Protein Quaternary Fold Recognition Using Conditional Graphical Models
![Conditional Random Fields • Hidden Markov model (HMM) [Rabiner, 1989] • Conditional random fields Conditional Random Fields • Hidden Markov model (HMM) [Rabiner, 1989] • Conditional random fields](https://slidetodoc.com/presentation_image/255be733c05c068a545e989ad7af2fc6/image-7.jpg)
![Experiments: Target Quaternary Fold • Triple beta-spirals [van Raaij et al. Nature 1999] Ø Experiments: Target Quaternary Fold • Triple beta-spirals [van Raaij et al. Nature 1999] Ø](https://slidetodoc.com/presentation_image/255be733c05c068a545e989ad7af2fc6/image-12.jpg)



- Slides: 22

Protein Quaternary Fold Recognition Using Conditional Graphical Models Yan Liu, Jaime Carbonell Vanathi Gopalakrishnan (U Pitt), Peter Weigele (MIT) Language Technologies Institute School of Computer Science Carnegie Mellon University IJCAI-2007 – Hyderabad, India Carnegie Mellon School of Computer Science 1

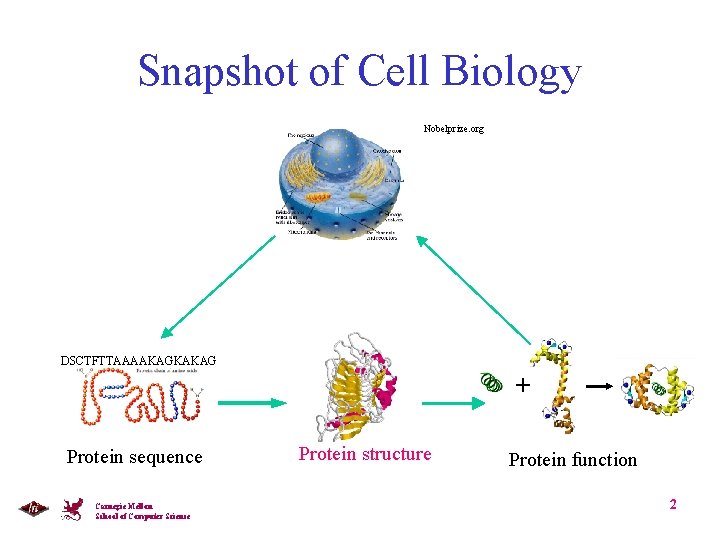
Snapshot of Cell Biology Nobelprize. org DSCTFTTAAAAKAGKAKAG + Protein sequence Carnegie Mellon School of Computer Science Protein structure Protein function 2

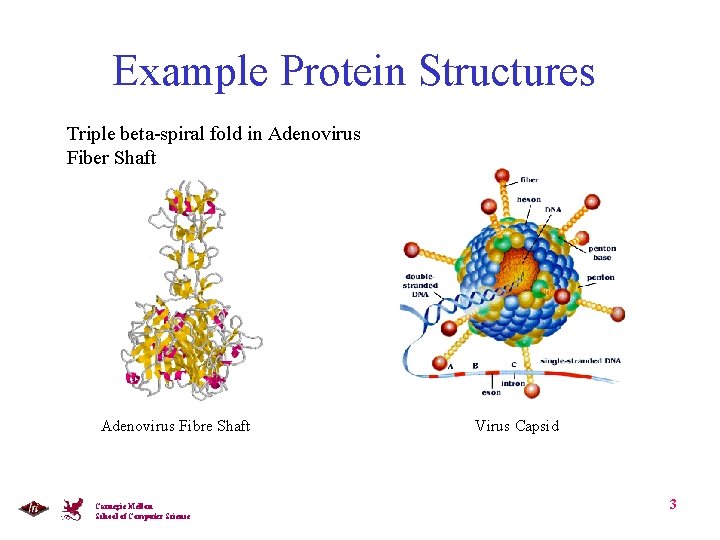
Example Protein Structures Triple beta-spiral fold in Adenovirus Fiber Shaft Adenovirus Fibre Shaft Carnegie Mellon School of Computer Science Virus Capsid 3

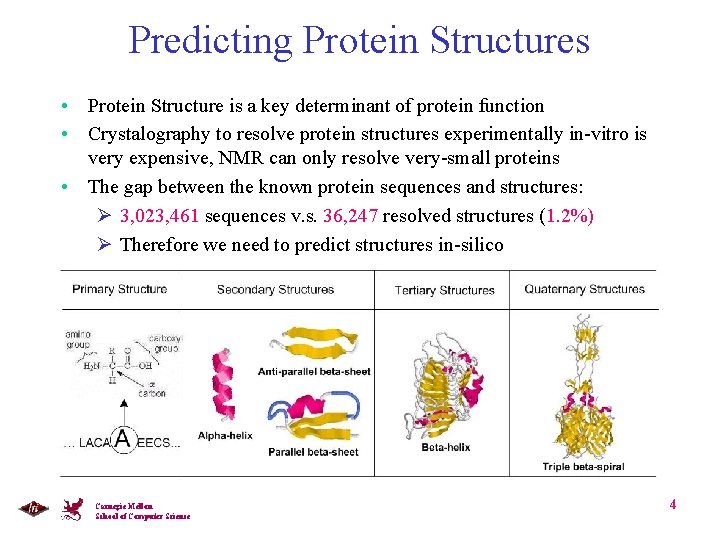
Predicting Protein Structures • Protein Structure is a key determinant of protein function • Crystalography to resolve protein structures experimentally in-vitro is very expensive, NMR can only resolve very-small proteins • The gap between the known protein sequences and structures: Ø 3, 023, 461 sequences v. s. 36, 247 resolved structures (1. 2%) Ø Therefore we need to predict structures in-silico Carnegie Mellon School of Computer Science 4

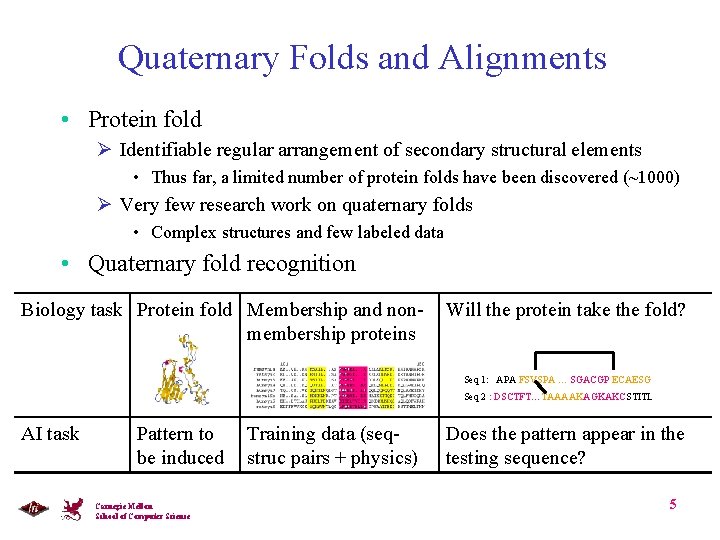
Quaternary Folds and Alignments • Protein fold Ø Identifiable regular arrangement of secondary structural elements • Thus far, a limited number of protein folds have been discovered (~1000) Ø Very few research work on quaternary folds • Complex structures and few labeled data • Quaternary fold recognition Biology task Protein fold Membership and nonmembership proteins Will the protein take the fold? Seq 1: APA FSVSPA … SGACGP ECAESG Seq 2 : DSCTFT…TAAAAKAGKAKCSTITL AI task Pattern to be induced Carnegie Mellon School of Computer Science Training data (seqstruc pairs + physics) Does the pattern appear in the testing sequence? 5

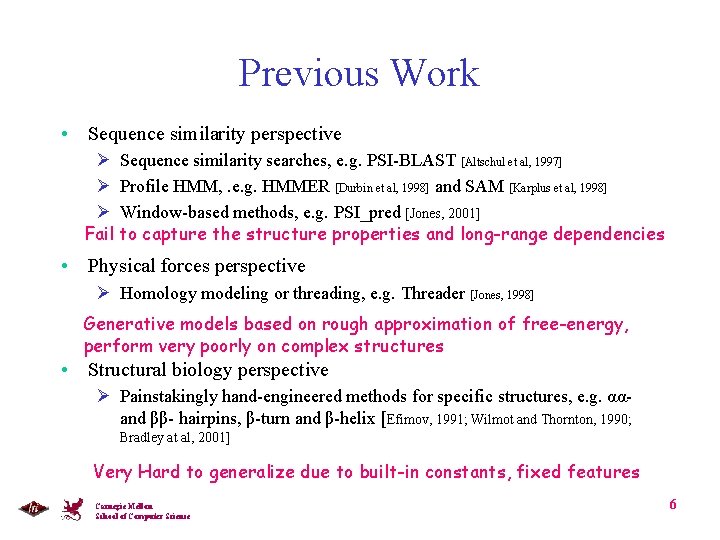
Previous Work • Sequence similarity perspective Ø Sequence similarity searches, e. g. PSI-BLAST [Altschul et al, 1997] Ø Profile HMM, . e. g. HMMER [Durbin et al, 1998] and SAM [Karplus et al, 1998] Ø Window-based methods, e. g. PSI_pred [Jones, 2001] Fail to capture the structure properties and long-range dependencies • Physical forces perspective Ø Homology modeling or threading, e. g. Threader [Jones, 1998] Generative models based on rough approximation of free-energy, perform very poorly on complex structures • Structural biology perspective Ø Painstakingly hand-engineered methods for specific structures, e. g. ααand ββ- hairpins, β-turn and β-helix [Efimov, 1991; Wilmot and Thornton, 1990; Bradley at al, 2001] Very Hard to generalize due to built-in constants, fixed features Carnegie Mellon School of Computer Science 6
![Conditional Random Fields Hidden Markov model HMM Rabiner 1989 Conditional random fields Conditional Random Fields • Hidden Markov model (HMM) [Rabiner, 1989] • Conditional random fields](https://slidetodoc.com/presentation_image/255be733c05c068a545e989ad7af2fc6/image-7.jpg)
Conditional Random Fields • Hidden Markov model (HMM) [Rabiner, 1989] • Conditional random fields (CRFs) [Lafferty et al, 2001] Ø Model conditional probability directly (discriminative models, directly optimizable) Ø Allow arbitrary dependencies in observation Ø Adaptive to different loss functions and regularizers Ø Promising results in multiple applications Ø But, need to scale up (computationally) and extend to longdistance dependencies Carnegie Mellon School of Computer Science 7

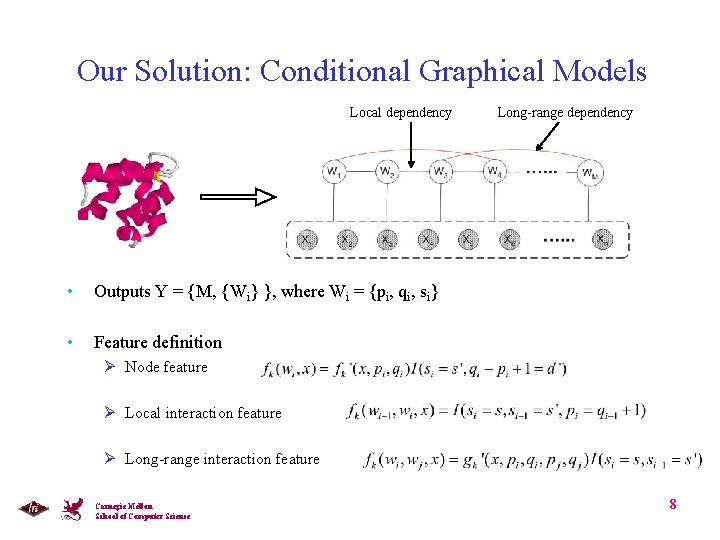
Our Solution: Conditional Graphical Models Local dependency • Outputs Y = {M, {Wi} }, where Wi = {pi, qi, si} • Feature definition Long-range dependency Ø Node feature Ø Local interaction feature Ø Long-range interaction feature Carnegie Mellon School of Computer Science 8

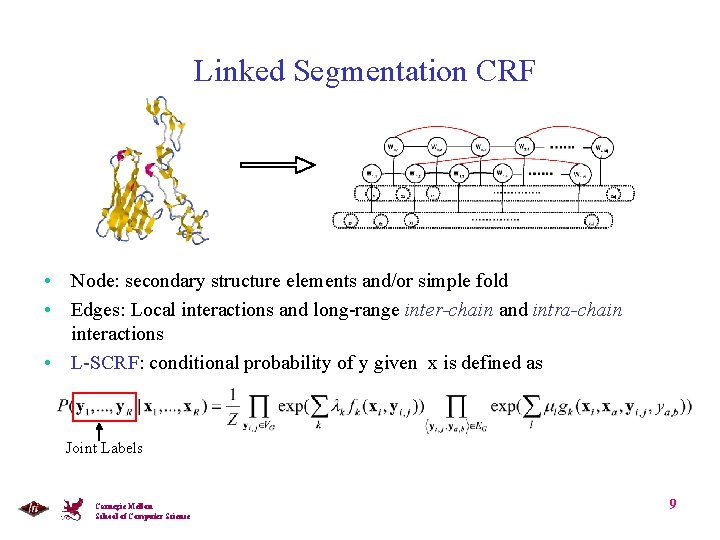
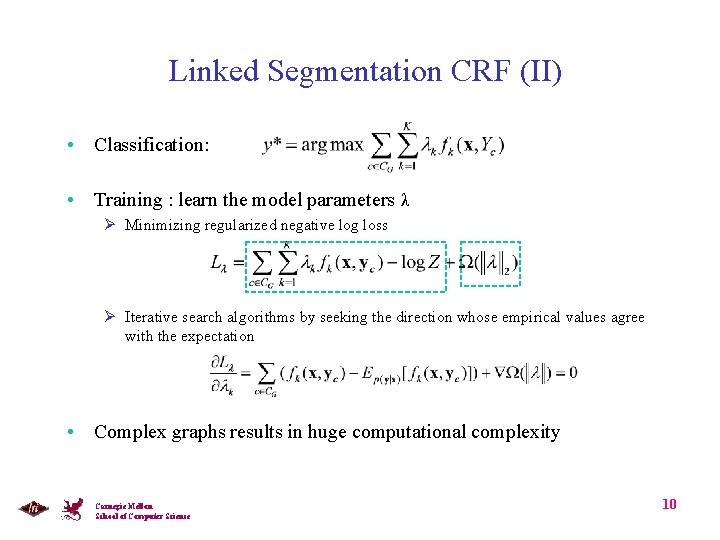
Linked Segmentation CRF • Node: secondary structure elements and/or simple fold • Edges: Local interactions and long-range inter-chain and intra-chain interactions • L-SCRF: conditional probability of y given x is defined as Joint Labels Carnegie Mellon School of Computer Science 9

Linked Segmentation CRF (II) • Classification: • Training : learn the model parameters λ Ø Minimizing regularized negative log loss Ø Iterative search algorithms by seeking the direction whose empirical values agree with the expectation • Complex graphs results in huge computational complexity Carnegie Mellon School of Computer Science 10

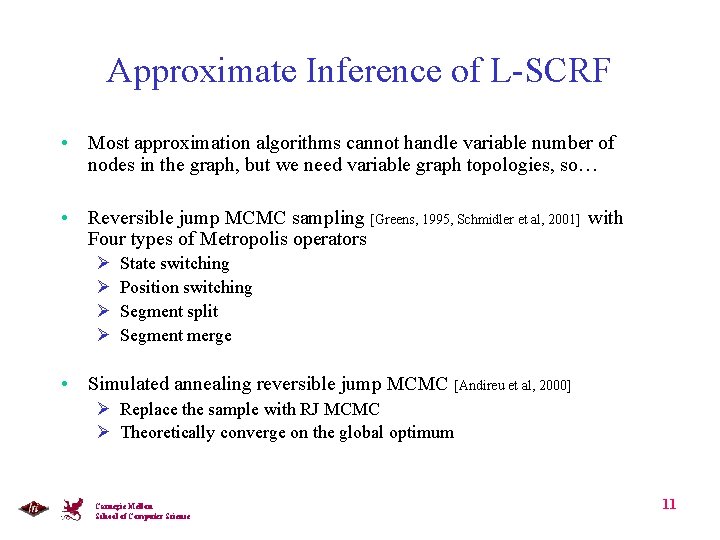
Approximate Inference of L-SCRF • Most approximation algorithms cannot handle variable number of nodes in the graph, but we need variable graph topologies, so… • Reversible jump MCMC sampling [Greens, 1995, Schmidler et al, 2001] with Four types of Metropolis operators Ø Ø State switching Position switching Segment split Segment merge • Simulated annealing reversible jump MCMC [Andireu et al, 2000] Ø Replace the sample with RJ MCMC Ø Theoretically converge on the global optimum Carnegie Mellon School of Computer Science 11
![Experiments Target Quaternary Fold Triple betaspirals van Raaij et al Nature 1999 Ø Experiments: Target Quaternary Fold • Triple beta-spirals [van Raaij et al. Nature 1999] Ø](https://slidetodoc.com/presentation_image/255be733c05c068a545e989ad7af2fc6/image-12.jpg)
Experiments: Target Quaternary Fold • Triple beta-spirals [van Raaij et al. Nature 1999] Ø Virus fibers in adenovirus, reovirus and PRD 1 • Double barrel trimer [Benson et al, 2004] Ø Coat protein of adenovirus, PRD 1, STIV, PBCV Carnegie Mellon School of Computer Science 12

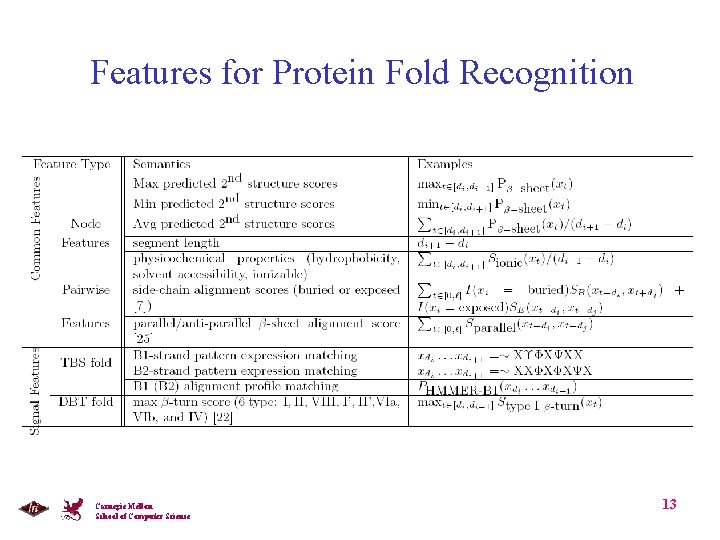
Features for Protein Fold Recognition Carnegie Mellon School of Computer Science 13

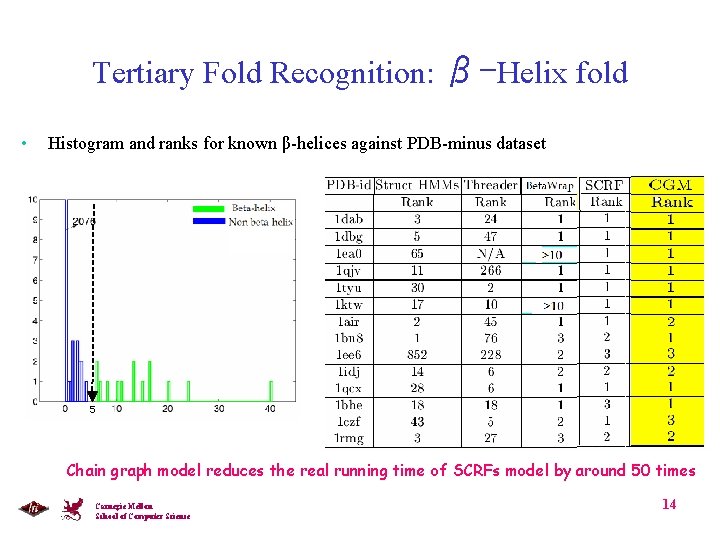
Tertiary Fold Recognition: β-Helix fold • Histogram and ranks for known β-helices against PDB-minus dataset 5 Chain graph model reduces the real running time of SCRFs model by around 50 times Carnegie Mellon School of Computer Science 14

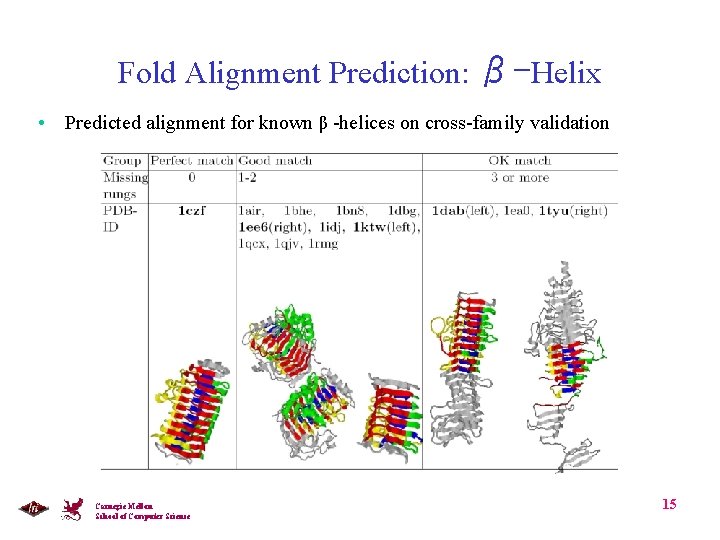
Fold Alignment Prediction: β-Helix • Predicted alignment for known β -helices on cross-family validation Carnegie Mellon School of Computer Science 15

Discovery of New Potential β-helices • Run structural predictor seeking potential β-helices from Uniprot (structurally unresolved) databases Ø Full list (98 new predictions) can be accessed at www. cs. cmu. edu/~yanliu/SCRF. html • Verification on 3 proteins with later experimentally resolved structures from different organisms Ø 1 YP 2: Potato Tuber ADP-Glucose Pyrophosphorylase Ø 1 PXZ: The Major Allergen From Cedar Pollen Ø GP 14 of Shigella bacteriophage as a β-helix protein Ø No single false positive! Carnegie Mellon School of Computer Science 16

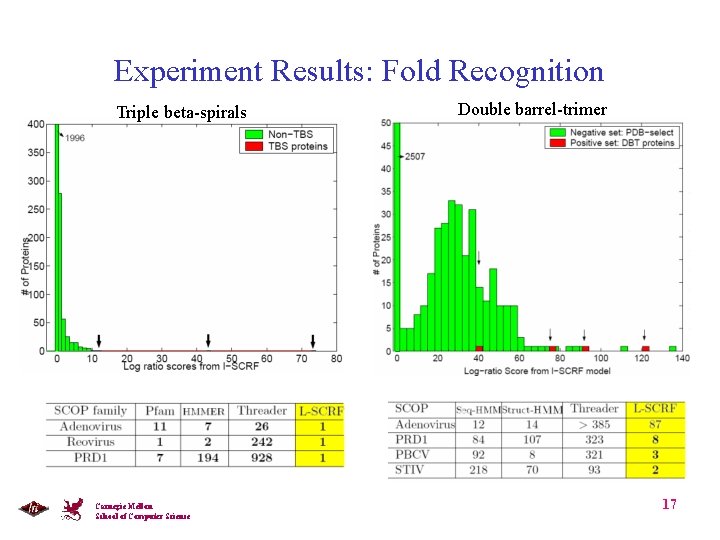
Experiment Results: Fold Recognition Triple beta-spirals Carnegie Mellon School of Computer Science Double barrel-trimer 17

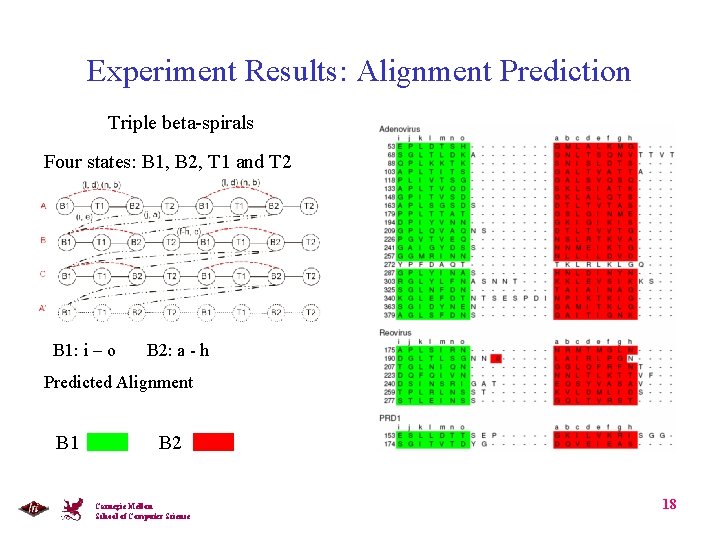
Experiment Results: Alignment Prediction Triple beta-spirals Four states: B 1, B 2, T 1 and T 2 Correct Alignment: B 1: i – o B 2: a - h Predicted Alignment B 1 B 2 Carnegie Mellon School of Computer Science 18

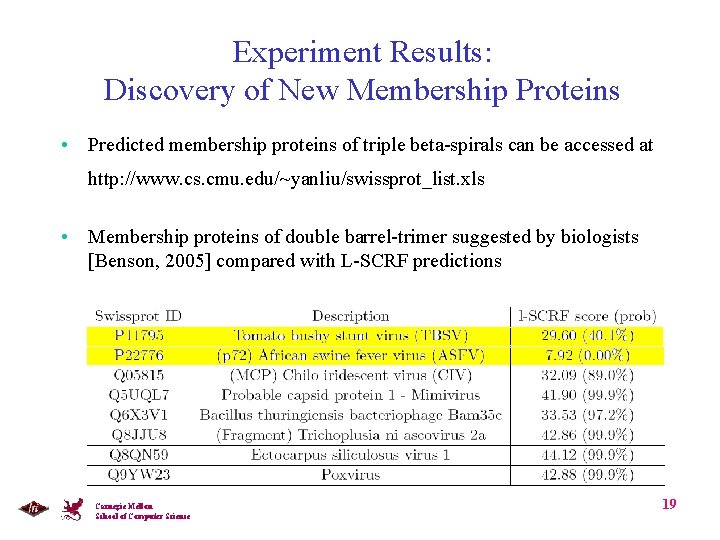
Experiment Results: Discovery of New Membership Proteins • Predicted membership proteins of triple beta-spirals can be accessed at http: //www. cs. cmu. edu/~yanliu/swissprot_list. xls • Membership proteins of double barrel-trimer suggested by biologists [Benson, 2005] compared with L-SCRF predictions Carnegie Mellon School of Computer Science 19

Conclusion • Conditional graphical models for protein structure prediction Ø Effective representation for protein structural properties Ø Feasibility to incorporate different kinds of informative features Ø Efficient inference algorithms for large-scale applications • A major extension compared with previous work Ø Knowledge representation through graphical models Ø Ability to handle long-range interactions within one chain and between chains • Future work Ø Automatic learning of graph topology Ø Applications to other domains Carnegie Mellon School of Computer Science 20

Carnegie Mellon School of Computer Science 21

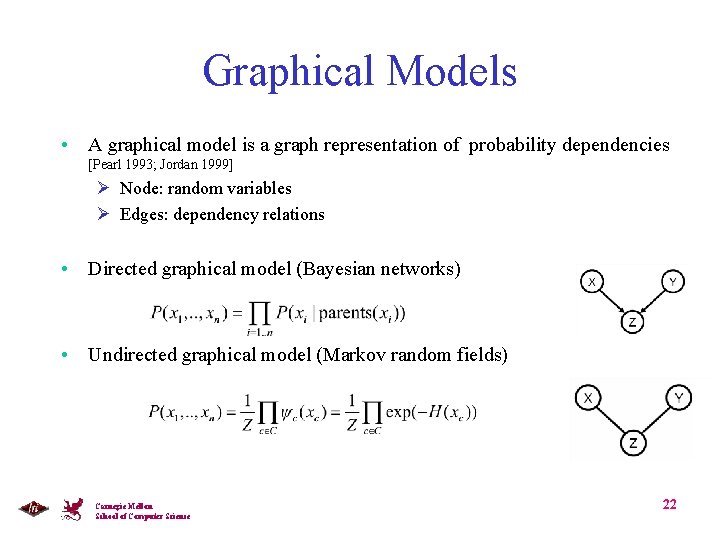
Graphical Models • A graphical model is a graph representation of probability dependencies [Pearl 1993; Jordan 1999] Ø Node: random variables Ø Edges: dependency relations • Directed graphical model (Bayesian networks) • Undirected graphical model (Markov random fields) Carnegie Mellon School of Computer Science 22