Statistical Genomics Lecture 23 Cross validation Zhiwu Zhang





![Choosing the top ten SNPs ntop=10 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1: Choosing the top ten SNPs ntop=10 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1:](https://slidetodoc.com/presentation_image/1542c4e2ea4201c1fae7d4315a4c18d3/image-8.jpg)
![Prediction with top 200 SNPs ntop=200 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1: Prediction with top 200 SNPs ntop=200 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1:](https://slidetodoc.com/presentation_image/1542c4e2ea4201c1fae7d4315a4c18d3/image-10.jpg)




- Slides: 24

Statistical Genomics Lecture 23: Cross validation Zhiwu Zhang Washington State University

Administration Homework 5, due April 12, Wednesday, 3: 10 PM Final exam: May 4 (Thursday), 120 minutes (3: 105: 10 PM), 50

Course evaluation and response Genomic selection methods with packages in R GS by GWAS rr. BLUP g. BLUP c. BLUP s. BLUP Bayesian: A, B, CPi LASSO

Outline GS by GWAS Over fitting Cross validation K-fold validation Jack knife Re-sampling Two ways of calculating accuracy Bias and correction

Setup GAPIT #source("http: //www. bioconductor. org/bioc. Lite. R") #bioc. Lite("multtest") #install. packages("gplots") #install. packages("scatterplot 3 d")#The downloaded link at: http: //cran. rproject. org/package=scatterplot 3 d library('MASS') # required for ginv library(multtest) library(gplots) library(compiler) #required for cmpfun library("scatterplot 3 d") source("http: //www. zzlab. net/GAPIT/emma. txt") source("http: //www. zzlab. net/GAPIT/gapit_functions. txt")

Import data and simulate phenotype my. GD=read. table(file="http: //zzlab. net/GAPIT/data/mdp_numeric. txt", head=T) my. GM=read. table(file="http: //zzlab. net/GAPIT/data/mdp_SNP_information. txt", head=T) my. CV=read. table(file="http: //zzlab. net/GAPIT/data/mdp_env. txt", head=T) #Simultate 10 QTN on the first half chromosomes X=my. GD[, -1] index 1 to 5=my. GM[, 2]<6 X 1 to 5 = X[, index 1 to 5] taxa=my. GD[, 1] set. seed(99164) GD. candidate=cbind(taxa, X 1 to 5) source("~/Dropbox/GAPIT/Functions/GAPIT. Phenotype. Simulation. R") my. Sim=GAPIT. Phenotype. Simulation(GD=GD. candidate, GM=my. GM[index 1 to 5, ], h 2=. 5, NQ TN=10, effectunit =. 95, QTNDist="normal", CV=my. CV, cveff=c(. 51, . 51)) setwd("~/Desktop/temp")

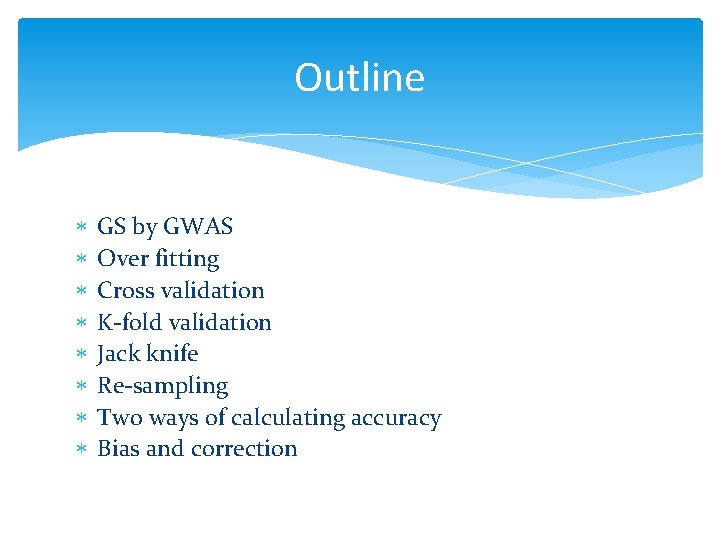
Prediction with PC and ENV my. GAPIT <- GAPIT( Y=my. Sim$Y, GD=my. GD, GM=my. GM, PCA. total=3, CV=my. CV, group. from=1, group. to=1, group. by=10, QTN. position=my. Sim$QTN. position, #SNP. test=FALSE, memo="GLM", ) ry 2=cor(my. GAPIT$Pred[, 8], my. Sim$Y[, 2])^2 ru 2=cor(my. GAPIT$Pred[, 8], my. Sim$u)^2 par(mfrow=c(2, 1), mar = c(3, 4, 1, 1)) plot(my. GAPIT$Pred[, 8], my. Sim$Y[, 2]) mtext(paste("R square=", ry 2, sep=""), side = 3) plot(my. GAPIT$Pred[, 8], my. Sim$u) mtext(paste("R square=", ru 2, sep=""), side = 3)
![Choosing the top ten SNPs ntop10 indexordermy GAPITP topindex1 ntop my QTNcbindmy GAPITPCA 1 Choosing the top ten SNPs ntop=10 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1:](https://slidetodoc.com/presentation_image/1542c4e2ea4201c1fae7d4315a4c18d3/image-8.jpg)
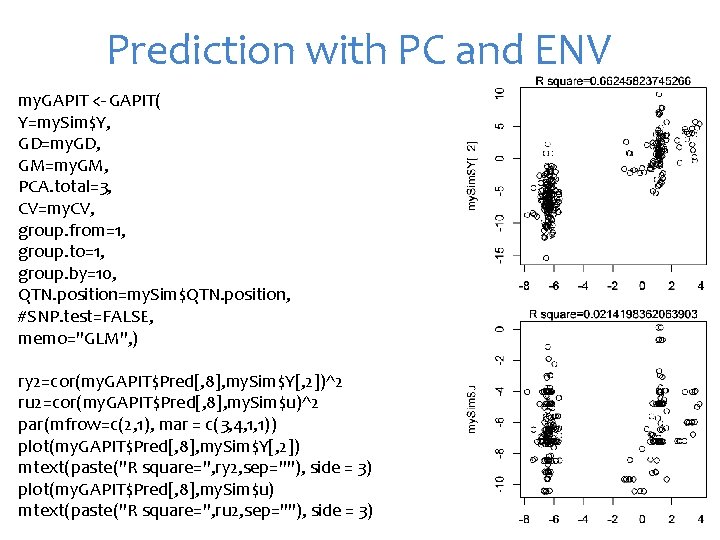
Choosing the top ten SNPs ntop=10 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1: 4], my. CV[, 2: 3], my. GD[, c(top+1)])

Prediction with top ten SNPs my. GAPIT 2<- GAPIT( Y=my. Sim$Y, GD=my. GD, GM=my. GM, #PCA. total=3, CV=my. QTN, group. from=1, group. to=1, group. by=10, QTN. position=my. Sim$QTN. position, SNP. test=FALSE, memo="GLM+QTN", ) d e v o pr Im Im ry 2=cor(my. GAPIT 2$Pred[, 8], my. Sim$Y[, 2])^2 pro ved ru 2=cor(my. GAPIT 2$Pred[, 8], my. Sim$u)^2 par(mfrow=c(2, 1), mar = c(3, 4, 1, 1)) plot(my. GAPIT 2$Pred[, 8], my. Sim$Y[, 2]) mtext(paste("R square=", ry 2, sep=""), side = 3) plot(my. GAPIT 2$Pred[, 8], my. Sim$u) mtext(paste("R square=", ru 2, sep=""), side = 3)
![Prediction with top 200 SNPs ntop200 indexordermy GAPITP topindex1 ntop my QTNcbindmy GAPITPCA 1 Prediction with top 200 SNPs ntop=200 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1:](https://slidetodoc.com/presentation_image/1542c4e2ea4201c1fae7d4315a4c18d3/image-10.jpg)
Prediction with top 200 SNPs ntop=200 index=order(my. GAPIT$P) top=index[1: ntop] my. QTN=cbind(my. GAPIT$PCA[, 1: 4], my. CV[, 2: 3], my. GD[, c(top+1)]) my. GAPIT 2<- GAPIT( Y=my. Sim$Y, GD=my. GD, GM=my. GM, #PCA. total=3, CV=my. QTN, group. from=1, group. to=1, group. by=10, QTN. position=my. Sim$QTN. position, SNP. test=FALSE, memo="GLM+QTN", ) d e v o pr Im No Im pro v e

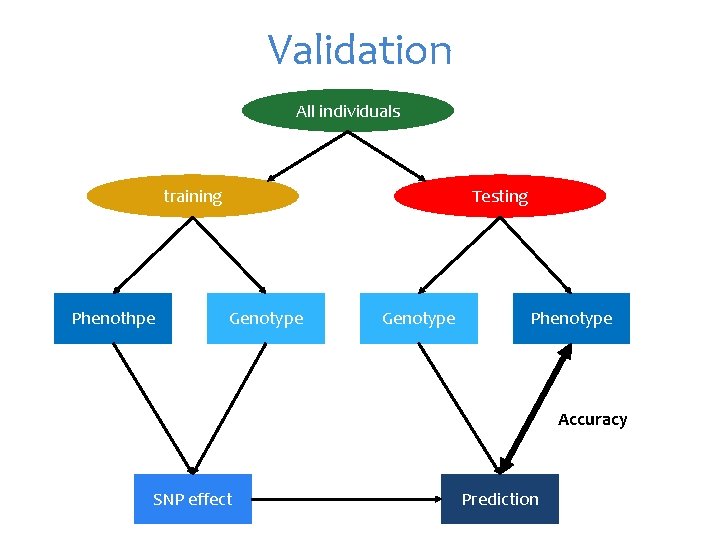
Validation All individuals training Phenothpe Testing Genotype Phenotype Accuracy SNP effect Prediction

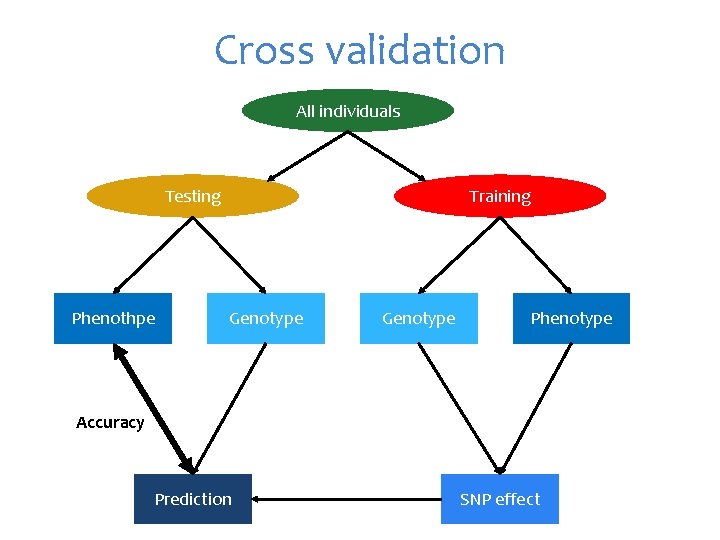
Cross validation All individuals Testing Phenothpe Training Genotype Phenotype Accuracy Prediction SNP effect

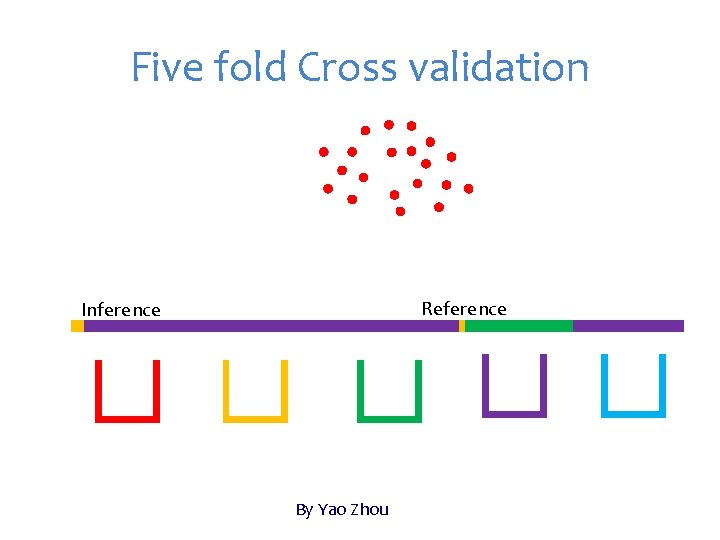
Five fold Cross validation Reference Inference By Yao Zhou

Jack Knife Until every individuals get predicted Inference

Jack Knife: extreme case of K=N N: number of individuals K: number of folds Leave-one-out cross-validation Inference (training) contain only one individuals Not possible to calculate correlation between observed and predicted within inference Evaluation of accuracy must be hold until every individuals receive predictions. Resampling is not available

Re-sampling Sample partial population, e. g. , 20%, as inference (testing), and leave the rest as reference (Training) Instantly evaluate accuracy of inference Repeated for multiple times Average accuracy across replicates Some individuals may never be in the testing

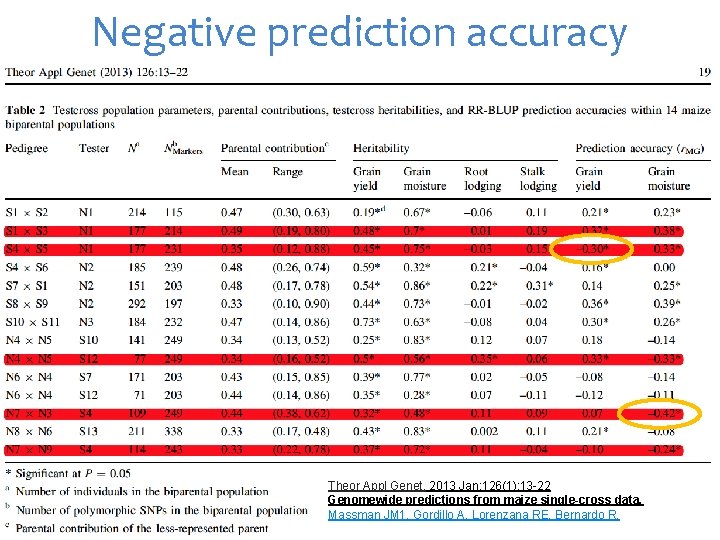
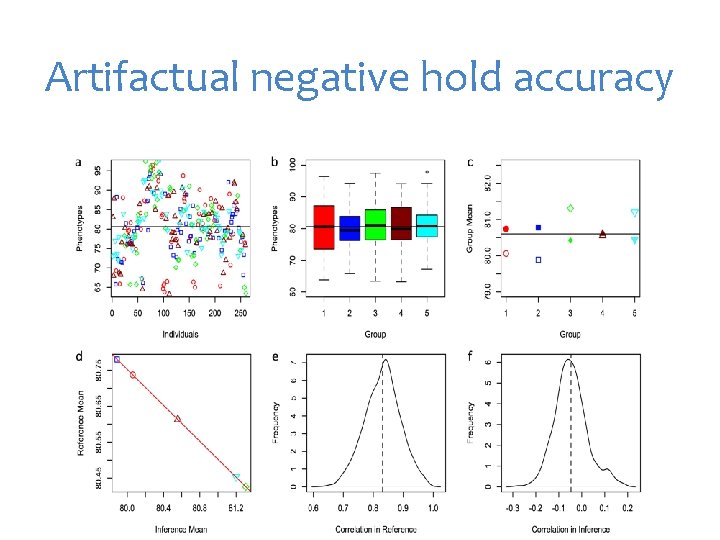
Negative prediction accuracy Theor Appl Genet. 2013 Jan; 126(1): 13 -22 Genomewide predictions from maize single-cross data. Massman JM 1, Gordillo A, Lorenzana RE, Bernardo R.

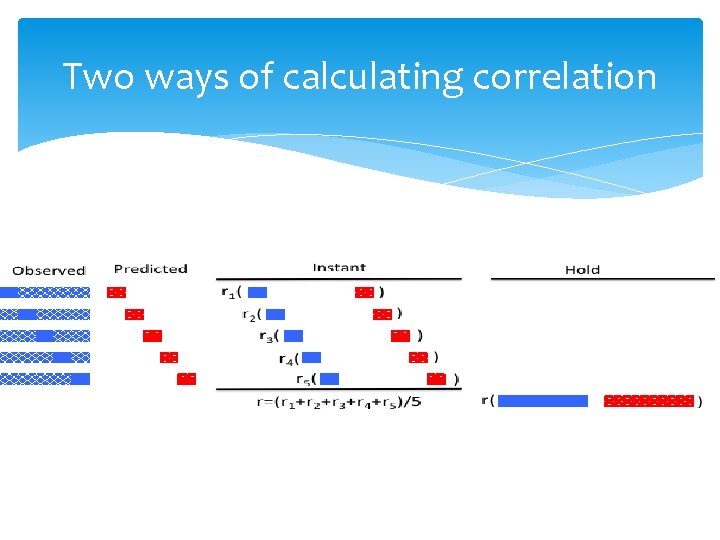
Two ways of calculating correlation

Artifactual negative hold accuracy

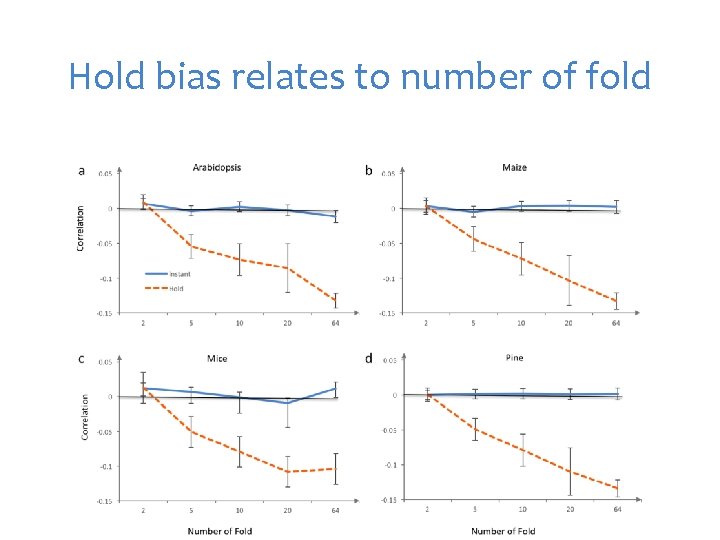
Hold bias relates to number of fold

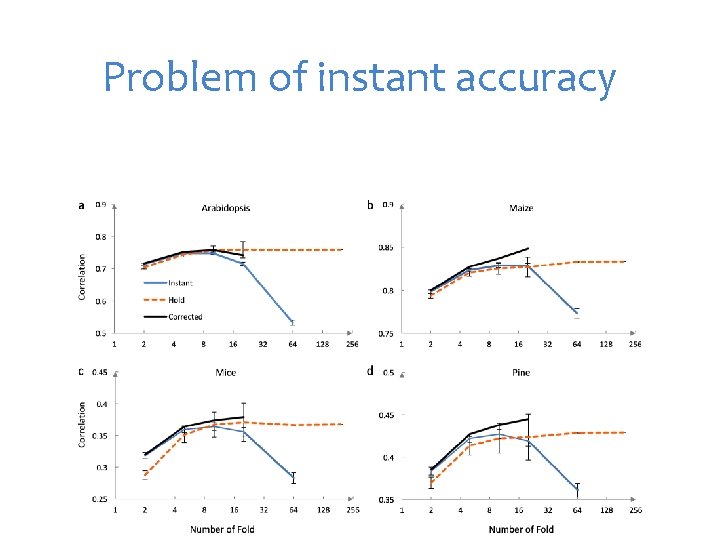
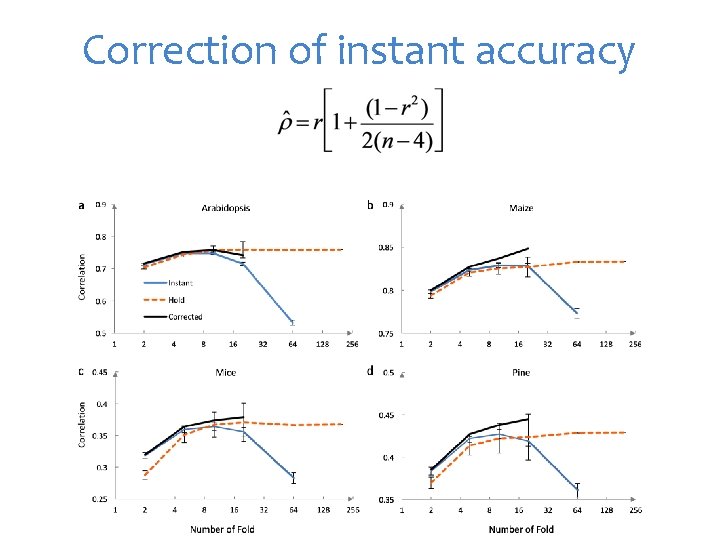
Problem of instant accuracy

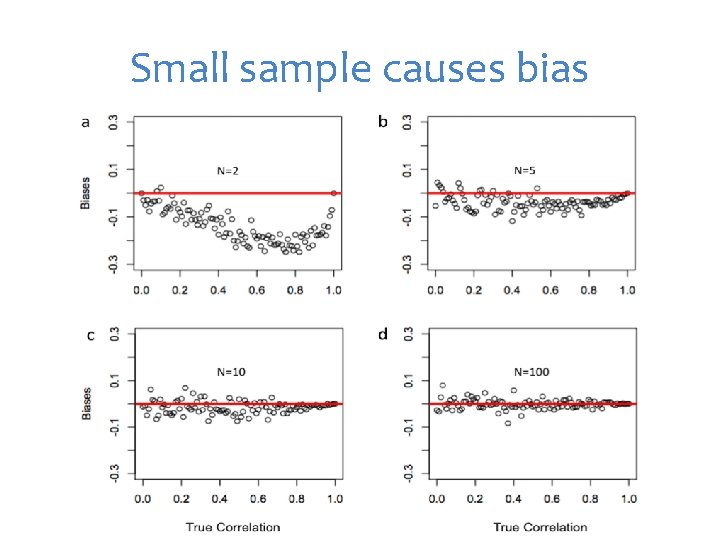
Small sample causes bias

Correction of instant accuracy

Highlight GS by GWAS Over fitting Cross validation K-fold validation Jack knife Re-sampling Two ways of calculating accuracy Bias and correction