Potential Role for Endocrine Disruptor Expert Systems in



















- Slides: 53

Potential Role for Endocrine Disruptor Expert Systems in Investigating ‘Endocrine System-Epigenome’ Interactions P. Schmieder EPA, ORD, NHEERL, MED-Duluth, MN 2 nd Mc. Kim Cancer Workshop

Effects-based Expert System Automated rule-based decision trees to predict which chemicals have the potential to disrupt endocrine systems. This is done by: • testing key chemicals within a chemical class to set boundaries on biological/toxicological activity, to predict activity of other members of the “class” • Grouping chemicals by a common biological activity, then determining what is similar about the chemical structures and properties that explain their activity • writing rules that help categorize similar but untested chemicals. The Program Offices use these tools to decide which, of the hundreds of chemicals on Agency chemical lists, should be evaluated first for likelihood of disrupting endocrine-mediated pathways the results in hormonal imbalances.

ER-mediated AOP Significant evidence exists linking ER binding chemicals to adverse outcomes related to reproductive effects (ER-mediated reproductive impairment AOP): hh - drug design of anti-estrogens for breast cancer research and treatment env - multiple studies linking chemical ER binders to adverse effects -potent pharmaceuticals - e. g. , EE 2 -weak affinity environmental chemicals - e. g. , APs If there is evidence, or evidence found in future, for NR mediated AOPs linked to cancer, then the approach described here will be relevant for hypothesizing what chemical might do this. This presentation will focus on: using an ER-mediated AOP to form chemical categories for addressing a specific risk assessment application

ER-mediated AOP The risk context to which AOP is applied influences the approach: This example: Risk context – USEPA needs to evaluate large lists of data-limited chemicals for ED potential; how can predictive tools be used to identify which of these chemicals have the greatest potential to cause an adverse effect because of their estrogenic potential; Goal - Given limited testing resources, prioritize chemicals on targeted inventories, so that those with the highest likelihood of producing an adverse outcome are tested first Targeted chemical inventories: -inert ingredients in pesticides used on food crops -antimicrobial active ingredients -inert ingredients in pesticides not used on food crops

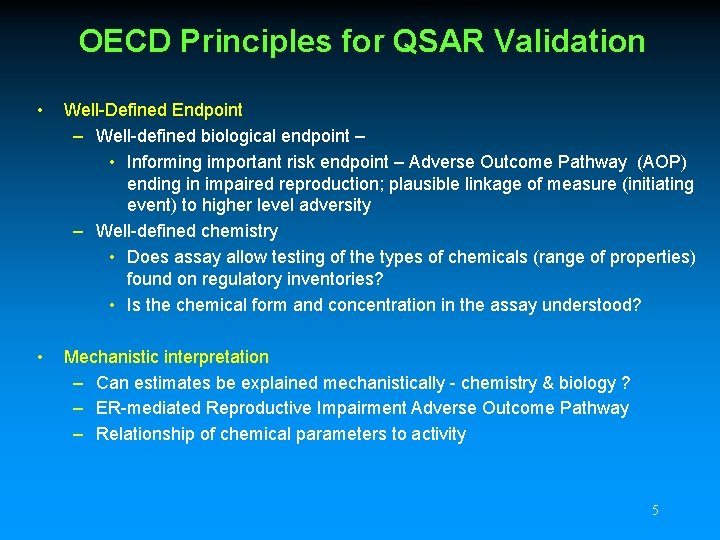
OECD Principles for QSAR Validation • Well-Defined Endpoint – Well-defined biological endpoint – • Informing important risk endpoint – Adverse Outcome Pathway (AOP) ending in impaired reproduction; plausible linkage of measure (initiating event) to higher level adversity – Well-defined chemistry • Does assay allow testing of the types of chemicals (range of properties) found on regulatory inventories? • Is the chemical form and concentration in the assay understood? • Mechanistic interpretation – Can estimates be explained mechanistically - chemistry & biology ? – ER-mediated Reproductive Impairment Adverse Outcome Pathway – Relationship of chemical parameters to activity 5

OECD Principles for QSAR Validation (cont. ) • Defined Model Applicability Domain – Well-defined application • Is the regulatory question well-defined – priority setting is different than risk assessment? • Is the QSAR model domain coverage well-defined? • Does the QSAR chemical domain adequately cover the regulatory chemical domain i. e. , the regulatory question? • Appropriate measures of goodness of fit, robustness, ability to predict – Measures appropriate for a regression model likely not appropriate to evaluate an expert system • Unambiguous algorithm – Expert Systems – logic tree, rules/queries, supporting information 6

Application of OECD Principles to Forming Chemical Categories: Key Questions Transparency -How reasonable is the estimate compared with data for similar chemicals ? -Can the QSAR estimate be explained mechanistically? Usefulness - Are the predictions applicable to all the chemicals of regulatory concern? - Does the model/expert system answer the regulatory question?

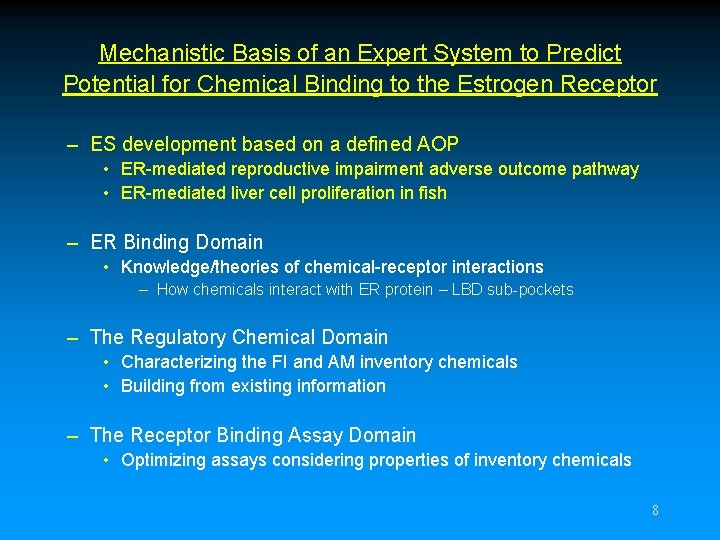
Mechanistic Basis of an Expert System to Predict Potential for Chemical Binding to the Estrogen Receptor – ES development based on a defined AOP • ER-mediated reproductive impairment adverse outcome pathway • ER-mediated liver cell proliferation in fish – ER Binding Domain • Knowledge/theories of chemical-receptor interactions – How chemicals interact with ER protein – LBD sub-pockets – The Regulatory Chemical Domain • Characterizing the FI and AM inventory chemicals • Building from existing information – The Receptor Binding Assay Domain • Optimizing assays considering properties of inventory chemicals 8

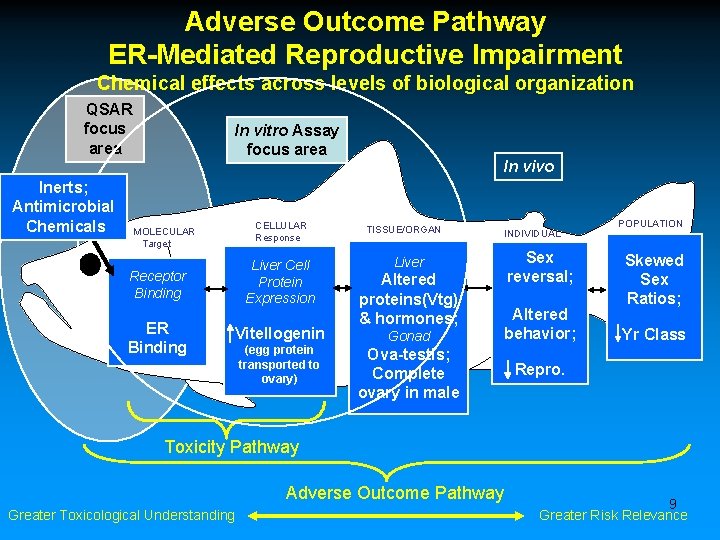
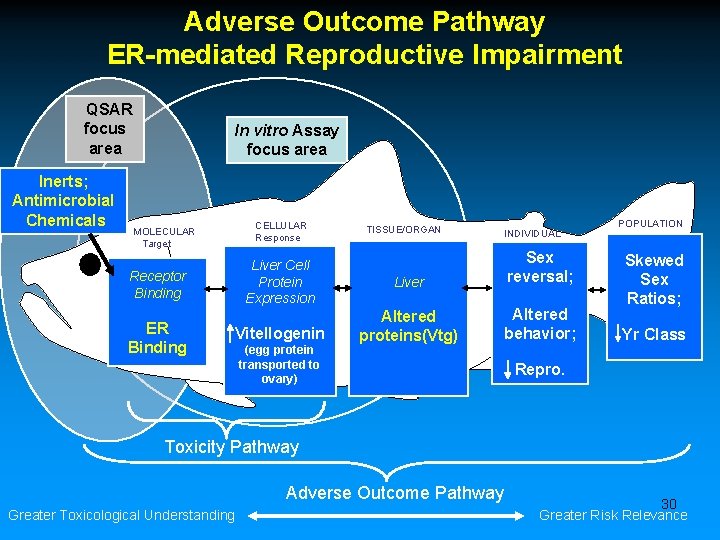
Adverse Outcome Pathway ER-Mediated Reproductive Impairment Chemical effects across levels of biological organization QSAR focus area Inerts; Antimicrobial Chemicals In vitro Assay focus area CELLULAR Response MOLECULAR Target Liver Cell Protein Expression Receptor Binding ER Binding Vitellogenin (egg protein transported to ovary) In vivo TISSUE/ORGAN Liver Altered proteins(Vtg) & hormones; Gonad Ova-testis; Complete ovary in male INDIVIDUAL Sex reversal; Altered behavior; POPULATION Skewed Sex Ratios; Yr Class Repro. Toxicity Pathway Adverse Outcome Pathway Greater Toxicological Understanding 9 Greater Risk Relevance

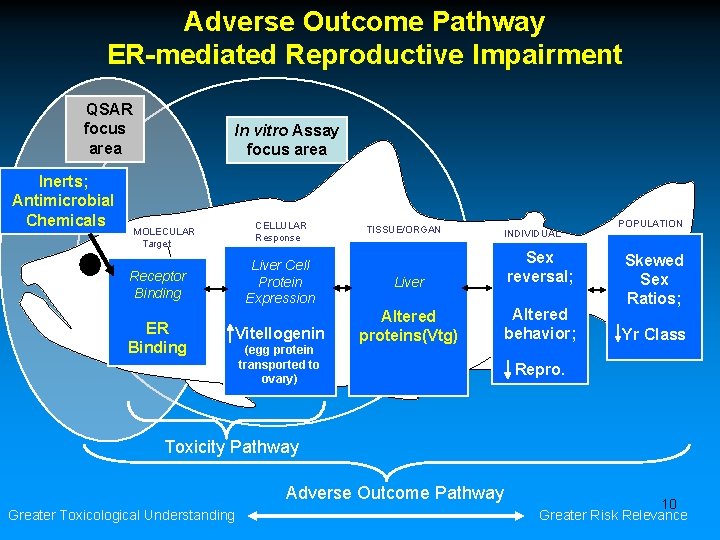
Adverse Outcome Pathway ER-mediated Reproductive Impairment QSAR focus area Inerts; Antimicrobial Chemicals In vitro Assay focus area CELLULAR Response MOLECULAR Target Liver Cell Protein Expression Receptor Binding ER Binding Vitellogenin (egg protein transported to ovary) TISSUE/ORGAN INDIVIDUAL Liver Sex reversal; Altered proteins(Vtg) Altered behavior; POPULATION Skewed Sex Ratios; Yr Class Repro. Toxicity Pathway Adverse Outcome Pathway Greater Toxicological Understanding 10 Greater Risk Relevance

Mechanistic Basis of the Expert System ER Binding Affinity: An Indicator of Potential Reproductive Effects • ER-mediated reproductive impairment adverse outcome pathway ER Binding Domain • Knowledge/theories of chemical-receptor interactions – ER sub-pockets The Regulatory Chemical Domain • Characterizing the FI and AM inventory chemicals • Building from existing information The Receptor Binding Assay Domain • Optimizing assays considering properties of inventory chemicals 11

ER Binding Domain - Bioassays for ER binding were available from drug-design, although extant methods were focused on potent anti-estrogens - Drug-design research provides mechanistic insights on multiple types of interaction within the ER binding site 12

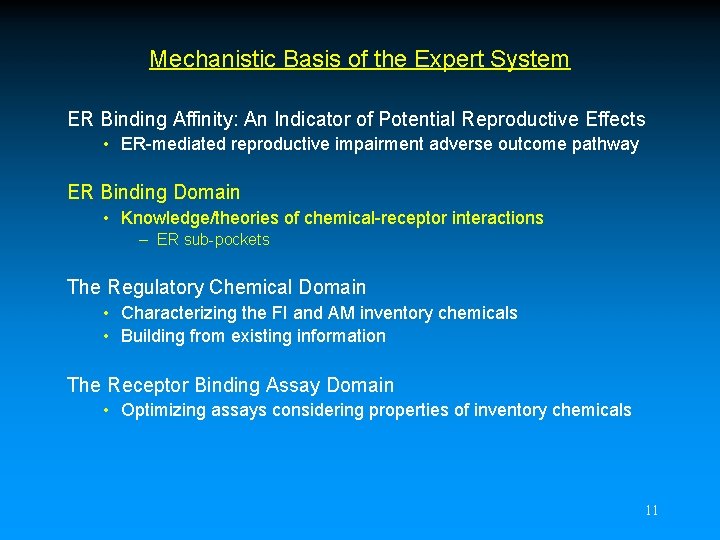
A_B Interaction T 347 C QOxygen= -0. 25 E 353 QOxygen= -0. 32 H A H 524 CH 3 H HO OH B R 394 H H Distance = 10. 8 for 17 -Estradiol J. Katzenellenbogen 13

Mechanistic Basis of the Expert System ER Binding Affinity: An Indicator of Potential Reproductive Effects • ER-mediated reproductive impairment adverse outcome pathway ER Binding Domain • Knowledge/theories of chemical-receptor interactions – ER sub-pockets The Regulatory Chemical Domain • Characterizing the FI and AM inventory chemicals • Building from existing information The Receptor Binding Assay Domain • Optimizing assays considering properties of inventory chemicals 14

Apply knowledge/theory of ER Binding Domain to The Regulatory Chemical Domain (continuing to seek MECHANISTIC understanding) Hypothesize ER interactions of Inventory Chemicals Inert ingredients and antimicrobial pesticides are non-steroidal and do not contain multiple H-bonding groups at distance needed for steroid-like interactions Hypotheses: - Any pesticide inert or antimicrobial that does bind ER will do so through an interaction mechanism that results in low affinity binding - Only a small % of these chemicals are likely to bind ER - A chemical group approach will facilitate regulatory application - Chemicals can be grouped based on how they interact with the 15 ER (within specific ER sub-pockets)

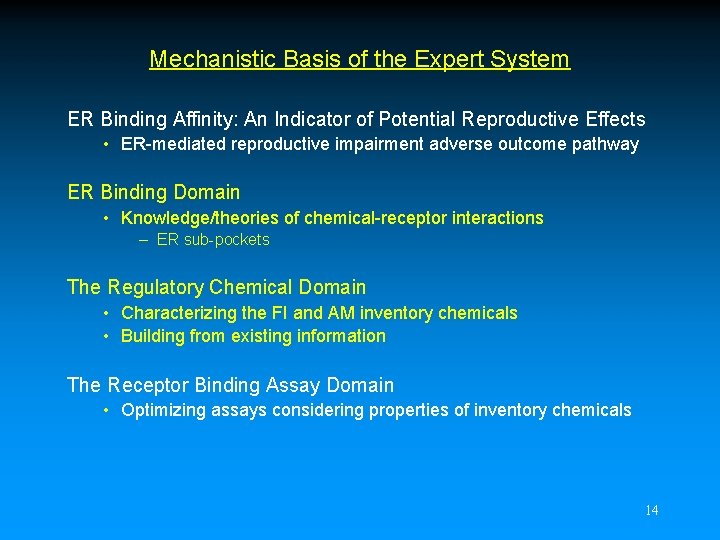
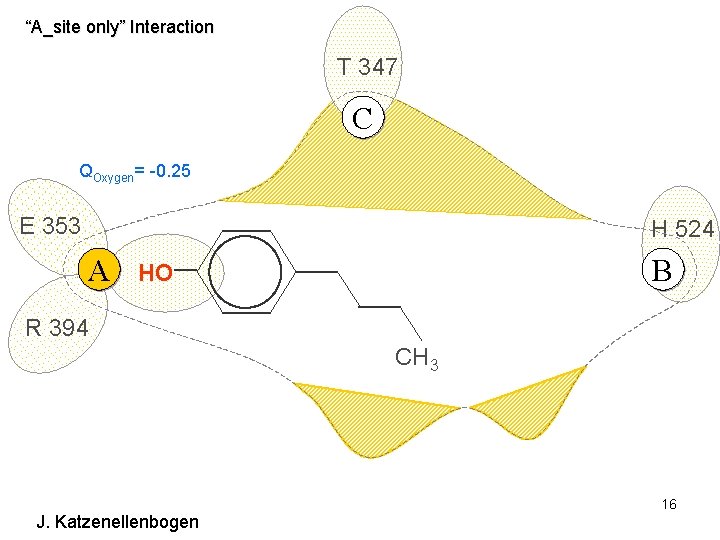
“A_site only” Interaction T 347 C QOxygen= -0. 25 E 353 H 524 A B HO R 394 CH 3 J. Katzenellenbogen 16

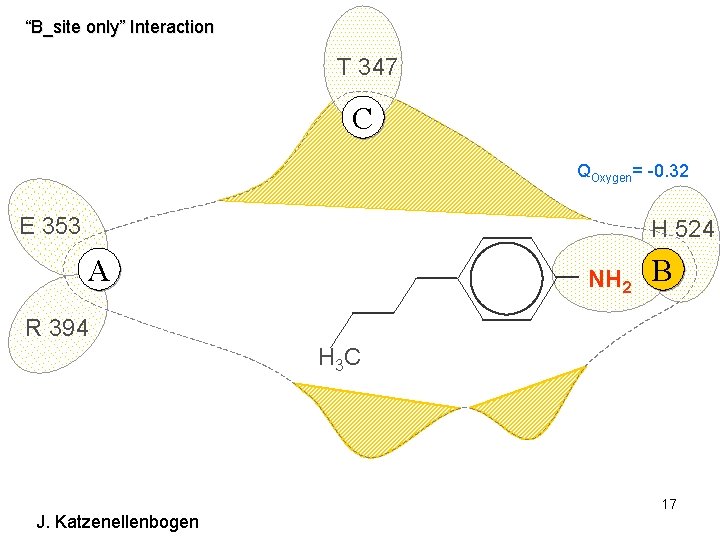
“B_site only” Interaction T 347 C QOxygen= -0. 32 E 353 H 524 A NH 2 B R 394 H 3 C J. Katzenellenbogen 17

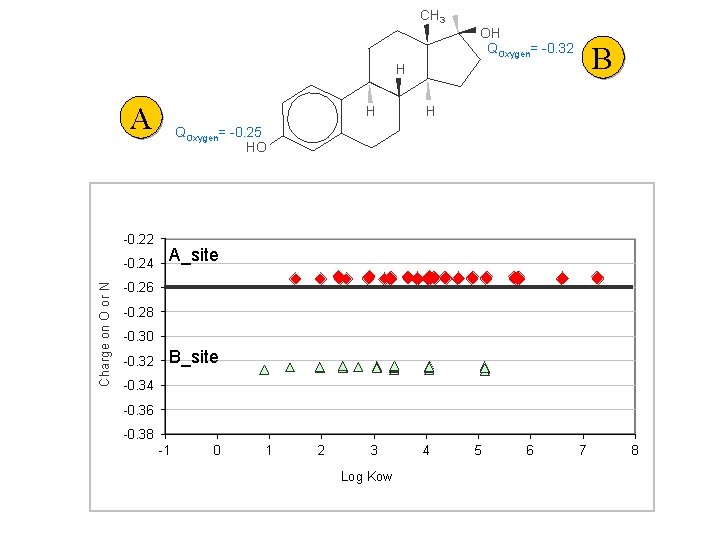
CH 3 OH QOxygen= -0. 32 B H A -0. 22 Charge on O or N -0. 24 H H QOxygen= -0. 25 HO A_site -0. 26 -0. 28 -0. 30 -0. 32 B_site -0. 34 -0. 36 -0. 38 -1 0 1 2 3 Log Kow 4 5 6 7 8

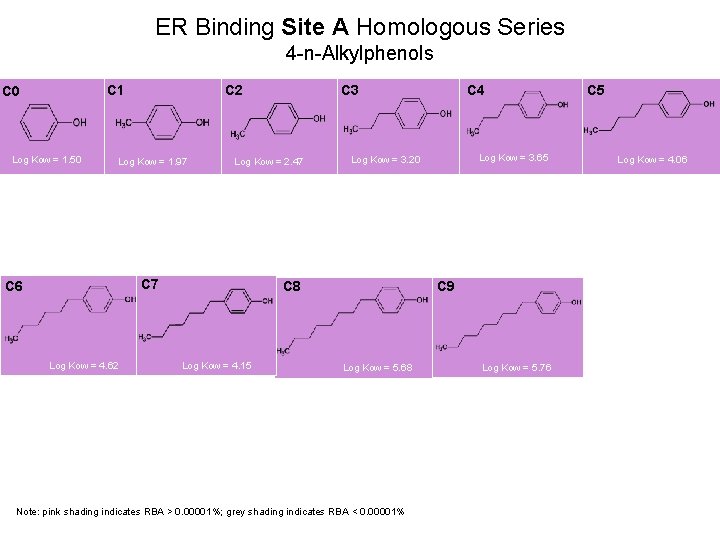
ER Binding Site A Homologous Series 4 -n-Alkylphenols C 1 C 0 Log Kow = 1. 50 C 2 Log Kow = 1. 97 Log Kow = 2. 47 C 6 Log Kow = 4. 62 C 3 Log Kow = 3. 65 Log Kow = 3. 20 C 8 Log Kow = 4. 15 C 4 C 5 Log Kow = 4. 06 C 9 Log Kow = 5. 68 Note: pink shading indicates RBA > 0. 00001%; grey shading indicates RBA < 0. 00001% Log Kow = 5. 76 19

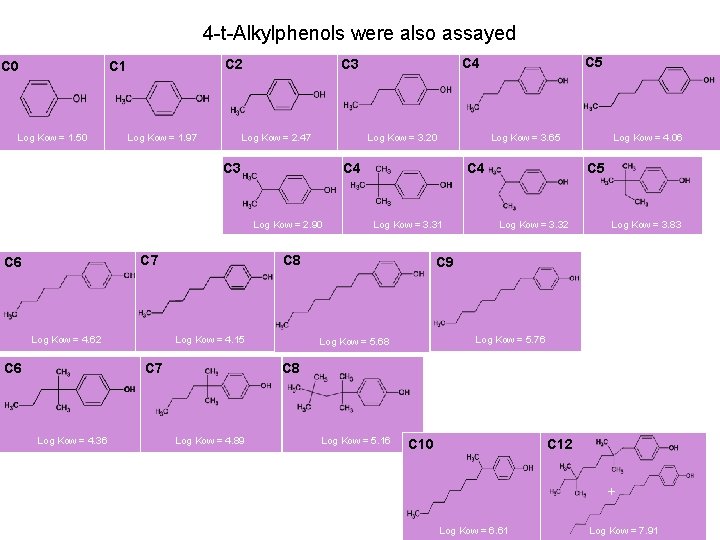
4 -t-Alkylphenols were also assayed C 0 C 2 C 1 Log Kow = 1. 50 Log Kow = 1. 97 Log Kow = 2. 47 Log Kow = 3. 20 C 3 Log Kow = 4. 62 C 6 Log Kow = 4. 15 C 7 Log Kow = 4. 36 C 4 Log Kow = 3. 31 C 8 C 7 C 6 Log Kow = 3. 65 C 4 Log Kow = 2. 90 C 5 C 4 C 3 Log Kow = 4. 06 C 5 Log Kow = 3. 32 Log Kow = 3. 83 C 9 Log Kow = 5. 76 Log Kow = 5. 68 C 8 Log Kow = 4. 89 Log Kow = 5. 16 C 10 C 12 + Log Kow = 6. 61 Log Kow = 7. 91

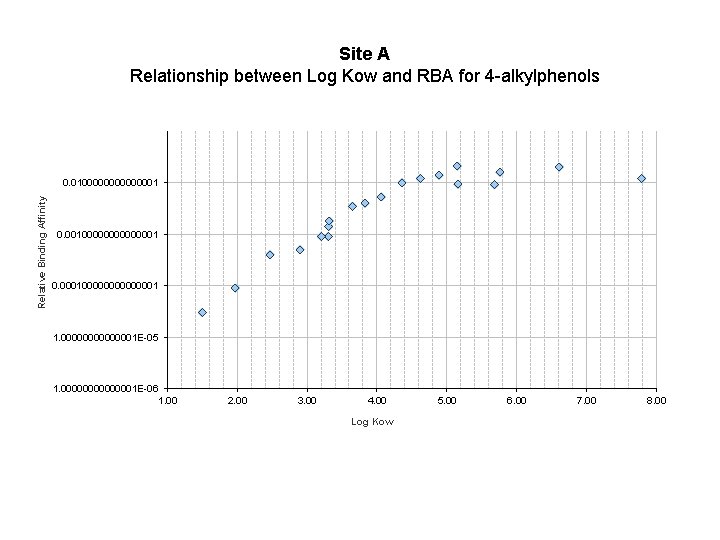
Site A Relationship between Log Kow and RBA for 4 -alkylphenols Relative Binding Affinity 0. 0100000000000001 0. 000100000001 1. 00000001 E-05 1. 00000001 E-06 1. 00 2. 00 3. 00 4. 00 5. 00 6. 00 7. 00 8. 00 Log Kow 21

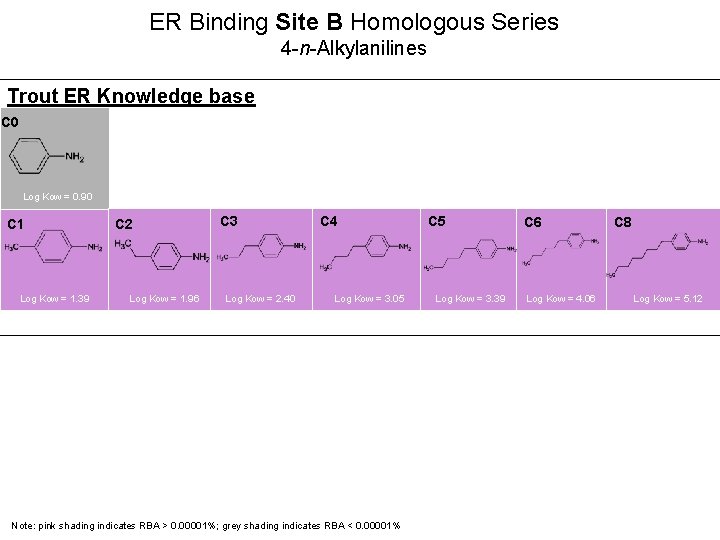
ER Binding Site B Homologous Series 4 -n-Alkylanilines Trout ER Knowledge base C 0 Log Kow = 0. 90 C 1 Log Kow = 1. 39 C 2 Log Kow = 1. 96 C 3 Log Kow = 2. 40 C 4 Log Kow = 3. 05 C 5 Log Kow = 3. 39 C 6 Log Kow = 4. 06 C 8 Log Kow = 5. 12 22 Note: pink shading indicates RBA > 0. 00001%; grey shading indicates RBA < 0. 00001%

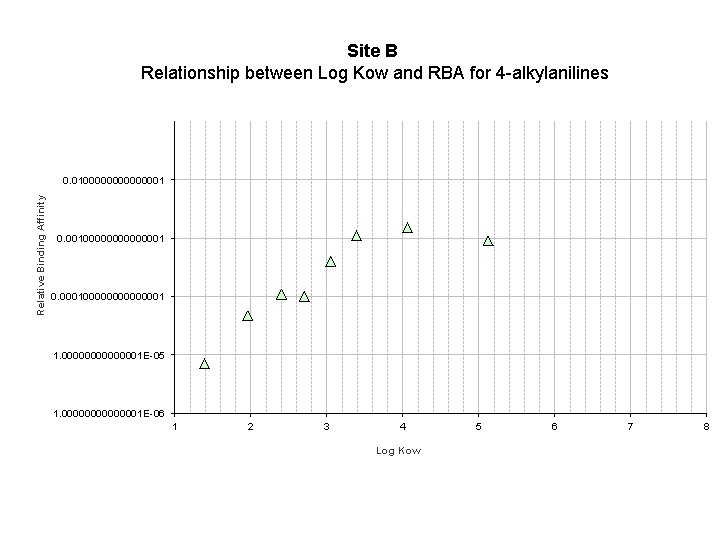
Site B Relationship between Log Kow and RBA for 4 -alkylanilines Relative Binding Affinity 0. 0100000000000001 0. 000100000001 1. 00000001 E-05 1. 00000001 E-06 1 2 3 4 5 6 7 8 Log Kow 23

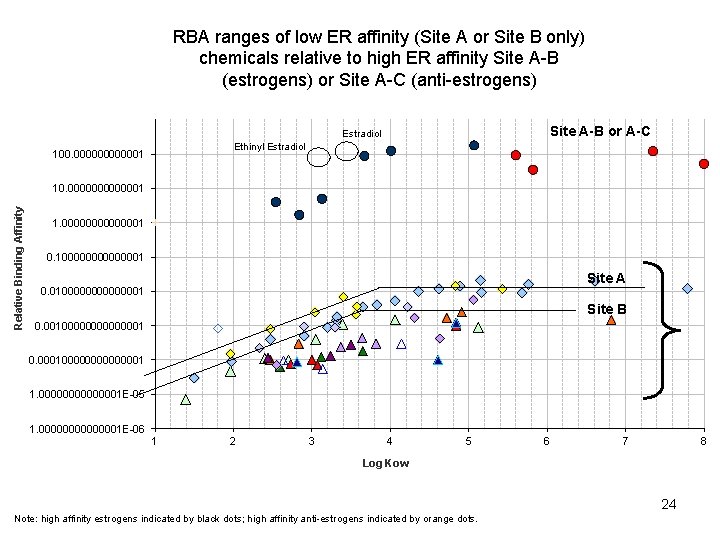
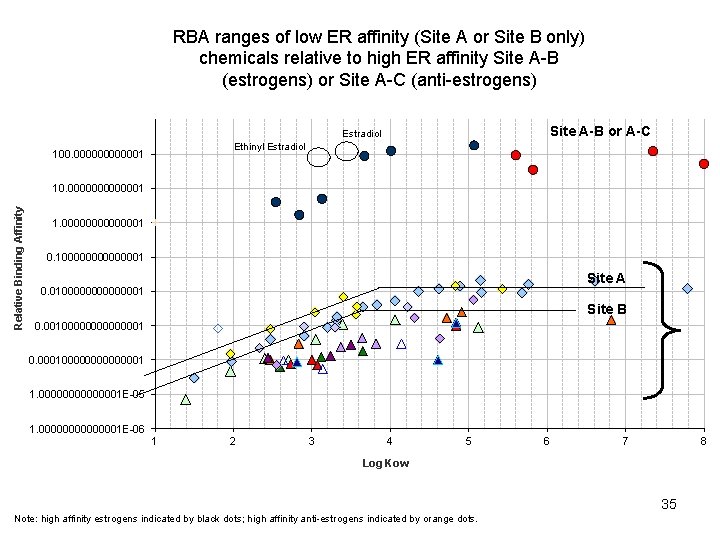
RBA ranges of low ER affinity (Site A or Site B only) chemicals relative to high ER affinity Site A-B (estrogens) or Site A-C (anti-estrogens) Site A-B or A-C Estradiol Ethinyl Estradiol 100. 0000001 Relative Binding Affinity 10. 0000001 1. 00000001 0. 100000001 Site A 0. 0100000001 Site B 0. 00100000001 0. 000100000001 1. 00000001 E-05 1. 00000001 E-06 1 2 3 4 5 6 7 8 Log Kow 24 Note: high affinity estrogens indicated by black dots; high affinity anti-estrogens indicated by orange dots.

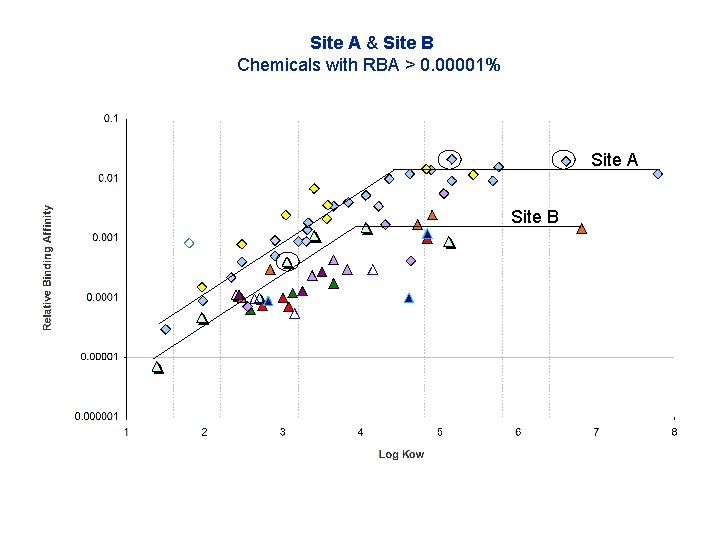
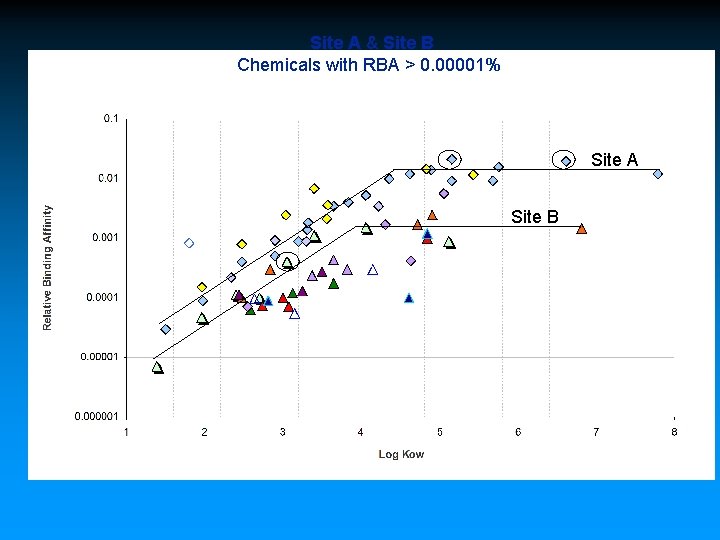
Site A & Site B Chemicals with RBA > 0. 00001% Site A Site B Symbols - Site A chemical groups - diamonds Symbols - Site B chemical groups - triangles

Mechanistic Basis of the Expert System to Predict Relative Estrogen Receptor Binding Affinity ER Binding Affinity: An Indicator of Potential Reproductive Effects • ER-mediated reproductive impairment adverse outcome pathway ER Binding Domain • Knowledge/theories of chemical-receptor interactions – ER sub-pockets The Regulatory Chemical Domain • Characterizing the FI and AM inventory chemicals • Building from existing information The Receptor Binding Assay Domain • Optimizing assays considering properties of inventory chemicals 26

Food use Inert Ingredients Inert chemicals in pesticides used on food crops The 2004 List included: 893 entries = 393 discrete chemicals + 500 non-discrete substances (44% discrete : 56% non-discrete) 393 discrete chemicals include: 366 organic chemicals (93%) 24 inorganic chemicals (6%) 3 organometallic compounds (1%) 500 non-discrete substances include: 147 polymers of mixed chain length 170 mixtures 183 undefined substances 27

Antimicrobial Pesticides Antimicrobials and sanitizers list included: 299 = 211 discrete chemicals + 88 non-discrete substances (71% discrete : 29% non-discrete) 211 discrete chemicals include: 153 organic chemicals (72%) 52 inorganic chemicals (25%) 6 acyclic organometallic compounds (3%) 88 non-discrete substances include: 25 polymers of mixed chain length 35 mixtures 28 undefined substances 28

Mechanistic Basis of the Expert System to Predict Relative Estrogen Receptor Binding Affinity ER Binding Affinity: An Indicator of Potential Reproductive Effects • ER-mediated reproductive impairment adverse outcome pathway ER Binding Domain • Knowledge/theories of chemical-receptor interactions – ER sub-pockets The Regulatory Chemical Domain • Characterizing the FI and AM inventory chemicals • Building on existing information The Receptor Binding Assay Domain • Optimizing assays considering properties of inventory chemicals 29

Adverse Outcome Pathway ER-mediated Reproductive Impairment QSAR focus area Inerts; Antimicrobial Chemicals In vitro Assay focus area CELLULAR Response MOLECULAR Target Liver Cell Protein Expression Receptor Binding ER Binding Vitellogenin (egg protein transported to ovary) TISSUE/ORGAN INDIVIDUAL Liver Sex reversal; Altered proteins(Vtg) Altered behavior; POPULATION Skewed Sex Ratios; Yr Class Repro. Toxicity Pathway Adverse Outcome Pathway Greater Toxicological Understanding 30 Greater Risk Relevance

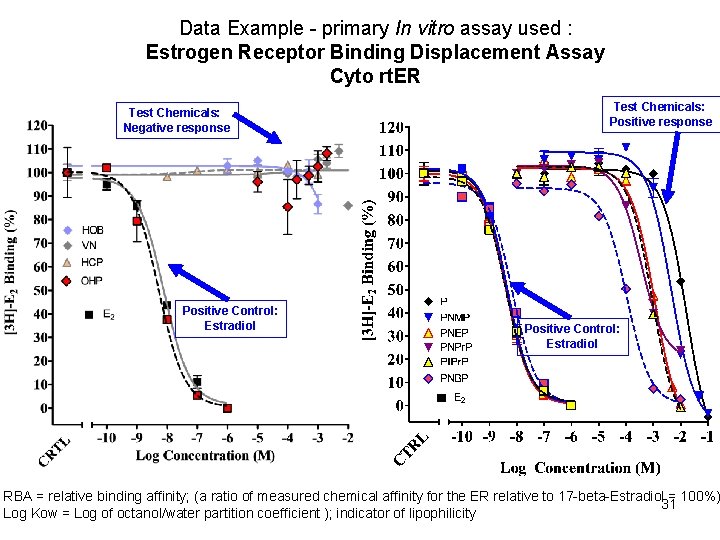
Data Example - primary In vitro assay used : Estrogen Receptor Binding Displacement Assay Cyto rt. ER Test Chemicals: Negative response Positive Control: Estradiol Test Chemicals: Positive response Positive Control: Estradiol RBA = relative binding affinity; (a ratio of measured chemical affinity for the ER relative to 17 -beta-Estradiol = 100%) 31 Log Kow = Log of octanol/water partition coefficient ); indicator of lipophilicity

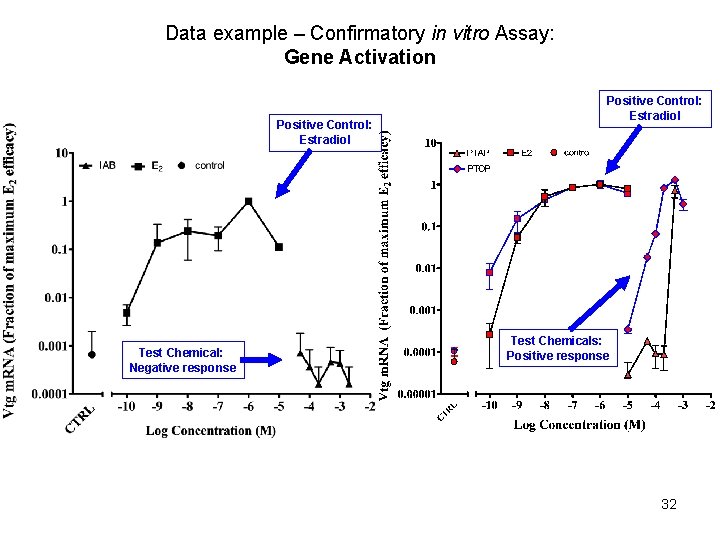
Data example – Confirmatory in vitro Assay: Gene Activation Positive Control: Estradiol Test Chemical: Negative response Positive Control: Estradiol Test Chemicals: Positive response 32

Site A & Site B Chemicals with RBA > 0. 00001% Site A Site B Symbols - Site A chemical groups - diamonds Symbols - Site B chemical groups - triangles

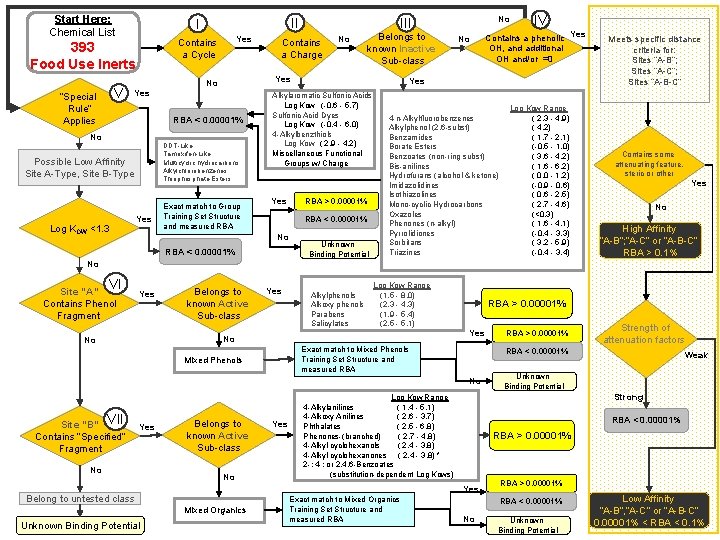
Start Here: Chemical List V Yes Contains a Cycle 393 Food Use Inerts “Special Rule” Applies II I Yes RBA < 0. 00001% No DDT-Like Tamoxifen-Like Multicyclic hydrocarbons Alkylchlorobenzenes Thiophosphate Esters Possible Low Affinity Site A-Type, Site B-Type Log KOW <1. 3 Yes Contains a Charge No Belongs to known Inactive Sub-class Yes No Exact match to Group Training Set Structure and measured RBA > 0. 00001% RBA < 0. 00001% No IV Contains a phenolic Yes OH, and additional OH and/or =0 Yes Alkylaromatic Sulfonic Acids Log Kow (-0. 6 - 5. 7) Sulfonic Acid Dyes Log Kow (-0. 4 - 6. 0) 4 -Alkylbenzthiols Log Kow ( 2. 9 - 4. 2) Miscellaneous Functional Groups w/ Charge Yes No III Unknown Binding Potential 4 -n-Alkylfluorobenzenes Alkylphenol (2, 6 -subst) Benzamides Borate Esters Benzoates (non-ring subst) Bis-anilines Hydrofurans (alcohol & ketone) Imidazolidines Isothiazolines Mono-cyclic Hydrocarbons Oxazoles Phenones (n-alkyl) Pyrrolidiones Sorbitans Triazines Log Kow Range ( 2. 3 - 4. 9) ( 4. 2) ( 1. 7 - 2. 1) (-0. 5 - 1. 0) ( 3. 6 - 4. 2) ( 1. 6 - 6. 2) ( 0. 0 - 1. 2) (-0. 9 - 0. 6) ( 0. 6 - 2. 5) ( 2. 7 - 4. 6) (<0. 3) ( 1. 6 - 4. 1) (-0. 4 - 3. 3) ( 3. 2 - 5. 9) (-0. 4 - 3. 4) Meets specific distance criteria for: Sites “A-B”; Sites “A-C”; Sites “A-B-C” Contains some attenuating feature, steric or other Yes No High Affinity “A-B”; “A-C” or “A-B-C” RBA > 0. 1% No VI Site “A” Contains Phenol Fragment Yes Belongs to known Active Sub-class Yes Alkylphenols Alkoxy phenols Parabens Salicylates Log Kow Range (1. 5 - 8. 0) (2. 3 - 4. 3) (1. 9 - 5. 4) (2. 5 - 5. 1) Yes No No RBA > 0. 00001% Exact match to Mixed Phenols Training Set Structure and measured RBA Mixed Phenols Yes No Belongs to known Active Sub-class No Mixed Organics Exact match to Mixed Organics Training Set Structure and measured RBA Weak Unknown Binding Potential Strong Log Kow Range 4 -Alkylanilines ( 1. 4 - 5. 1) 4 -Alkoxy Anilines ( 2. 6 - 3. 7) Phthalates ( 2. 5 - 6. 8) Phenones-(branched) ( 2. 7 - 4. 8) 4 -Alkyl cyclohexanols ( 2. 4 - 3. 8) 4 -Alkyl cyclohexanones ( 2. 4 - 3. 8) * 2 -; 4 -; or 2, 4, 6 -Benzoates (substitution-dependent Log Kows) RBA < 0. 00001% RBA > 0. 00001% Yes Belong to untested class Unknown Binding Potential Yes Strength of attenuation factors RBA < 0. 00001% No Site “B” VII Contains “Specified” Fragment RBA > 0. 00001% RBA < 0. 00001% No Unknown Binding Potential Low Affinity “A-B”, “A-C” or 34 “A-B-C” 0. 00001% < RBA < 0. 1%

RBA ranges of low ER affinity (Site A or Site B only) chemicals relative to high ER affinity Site A-B (estrogens) or Site A-C (anti-estrogens) Site A-B or A-C Estradiol Ethinyl Estradiol 100. 0000001 Relative Binding Affinity 10. 0000001 1. 00000001 0. 100000001 Site A 0. 0100000001 Site B 0. 00100000001 0. 000100000001 1. 00000001 E-05 1. 00000001 E-06 1 2 3 4 5 6 7 8 Log Kow 35 Note: high affinity estrogens indicated by black dots; high affinity anti-estrogens indicated by orange dots.

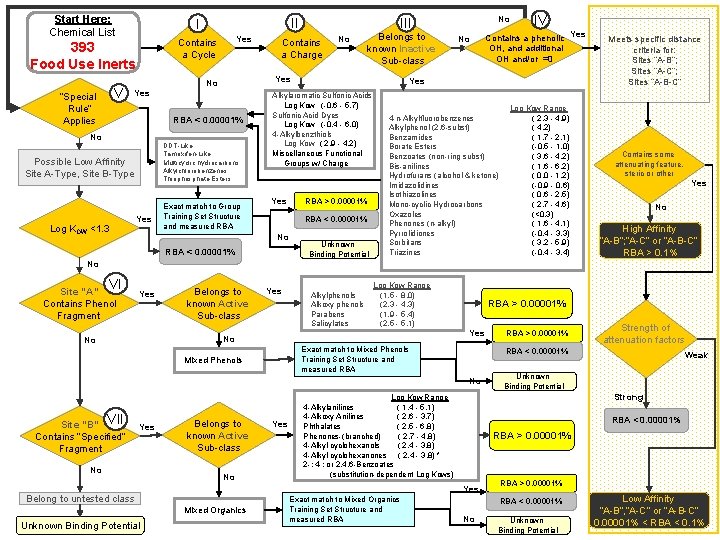
Start Here: Chemical List V Yes Contains a Cycle 393 Food Use Inerts “Special Rule” Applies II I Yes RBA < 0. 00001% No DDT-Like Tamoxifen-Like Multicyclic hydrocarbons Alkylchlorobenzenes Thiophosphate Esters Possible Low Affinity Site A-Type, Site B-Type Log KOW <1. 3 Yes Contains a Charge No Belongs to known Inactive Sub-class Yes No Exact match to Group Training Set Structure and measured RBA > 0. 00001% RBA < 0. 00001% No IV Contains a phenolic Yes OH, and additional OH and/or =0 Yes Alkylaromatic Sulfonic Acids Log Kow (-0. 6 - 5. 7) Sulfonic Acid Dyes Log Kow (-0. 4 - 6. 0) 4 -Alkylbenzthiols Log Kow ( 2. 9 - 4. 2) Miscellaneous Functional Groups w/ Charge Yes No III Unknown Binding Potential 4 -n-Alkylfluorobenzenes Alkylphenol (2, 6 -subst) Benzamides Borate Esters Benzoates (non-ring subst) Bis-anilines Hydrofurans (alcohol & ketone) Imidazolidines Isothiazolines Mono-cyclic Hydrocarbons Oxazoles Phenones (n-alkyl) Pyrrolidiones Sorbitans Triazines Log Kow Range ( 2. 3 - 4. 9) ( 4. 2) ( 1. 7 - 2. 1) (-0. 5 - 1. 0) ( 3. 6 - 4. 2) ( 1. 6 - 6. 2) ( 0. 0 - 1. 2) (-0. 9 - 0. 6) ( 0. 6 - 2. 5) ( 2. 7 - 4. 6) (<0. 3) ( 1. 6 - 4. 1) (-0. 4 - 3. 3) ( 3. 2 - 5. 9) (-0. 4 - 3. 4) Meets specific distance criteria for: Sites “A-B”; Sites “A-C”; Sites “A-B-C” Contains some attenuating feature, steric or other Yes No High Affinity “A-B”; “A-C” or “A-B-C” RBA > 0. 1% No VI Site “A” Contains Phenol Fragment Yes Belongs to known Active Sub-class Yes Alkylphenols Alkoxy phenols Parabens Salicylates Log Kow Range (1. 5 - 8. 0) (2. 3 - 4. 3) (1. 9 - 5. 4) (2. 5 - 5. 1) Yes No No RBA > 0. 00001% Exact match to Mixed Phenols Training Set Structure and measured RBA Mixed Phenols Yes No Belongs to known Active Sub-class No Mixed Organics Exact match to Mixed Organics Training Set Structure and measured RBA Weak Unknown Binding Potential Strong Log Kow Range 4 -Alkylanilines ( 1. 4 - 5. 1) 4 -Alkoxy Anilines ( 2. 6 - 3. 7) Phthalates ( 2. 5 - 6. 8) Phenones-(branched) ( 2. 7 - 4. 8) 4 -Alkyl cyclohexanols ( 2. 4 - 3. 8) 4 -Alkyl cyclohexanones ( 2. 4 - 3. 8) * 2 -; 4 -; or 2, 4, 6 -Benzoates (substitution-dependent Log Kows) RBA < 0. 00001% RBA > 0. 00001% Yes Belong to untested class Unknown Binding Potential Yes Strength of attenuation factors RBA < 0. 00001% No Site “B” VII Contains “Specified” Fragment RBA > 0. 00001% RBA < 0. 00001% No Unknown Binding Potential Low Affinity “A-B”, “A-C” or 36 “A-B-C” 0. 00001% < RBA < 0. 1%

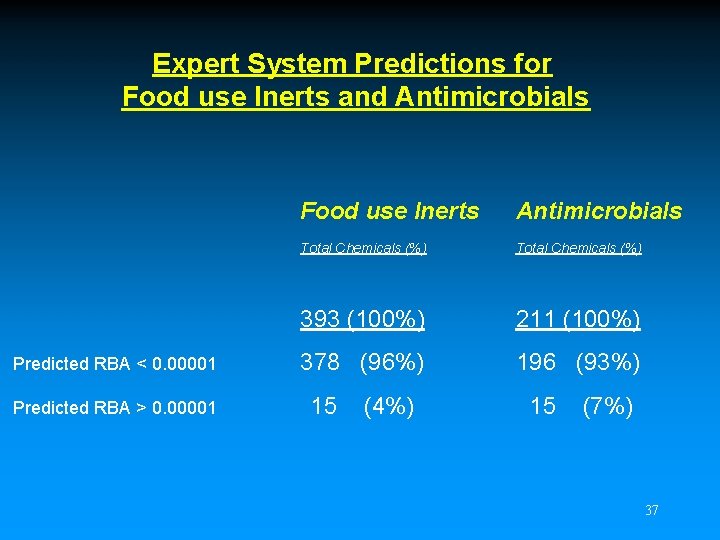
Expert System Predictions for Food use Inerts and Antimicrobials Predicted RBA < 0. 00001 Predicted RBA > 0. 00001 Food use Inerts Antimicrobials Total Chemicals (%) 393 (100%) 211 (100%) 378 (96%) 196 (93%) 15 (4%) 15 (7%) 37

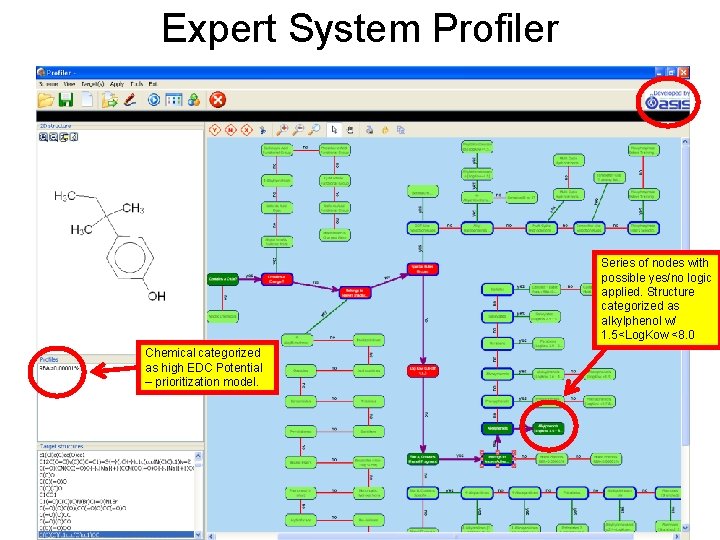
Expert System Profiler Series of nodes with possible yes/no logic applied. Structure categorized as alkylphenol w/ 1. 5<Log. Kow <8. 0 Chemical categorized as high EDC Potential – prioritization model.

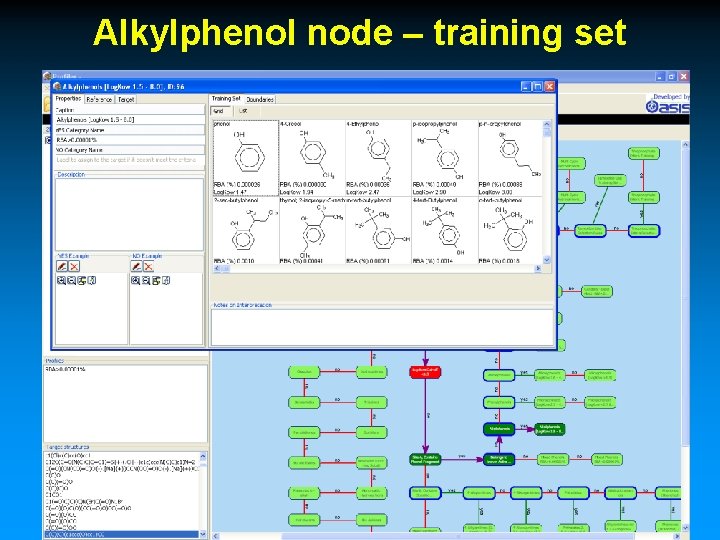
Alkylphenol node – training set

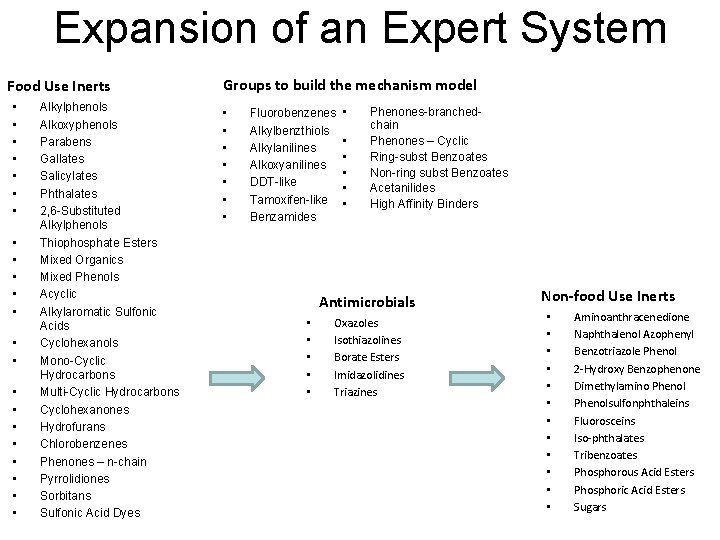
Expansion of an Expert System Non-Food Use Inert Ingredients Non-Food Use Inerts list included: 2888 = 1423 discrete chemicals + 1465 non-discrete substances (49% discrete : 51% non-discrete) 1423 discrete chemicals include: 1192 organic chemicals (84%) 205 inorganic chemicals (14%) 26 organometallic compounds (2%) 1465 non-discrete substances include: 596 polymers of mixed chain length 174 mixtures 695 undefined substances

Expansion of an Expert System Food Use Inerts • • • • • • Alkylphenols Alkoxyphenols Parabens Gallates Salicylates Phthalates 2, 6 -Substituted Alkylphenols Thiophosphate Esters Mixed Organics Mixed Phenols Acyclic Alkylaromatic Sulfonic Acids Cyclohexanols Mono-Cyclic Hydrocarbons Multi-Cyclic Hydrocarbons Cyclohexanones Hydrofurans Chlorobenzenes Phenones – n-chain Pyrrolidiones Sorbitans Sulfonic Acid Dyes Groups to build the mechanism model • • Fluorobenzenes Alkylbenzthiols Alkylanilines Alkoxyanilines DDT-like Tamoxifen-like Benzamides • • • Phenones-branchedchain Phenones – Cyclic Ring-subst Benzoates Non-ring subst Benzoates Acetanilides High Affinity Binders Antimicrobials • • • Oxazoles Isothiazolines Borate Esters Imidazolidines Triazines Non-food Use Inerts • • • Aminoanthracenedione Naphthalenol Azophenyl Benzotriazole Phenol 2 -Hydroxy Benzophenone Dimethylamino Phenolsulfonphthaleins Fluorosceins Iso-phthalates Tribenzoates Phosphorous Acid Esters Phosphoric Acid Esters Sugars

Human ER Binding Affinity and Gene Activation 42

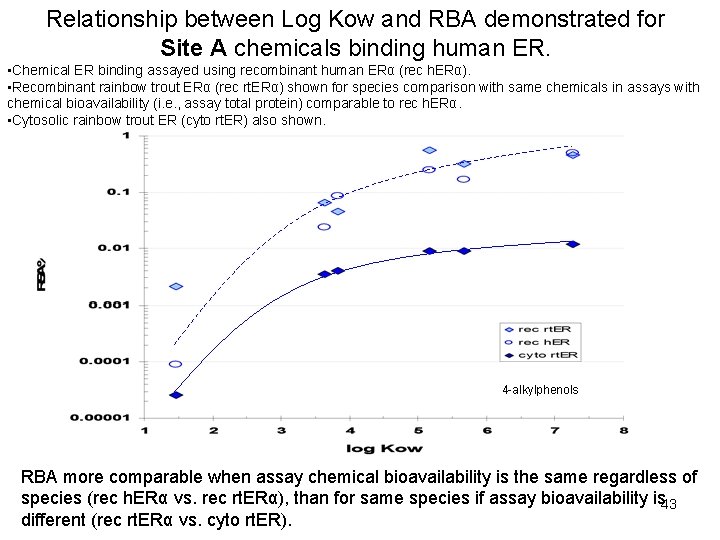
Relationship between Log Kow and RBA demonstrated for Site A chemicals binding human ER. • Chemical ER binding assayed using recombinant human ERα (rec h. ERα). • Recombinant rainbow trout ERα (rec rt. ERα) shown for species comparison with same chemicals in assays with chemical bioavailability (i. e. , assay total protein) comparable to rec h. ERα. • Cytosolic rainbow trout ER (cyto rt. ER) also shown. 4 -alkylphenols RBA more comparable when assay chemical bioavailability is the same regardless of species (rec h. ERα vs. rec rt. ERα), than for same species if assay bioavailability is 43 different (rec rt. ERα vs. cyto rt. ER).

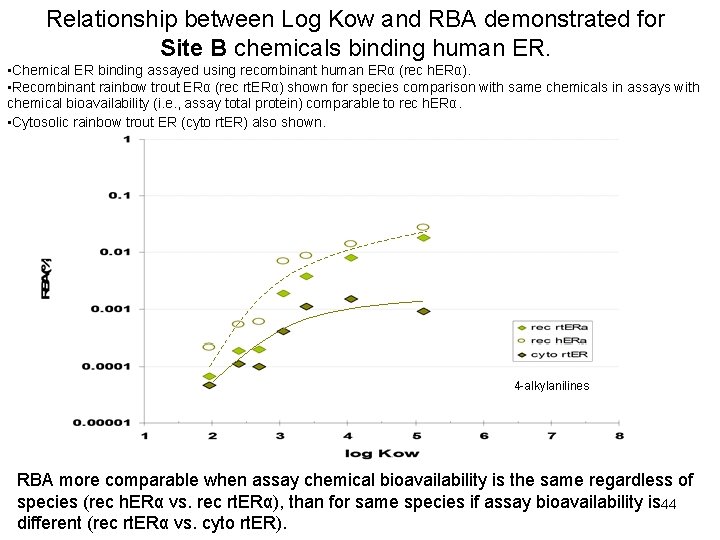
Relationship between Log Kow and RBA demonstrated for Site B chemicals binding human ER. • Chemical ER binding assayed using recombinant human ERα (rec h. ERα). • Recombinant rainbow trout ERα (rec rt. ERα) shown for species comparison with same chemicals in assays with chemical bioavailability (i. e. , assay total protein) comparable to rec h. ERα. • Cytosolic rainbow trout ER (cyto rt. ER) also shown. 4 -alkylanilines RBA more comparable when assay chemical bioavailability is the same regardless of species (rec h. ERα vs. rec rt. ERα), than for same species if assay bioavailability is 44 different (rec rt. ERα vs. cyto rt. ER).

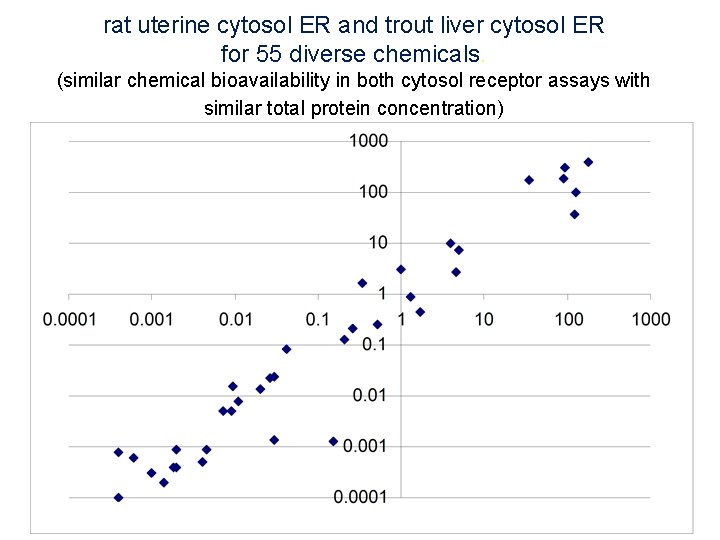
rat uterine cytosol ER and trout liver cytosol ER for 55 diverse chemicals. (similar chemical bioavailability in both cytosol receptor assays with similar total protein concentration) 45

Working with EPA NCCT to determine how mammalian model-based ER HTS assays correlate with the in vitro assay used to build ES Ø focus was on selection of chemicals to cover chemical classes used in development of ER Expert System Ø The NCCT ER assays are: • • Human ER, Bovine ER, Mouse ER-alpha competitive binding assays Human ER-alpha reporter gene assays: agonist & antagonist mode • • (2 vendors) ERE interaction assay Ø Future: plan to extend to AR activation in the future with goal to build class-based expert system Ø Goal: use h. ER data (low-, medium-, or high-throughput) to build ES with expanded chemical classes. 46

Conclusions • An Expert System based upon the ER-mediated AOP – Knowledge of common initiating event across chemical classes facilitates development of QSARs and read-across methods to predict toxicity potential of untested chemicals • OECD QSAR Validation Principles • ICPS MOA

Relevance of ER model to predicting xenobiotic epigenetic chemicals?

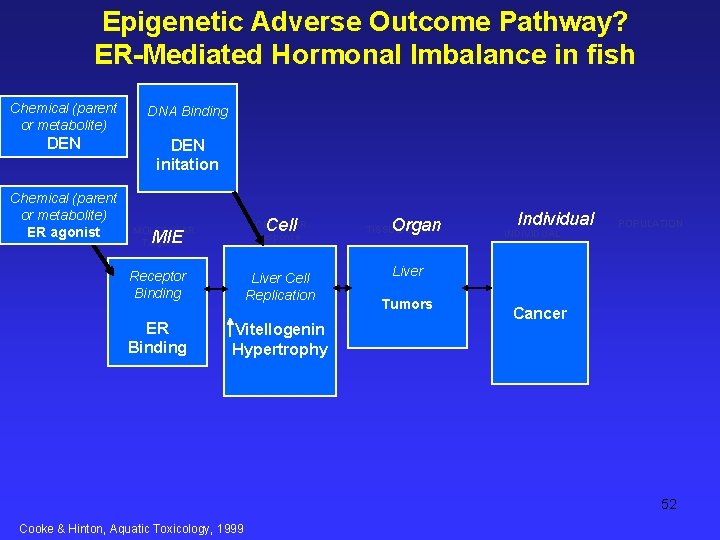
Promotion by 17 b-estradiol and b-hexachlorocyclohexane of hepatocellular tumors in medaka, Oryzias latipes. J. B. Cooke, D. E. Hinton. Aquatic Toxicology 45 (1999) 127– 145 Hypothesis: Many laboratory and field studies with various fish species show a higher prevalence Of hepatocellular neoplasia in females than in males. During female sexual maturation, endogenous estrogens stimulate substantial increases in synthetic activity (e. g. , vitellogenin, choriogenin productions) and hepatocytes proliferation. -tested hypothesis that estrogens promote growth of hepatic preneoplastic lesions and tumors. Medaka (Oryzias latipes) – -low dose of diethylnitrosamine (DEN; 200 mg /1 , 24 h) at 3 weeks of age -then fed purified casein-based diet daily from 1 to 7 months of age - with or w/o 17 b-estradiol (E 2; 0. 01– 10. 0 mg g 1 dry diet) -With xenoestrogen, b-hexachlorocyclohexane (b. HCH, 0. 01– 100. 0 mg g 1 dry diet). -Livers examined for foci of cellular alteration (FCA) & and hepatocellular tumors. -E 2 increased prevalences of hepatocellular adenoma or carcinoma -(26% in DEN plus 10 ppm E 2 group versus 4. 6% in DEN only group, PB 0. 01). -With incr E 2, avg# basophilic FCA (BF) rose; #eosinophilic FCA (EF) sharply declined. DEN plus b. HCH treatment groups > numbers of tumors in most, and greater numbers of BF in all

Promotion by 17 b-estradiol and b-hexachlorocyclohexane of hepatocellular tumors in medaka, Oryzias latipes. J. B. Cooke, D. E. Hinton. Aquatic Toxicology 45 (1999) 127– 145 Results: Livers examined for foci of cellular alteration (FCA) & and hepatocellular tumors. -E 2 increased prevalences of hepatocellular adenoma or carcinoma -(26% in DEN plus 10 ppm E 2 group versus 4. 6% in DEN only group, PB 0. 01). -With incr E 2, avg# basophilic FCA (BF) rose; #eosinophilic FCA (EF) sharply declined. DEN plus b. HCH treatment groups > numbers of tumors in most, and greater numbers of BF in all DEN only: For all DEN-treated groups, BF were more common in female medaka, and EF more common in males. No tumors were found in fish fed E 2 or b. HCH without DEN exposure Liver wts -control medaka, significantly larger in females -E 2 treatments (0. 1, 1. 0 or 10. 0 ppm E 2) elevated liver weights in males similar to that in females. -b. HCH had no effect on liver weights.

Promotion by 17 b-estradiol and b-hexachlorocyclohexane of hepatocellular tumors in medaka, Oryzias latipes. J. B. Cooke, D. E. Hinton. Aquatic Toxicology 45 (1999) 127– 145 Conclusions: -E 2 is a tumor promoter in medaka. -Because tumor increases were not statistically significant, b. HCH was considered a weakly positive modulator. -E 2 particularly promoted tumor development in male medaka, indicating xenobiotics with mechanism of action like that of E 2 may escalate growth in wild fish of previously initiated cells into tumors.

Epigenetic Adverse Outcome Pathway? ER-Mediated Hormonal Imbalance in fish Chemical (parent or metabolite) DNA Binding DEN initation Chemical (parent or metabolite) ER agonist Cell CELLULAR Response MOLECULAR Target MIE Receptor Binding ER Binding Liver Cell Replication Vitellogenin Hypertrophy Organ TISSUE/ORG Individual INDIVIDUAL POPULATION Liver Tumors Cancer 52 Cooke & Hinton, Aquatic Toxicology, 1999

ER model Relevance? Wildlife species: - reproductive impairment is more relevant than cancer from population perspective, so ER ES focused on repro impairment AOP Rodents: Rodent ER binding – good correlation with fish. ER - Estrogens and anabolic steroids can increase liver adenomas in rats/mice Mammals in general: Many of same chemicals bind ER and result in gene activation -possible role in promotion, progression? Liver is not considered one of the classic targets for hormonal carcinogenesis but liver cancer development is influenced by sex hormones (estrogen, testosterone).