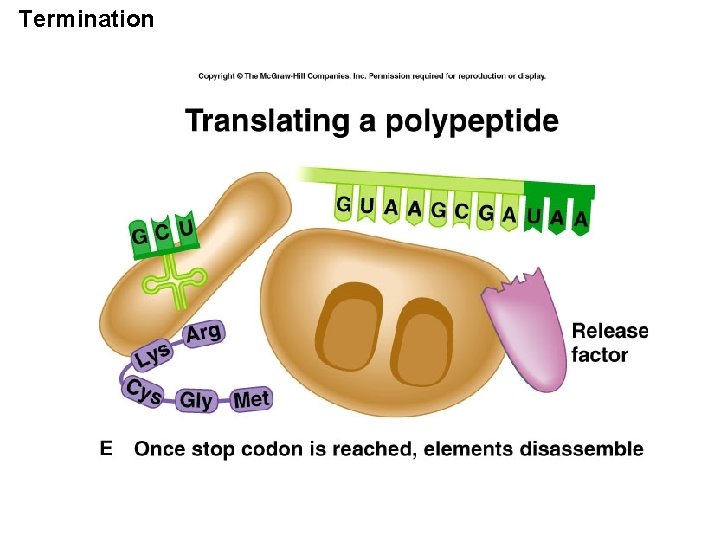
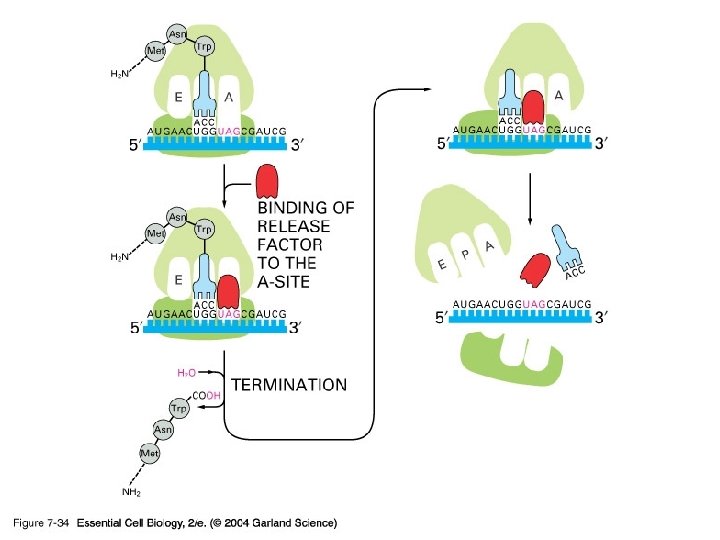
Termination

Termination

07_34_stop codon. jpg


Translation http: //www. youtube. com/watch? v=5 b. LEDd-PSTQ
 Rna polymerase 1 2 3
Rna polymerase 1 2 3 Transcription initiation in eukaryotes
Transcription initiation in eukaryotes Rna polymerase
Rna polymerase Ribosomem
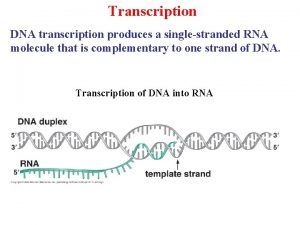
Ribosomem Dna to rna transcription
Dna to rna transcription Rna transcription
Rna transcription Dna to rna transcription
Dna to rna transcription Transcription and translation coloring
Transcription and translation coloring Overview of transcription and translation
Overview of transcription and translation Protein synthesis bbc bitesize
Protein synthesis bbc bitesize Dna transcription and translation
Dna transcription and translation Dna replication transcription and translation
Dna replication transcription and translation Translation protein synthesis
Translation protein synthesis Picture of protein synthesis
Picture of protein synthesis Transcription and translation
Transcription and translation Biology transcription and translation
Biology transcription and translation Rna matching
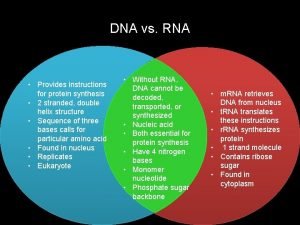
Rna matching What are the differences of dna and rna
What are the differences of dna and rna Messenger rna sequence
Messenger rna sequence 1201 broad rock blvd richmond va 23249
1201 broad rock blvd richmond va 23249 English 1201
English 1201 Introduction of the giver
Introduction of the giver Skilled trades 1201
Skilled trades 1201 The blue helmet chapter summary
The blue helmet chapter summary Transcription translation replication
Transcription translation replication Blood type chart
Blood type chart Dna meaning
Dna meaning Translation transcription
Translation transcription Transcription or translation
Transcription or translation Translation transcription
Translation transcription Central dogma
Central dogma Unlike dnarna contains
Unlike dnarna contains Template strand, new strand, base pair, and dna polymerase.
Template strand, new strand, base pair, and dna polymerase. Communicative vs semantic translation
Communicative vs semantic translation Left and right transformations
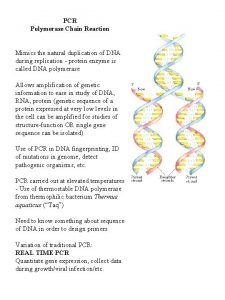
Left and right transformations Applicationof pcr
Applicationof pcr Lagging strand
Lagging strand Dna polymerase iii
Dna polymerase iii Replication
Replication Helicaee
Helicaee How many dna polymerase in eukaryotes
How many dna polymerase in eukaryotes Types of dna polymerase in eukaryotes
Types of dna polymerase in eukaryotes Dna polymerase
Dna polymerase Polymerase chain reaction
Polymerase chain reaction Dna polymerase
Dna polymerase Taqman gene expression assay
Taqman gene expression assay Polymerase chain reaction
Polymerase chain reaction Site:slidetodoc.com
Site:slidetodoc.com Pcr phases
Pcr phases Polilynker
Polilynker Adn polymérase
Adn polymérase Taq dna polymerase
Taq dna polymerase Dna polymerase
Dna polymerase Polymerase chain reaction
Polymerase chain reaction The three steps of polymerase chain reaction
The three steps of polymerase chain reaction What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Voice translation rules
Voice translation rules 10 noun phrases
10 noun phrases How to write 115 on a check
How to write 115 on a check Narrow transcription rules
Narrow transcription rules Dna and transcription tutorial
Dna and transcription tutorial Rna transfer
Rna transfer Places of articulation
Places of articulation Where did hiv come from
Where did hiv come from Speech phonetic transcription
Speech phonetic transcription Prokaryotic vs eukaryotic transcription
Prokaryotic vs eukaryotic transcription