Molecular Evolution Cdric Notredame 15022022 Molecular Evolution is













- Slides: 39

Molecular Evolution Cédric Notredame (15/02/2022)

Molecular Evolution is a rapidly developing field………. . Cédric Notredame (15/02/2022)

Phenotypic diversity in populations suggests underlying genetic diversity Cédric Notredame (15/02/2022)

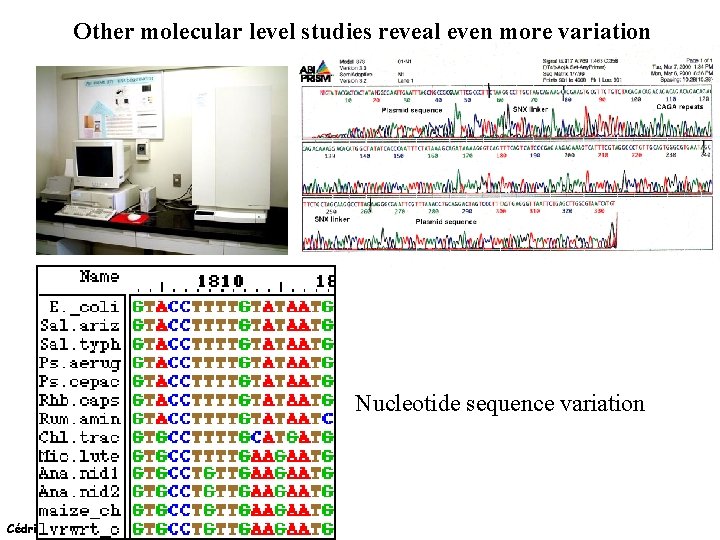
Other molecular level studies reveal even more variation Nucleotide sequence variation Cédric Notredame (15/02/2022)

Is Modern Genetic Data Compatible with Darwinism Cédric Notredame (15/02/2022)

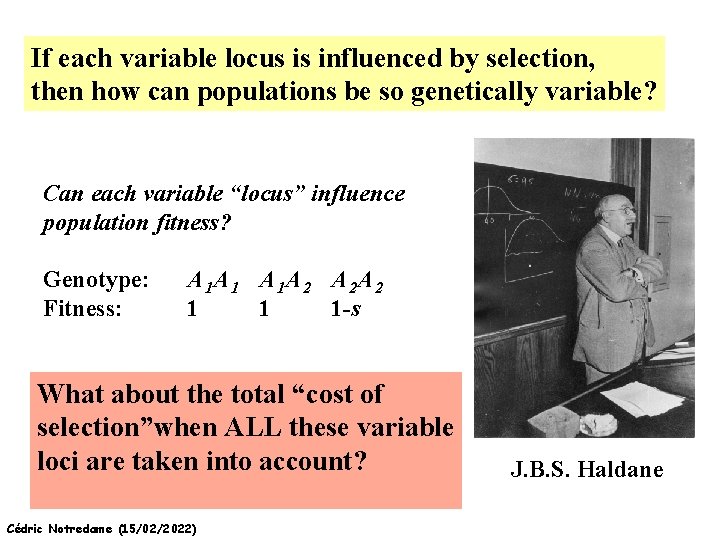
If each variable locus is influenced by selection, then how can populations be so genetically variable? Can each variable “locus” influence population fitness? Genotype: Fitness: A 1 A 1 A 1 A 2 A 2 A 2 1 1 1 -s What about the total “cost of selection”when ALL these variable loci are taken into account? Cédric Notredame (15/02/2022) J. B. S. Haldane

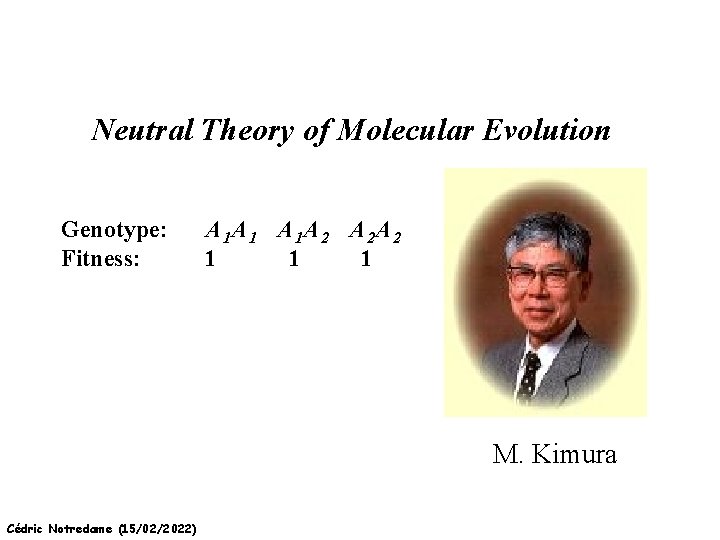
Neutral Theory of Molecular Evolution Genotype: Fitness: A 1 A 1 A 1 A 2 A 2 A 2 1 1 1 M. Kimura Cédric Notredame (15/02/2022)

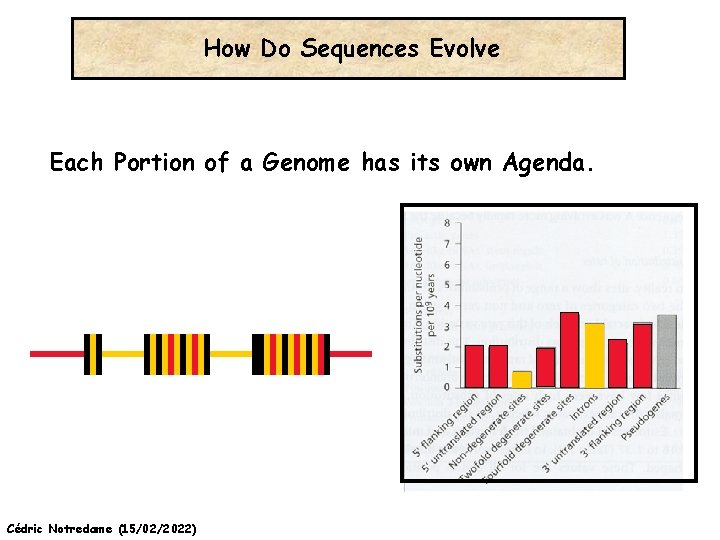
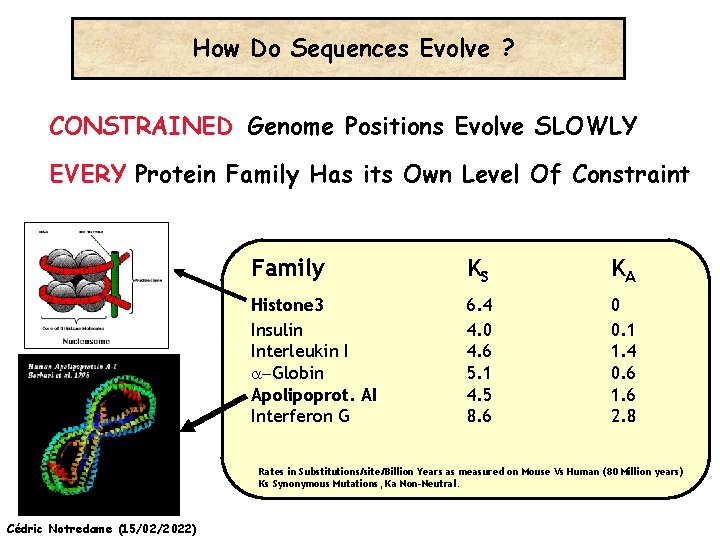
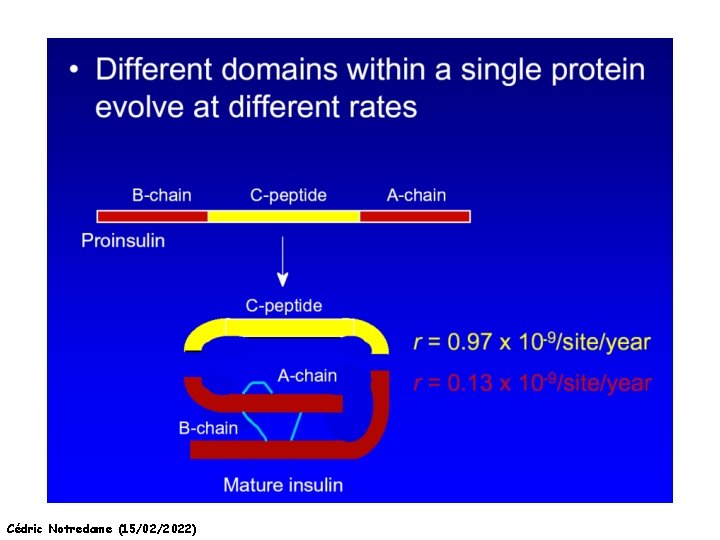
How Do Sequences Evolve Each Portion of a Genome has its own Agenda. Cédric Notredame (15/02/2022)

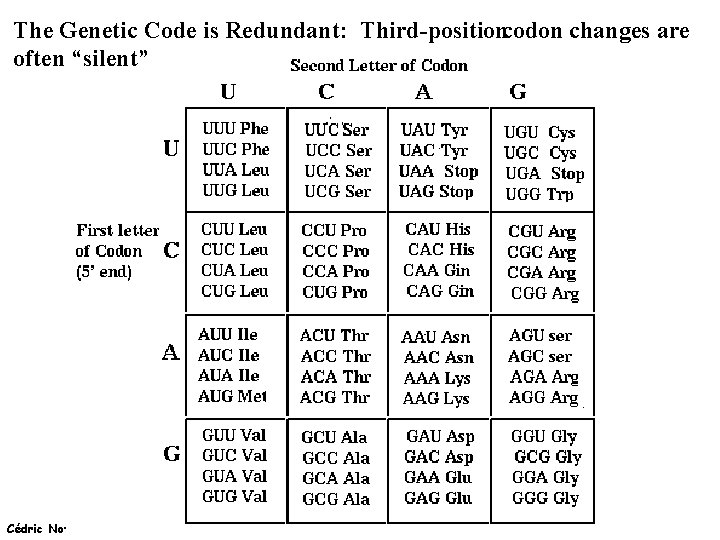
The Genetic Code is Redundant: Third-positioncodon changes are often “silent” Cédric Notredame (15/02/2022)

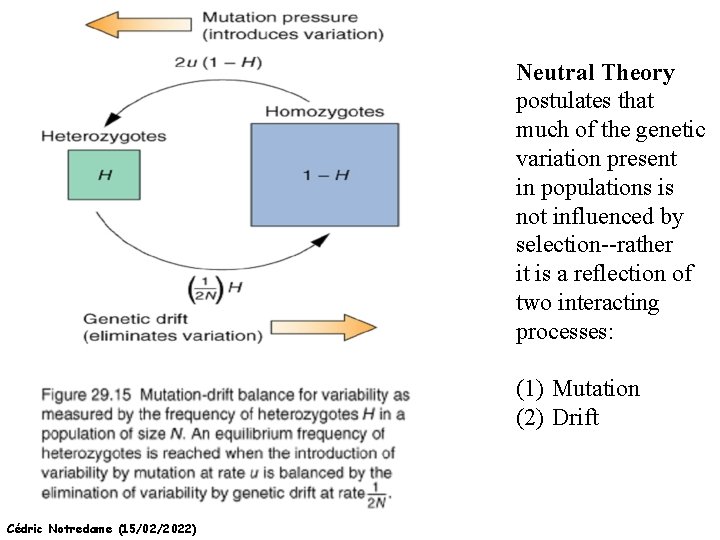
Neutral Theory postulates that much of the genetic variation present in populations is not influenced by selection--rather it is a reflection of two interacting processes: (1) Mutation (2) Drift Cédric Notredame (15/02/2022)

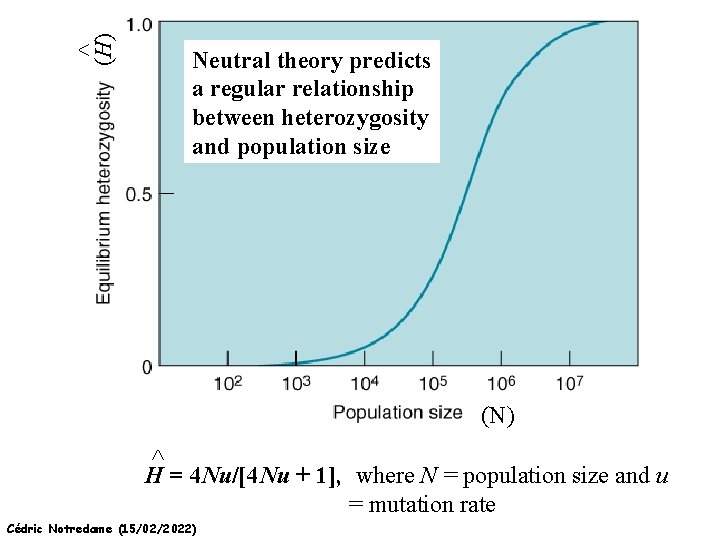
^ (H) Neutral theory predicts a regular relationship between heterozygosity and population size (N) ^ H = 4 Nu/[4 Nu + 1], where N = population size and u = mutation rate Cédric Notredame (15/02/2022)

The Molecular Clock Cédric Notredame (15/02/2022)

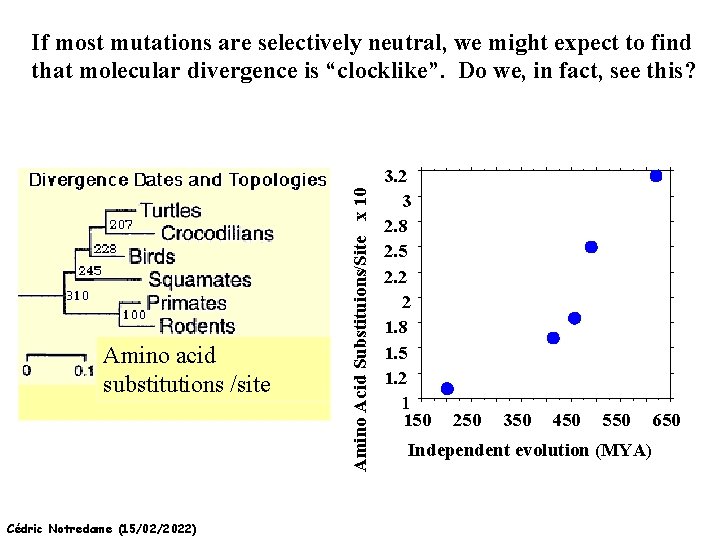
THE MOLECULAR CLOCK HYPOTHESIS Another prediction of Neutral Theory--Because mutation is regular (or “clocklike”) and because selection does not influence the rate of divergence, divergence of DNA and protein molecules in two separate lineages should occur in a REGULAR, clocklike manner i. e. , the longer the time separating two species from their common ancestor, the more divergent will be a given protein or DNA molecule (and this relationship should be LINEAR) Cédric Notredame (15/02/2022)

Amino acid substitutions /site Cédric Notredame (15/02/2022) Amino Acid Substituions/Site x 10 If most mutations are selectively neutral, we might expect to find that molecular divergence is “clocklike”. Do we, in fact, see this? 3. 2 3 2. 8 2. 5 2. 2 2 1. 8 1. 5 1. 2 1 150 250 350 450 550 650 Independent evolution (MYA)

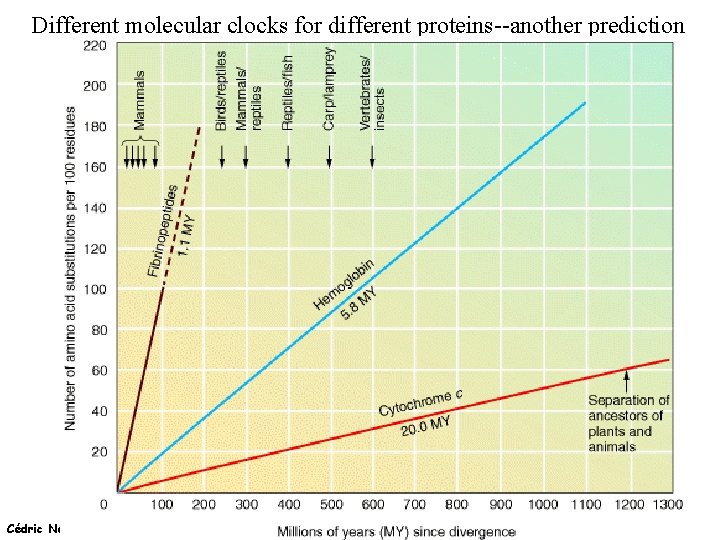
Different molecular clocks for different proteins--another prediction Cédric Notredame (15/02/2022)

How Do Sequences Evolve ? CONSTRAINED Genome Positions Evolve SLOWLY EVERY Protein Family Has its Own Level Of Constraint Family KS KA Histone 3 Insulin Interleukin I a-Globin Apolipoprot. AI Interferon G 6. 4 4. 0 4. 6 5. 1 4. 5 8. 6 0 0. 1 1. 4 0. 6 1. 6 2. 8 Rates in Substitutions/site/Billion Years as measured on Mouse Vs Human (80 Million years) Ks Synonymous Mutations, Ka Non-Neutral. Cédric Notredame (15/02/2022)

Cédric Notredame (15/02/2022)

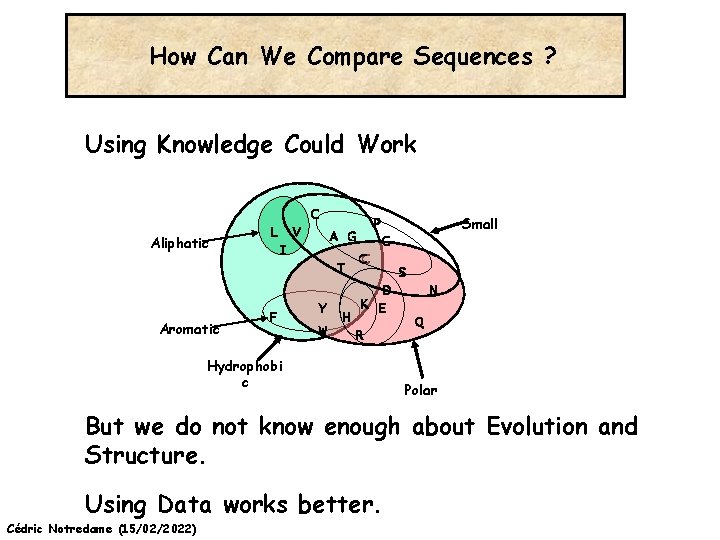
How Can We Compare Sequences ? Using Knowledge Could Work C Aliphatic L V I A G T Aromatic F Y W H Small P G CC D K E R Hydrophobi c S N Q Polar But we do not know enough about Evolution and Structure. Using Data works better. Cédric Notredame (15/02/2022)

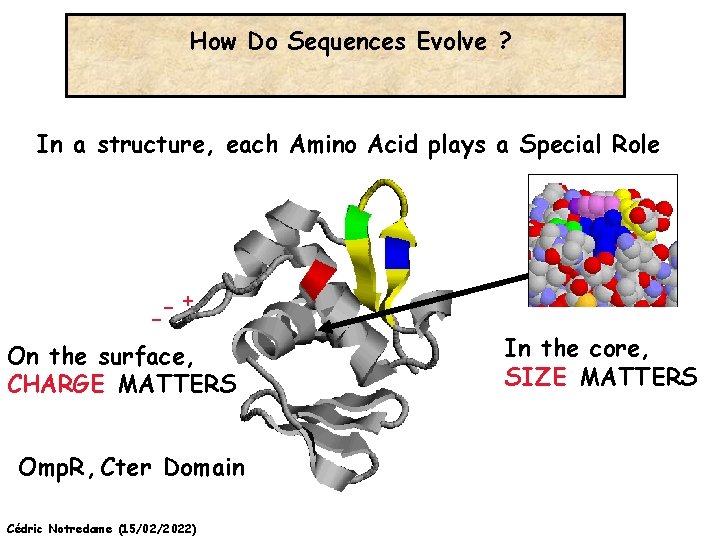
How Do Sequences Evolve ? In a structure, each Amino Acid plays a Special Role -+ On the surface, CHARGE MATTERS Omp. R, Cter Domain Cédric Notredame (15/02/2022) In the core, SIZE MATTERS

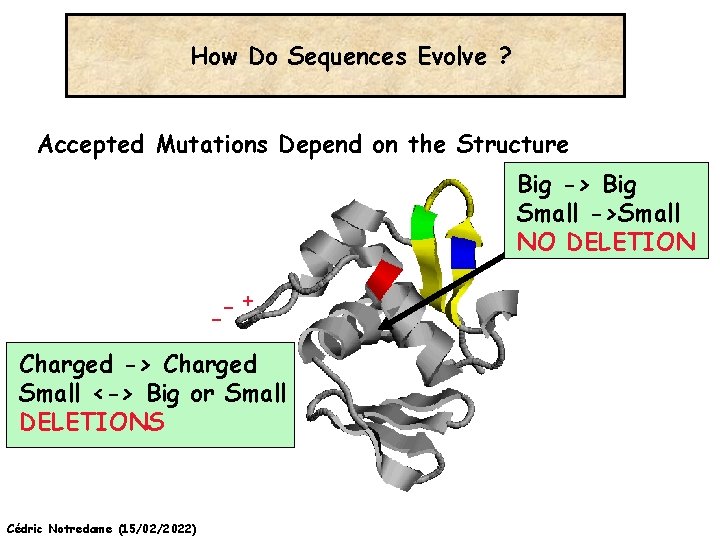
How Do Sequences Evolve ? Accepted Mutations Depend on the Structure Big -> Big Small ->Small NO DELETION + Charged -> Charged Small <-> Big or Small DELETIONS Cédric Notredame (15/02/2022)

Alignments and Evolution Cédric Notredame (15/02/2022)

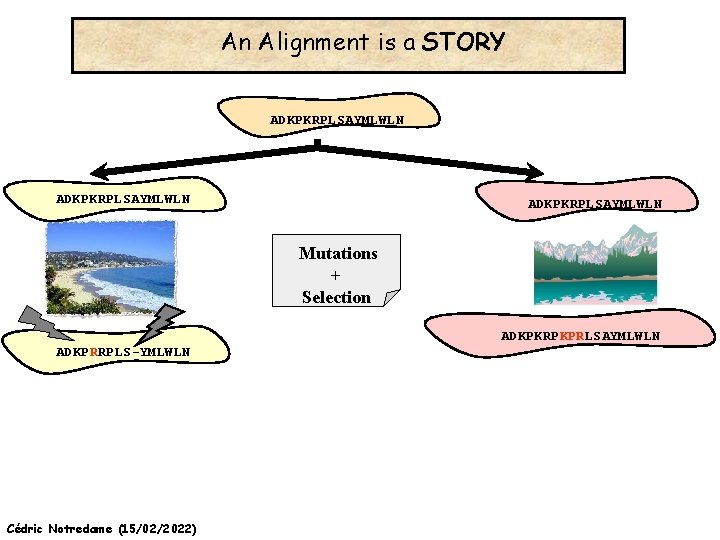
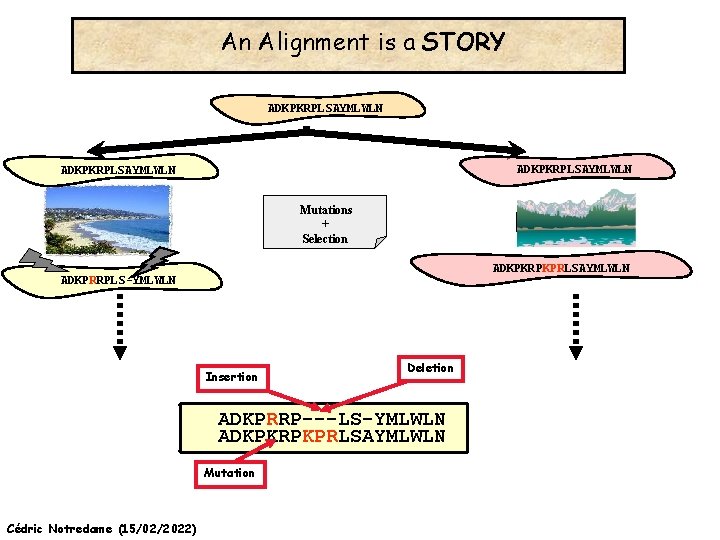
An Alignment is a STORY ADKPKRPLSAYMLWLN Mutations + Selection ADKPKRPKPRLSAYMLWLN ADKPRRPLS-YMLWLN Cédric Notredame (15/02/2022)

An Alignment is a STORY ADKPKRPLSAYMLWLN Mutations + Selection ADKPKRPKPRLSAYMLWLN ADKPRRPLS-YMLWLN Insertion Deletion ADKPRRP---LS-YMLWLN ADKPKRPKPRLSAYMLWLN Mutation Cédric Notredame (15/02/2022)

Comparing Is Reconstructing Evolution Cédric Notredame (15/02/2022)

Divergeant or Convergeant Evolution ? Cédric Notredame (15/02/2022)

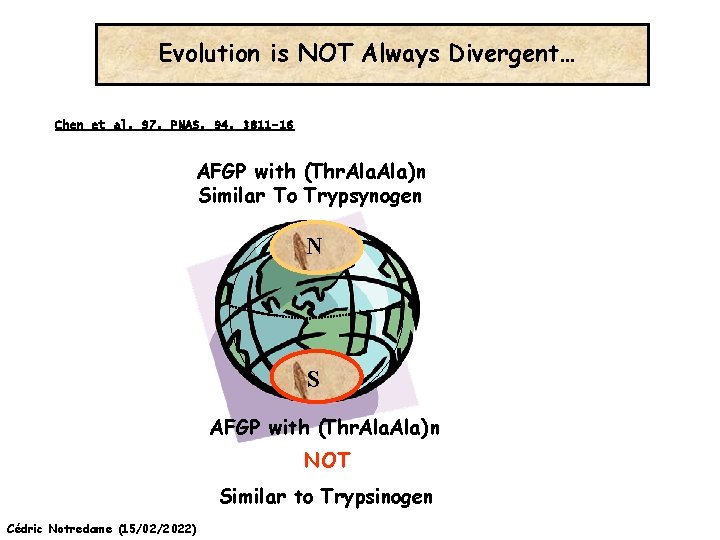
Evolution is NOT Always Divergent… Chen et al, 97, PNAS, 94, 3811 -16 AFGP with (Thr. Ala)n Similar To Trypsynogen N S AFGP with (Thr. Ala)n NOT Similar to Trypsinogen Cédric Notredame (15/02/2022)

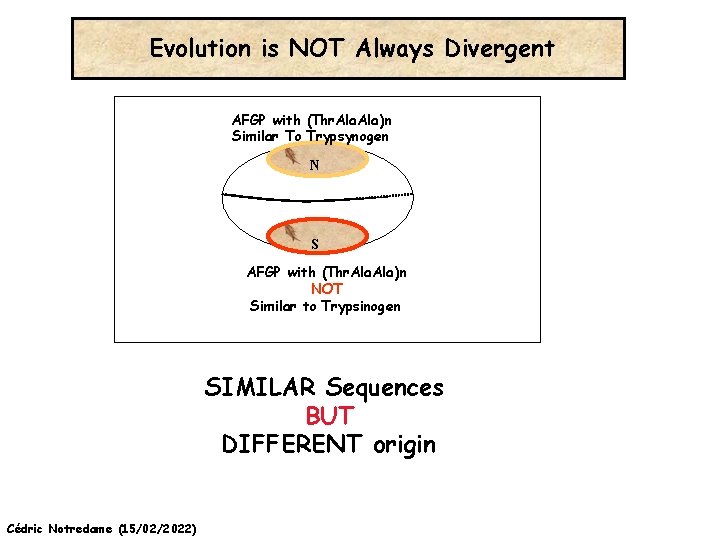
Evolution is NOT Always Divergent AFGP with (Thr. Ala)n Similar To Trypsynogen N S AFGP with (Thr. Ala)n NOT Similar to Trypsinogen SIMILAR Sequences BUT DIFFERENT origin Cédric Notredame (15/02/2022)

Orthologous And Paralogous Genes Cédric Notredame (15/02/2022)

Cédric Notredame (15/02/2022)

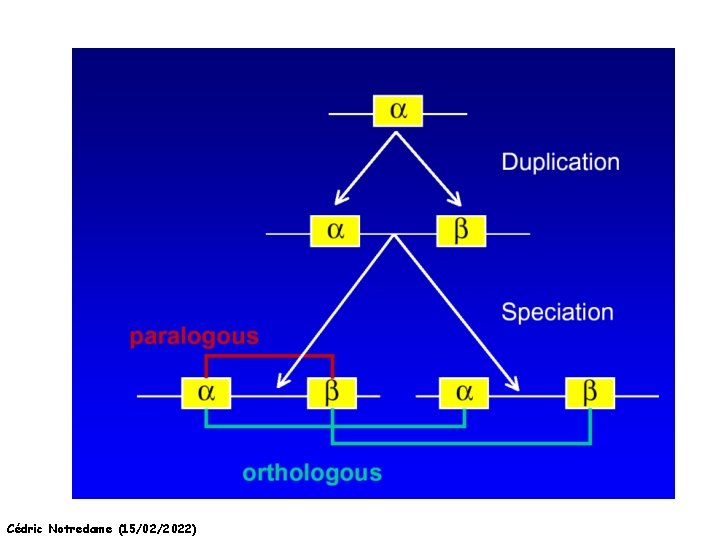
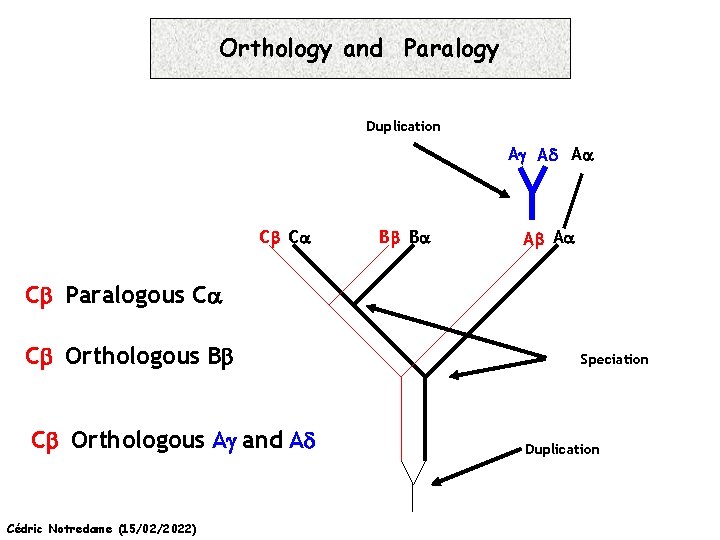
Orthology and Paralogy Duplication Ag Ad Aa Cb Ca Bb Ba Ab Aa Cb Paralogous Ca Cb Orthologous Bb Cb Orthologous Ag and Ad Cédric Notredame (15/02/2022) Speciation Duplication

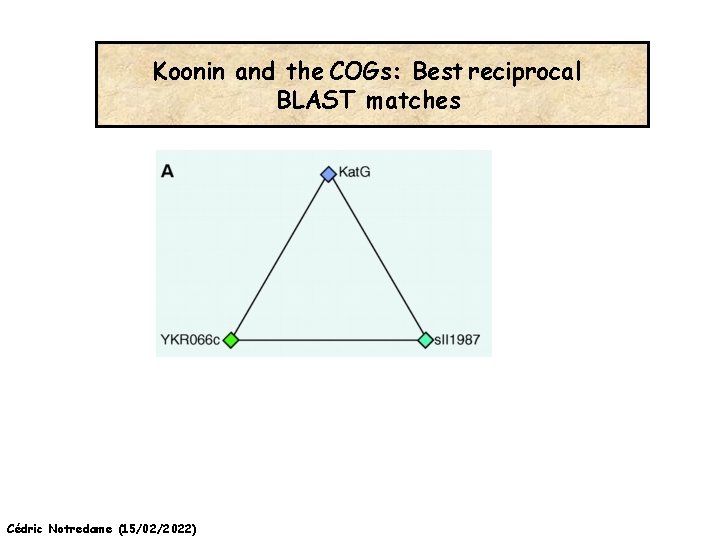
Koonin and the COGs: Best reciprocal BLAST matches Cédric Notredame (15/02/2022)

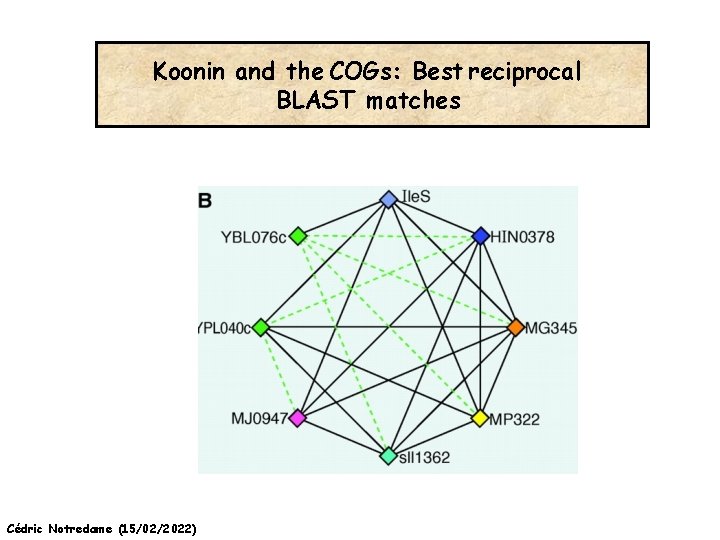
Koonin and the COGs: Best reciprocal BLAST matches Cédric Notredame (15/02/2022)

Cédric Notredame (15/02/2022)

Cédric Notredame (15/02/2022)

Cédric Notredame (15/02/2022)

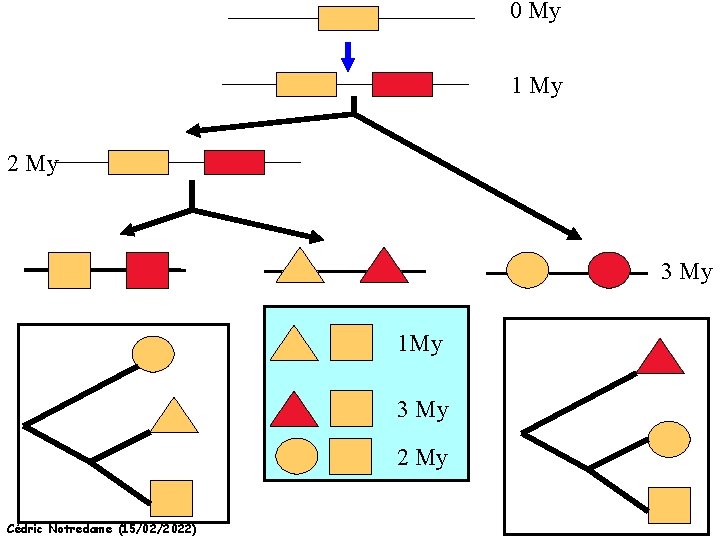
Mixing Orthologs and Paralogs leads to inexact Phylogenetic trees Cédric Notredame (15/02/2022)

0 My 1 My 2 My 3 My 1 My 3 My 2 My Cédric Notredame (15/02/2022)

Positions are not enough to infer Orthology Cédric Notredame (15/02/2022)

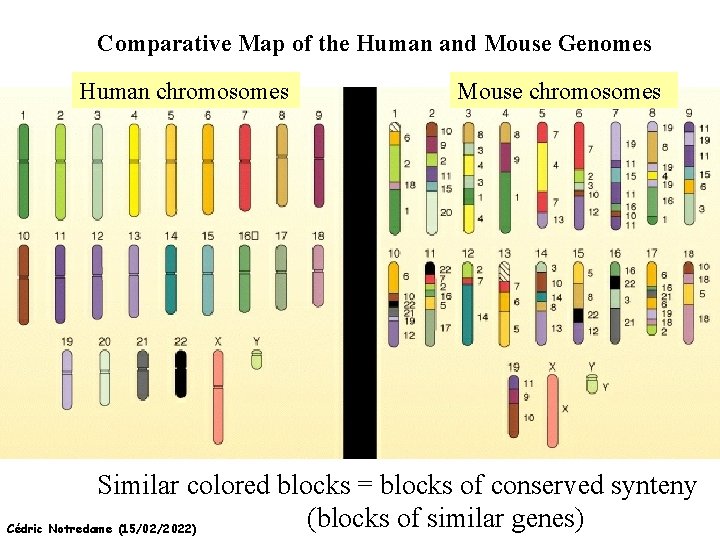
Comparative Map of the Human and Mouse Genomes Human chromosomes Mouse chromosomes Similar colored blocks = blocks of conserved synteny (blocks of similar genes) Cédric Notredame (15/02/2022)