Comparing Two Protein Sequences Cdric Notredame 21112020 Comparing







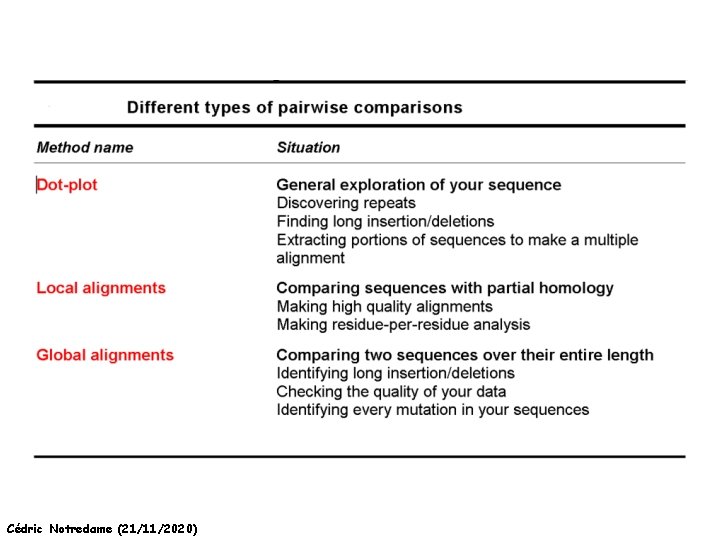






- Slides: 78

Comparing Two Protein Sequences Cédric Notredame (21/11/2020)

Comparing Two Protein Sequences Cédric Notredame (21/11/2020)

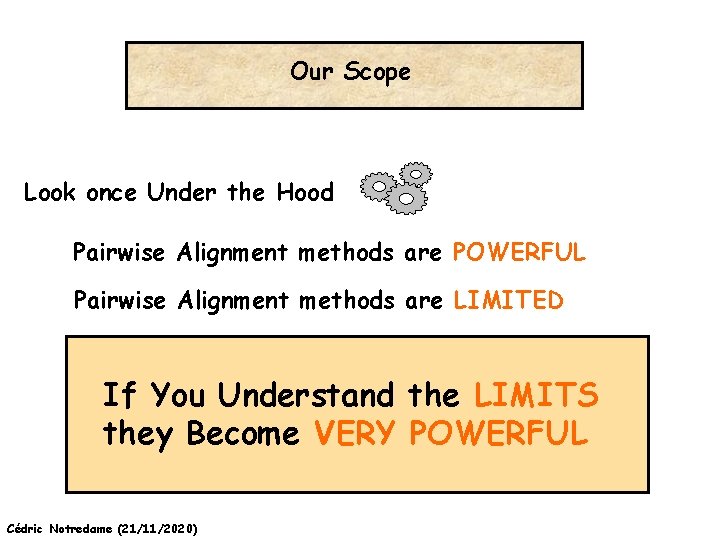
Our Scope Look once Under the Hood Pairwise Alignment methods are POWERFUL Pairwise Alignment methods are LIMITED If You Understand the LIMITS they Become VERY POWERFUL Cédric Notredame (21/11/2020)

Outline -WHY Does It Make Sense To Compare Sequences -HOW Can we Compare Two Sequences ? -HOW Can we Align Two Sequences ? -HOW can I Search a Database ? Cédric Notredame (21/11/2020)

Why Does It Make Sense To Compare Sequences ? Sequence Evolution Cédric Notredame (21/11/2020)

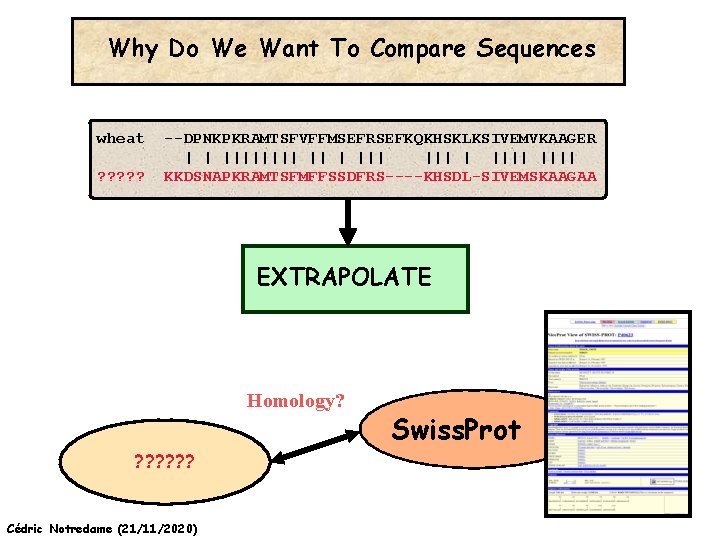
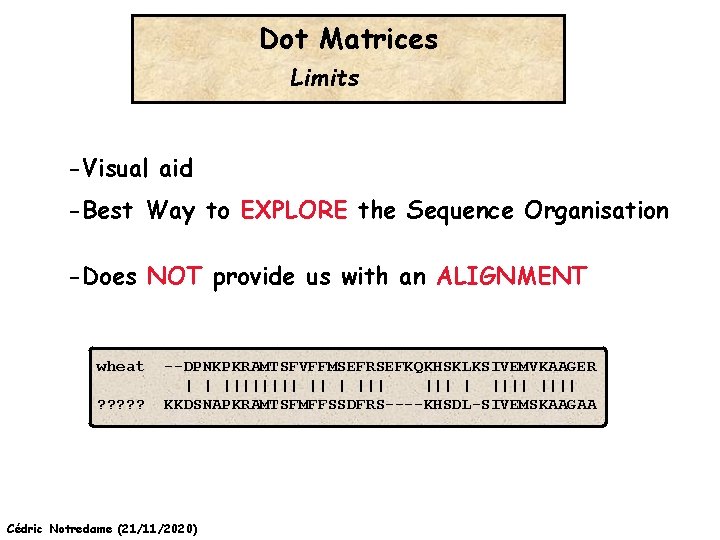
Why Do We Want To Compare Sequences wheat ? ? ? --DPNKPKRAMTSFVFFMSEFRSEFKQKHSKLKSIVEMVKAAGER | | |||| ||| | |||| KKDSNAPKRAMTSFMFFSSDFRS----KHSDL-SIVEMSKAAGAA EXTRAPOLATE Homology? ? ? ? Cédric Notredame (21/11/2020) Swiss. Prot

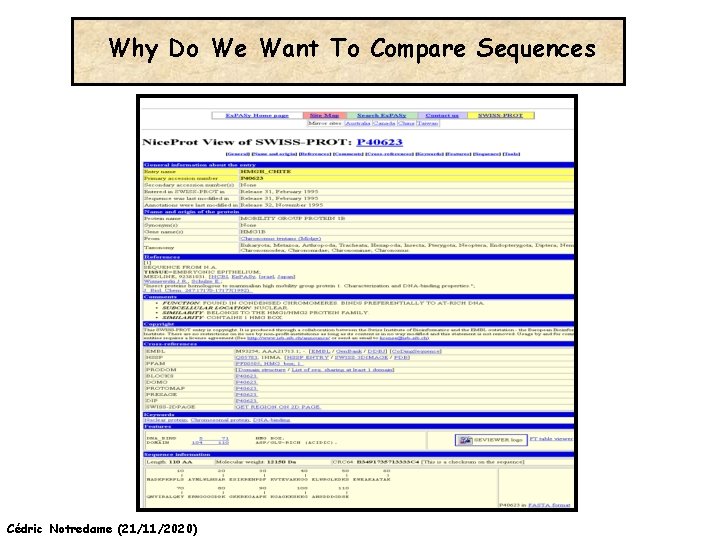
Why Do We Want To Compare Sequences Cédric Notredame (21/11/2020)

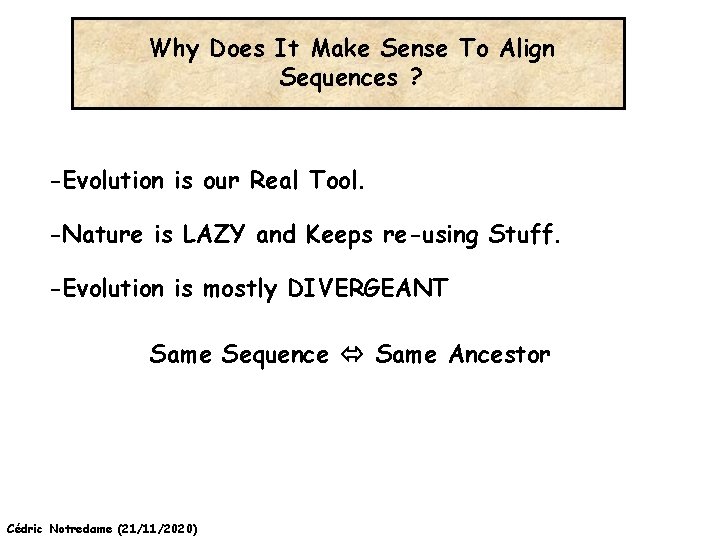
Why Does It Make Sense To Align Sequences ? -Evolution is our Real Tool. -Nature is LAZY and Keeps re-using Stuff. -Evolution is mostly DIVERGEANT Same Sequence Same Ancestor Cédric Notredame (21/11/2020)

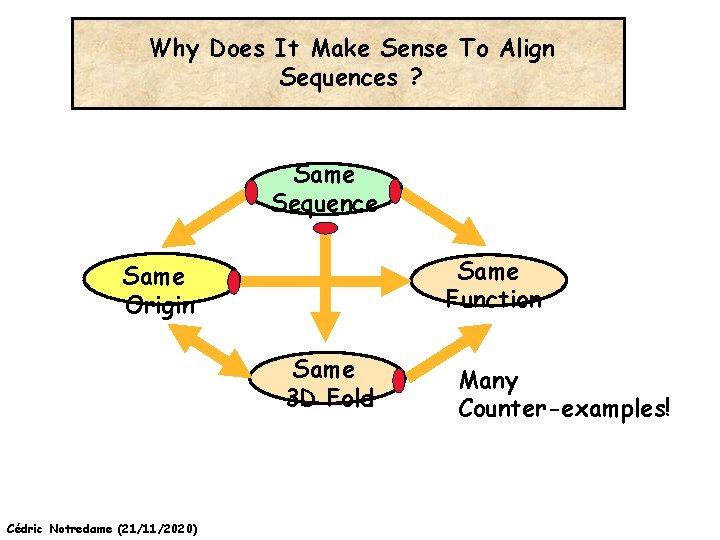
Why Does It Make Sense To Align Sequences ? Same Sequence Same Function Same Origin Same 3 D Fold Cédric Notredame (21/11/2020) Many Counter-examples!

Comparing Is Reconstructing Evolution Cédric Notredame (21/11/2020)

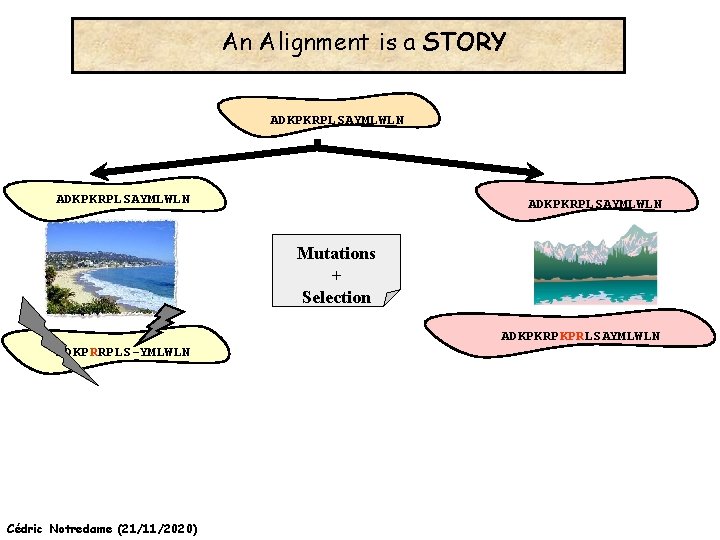
An Alignment is a STORY ADKPKRPLSAYMLWLN Mutations + Selection ADKPKRPKPRLSAYMLWLN ADKPRRPLS-YMLWLN Cédric Notredame (21/11/2020)

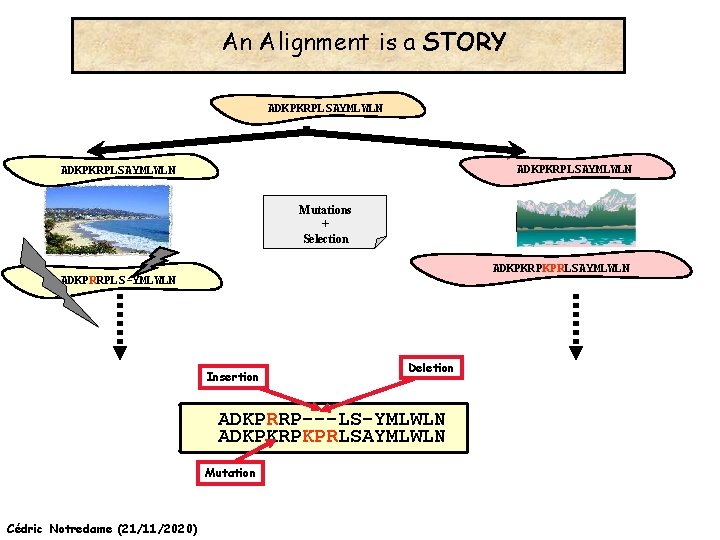
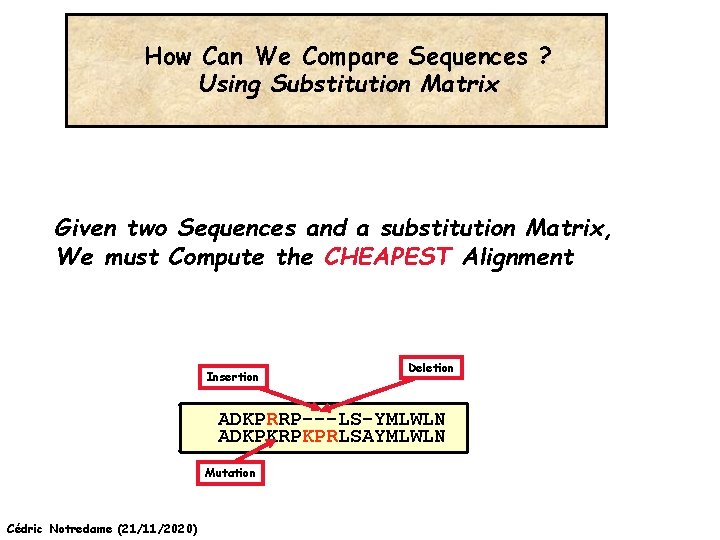
An Alignment is a STORY ADKPKRPLSAYMLWLN Mutations + Selection ADKPKRPKPRLSAYMLWLN ADKPRRPLS-YMLWLN Insertion Deletion ADKPRRP---LS-YMLWLN ADKPKRPKPRLSAYMLWLN Mutation Cédric Notredame (21/11/2020)

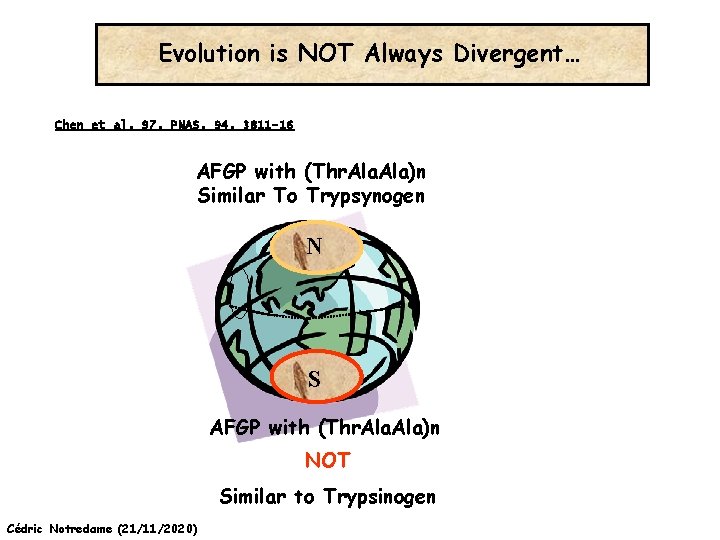
Evolution is NOT Always Divergent… Chen et al, 97, PNAS, 94, 3811 -16 AFGP with (Thr. Ala)n Similar To Trypsynogen N S AFGP with (Thr. Ala)n NOT Similar to Trypsinogen Cédric Notredame (21/11/2020)

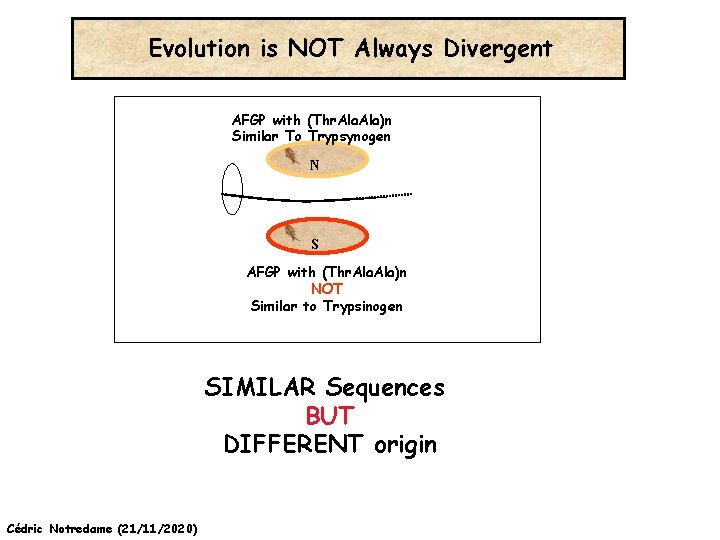
Evolution is NOT Always Divergent AFGP with (Thr. Ala)n Similar To Trypsynogen N S AFGP with (Thr. Ala)n NOT Similar to Trypsinogen SIMILAR Sequences BUT DIFFERENT origin Cédric Notredame (21/11/2020)

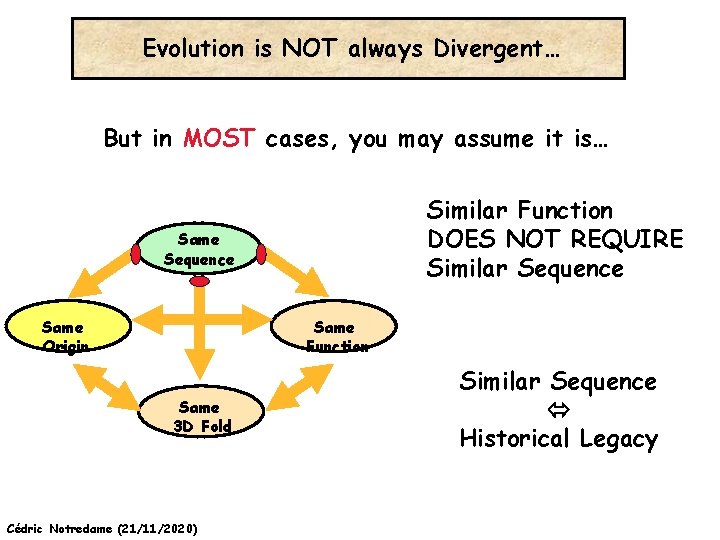
Evolution is NOT always Divergent… But in MOST cases, you may assume it is… Similar Function DOES NOT REQUIRE Similar Sequence Same Function Same Origin Same 3 D Fold Cédric Notredame (21/11/2020) Similar Sequence Historical Legacy

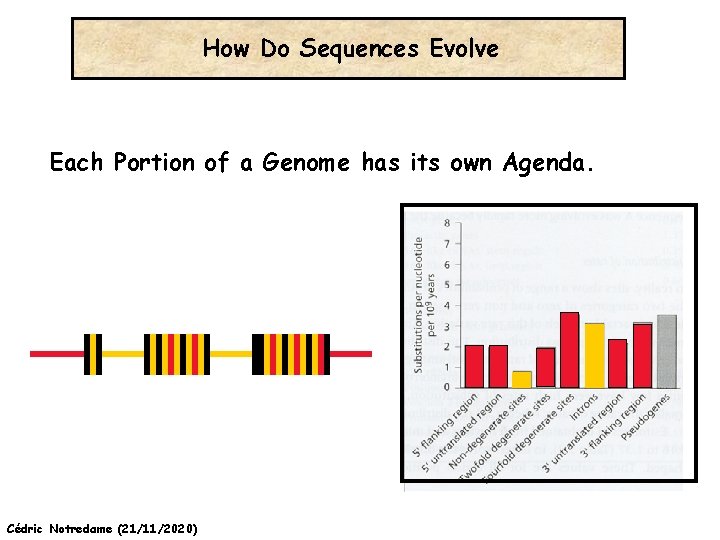
How Do Sequences Evolve Each Portion of a Genome has its own Agenda. Cédric Notredame (21/11/2020)

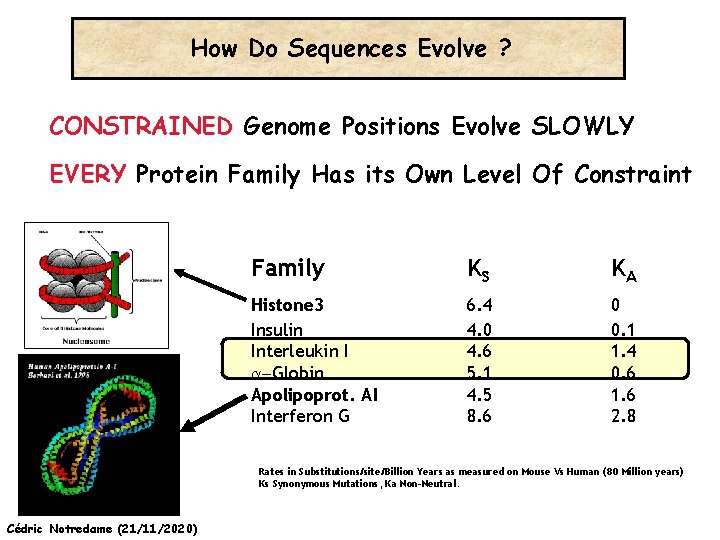
How Do Sequences Evolve ? CONSTRAINED Genome Positions Evolve SLOWLY EVERY Protein Family Has its Own Level Of Constraint Family KS KA Histone 3 Insulin Interleukin I a-Globin Apolipoprot. AI Interferon G 6. 4 4. 0 4. 6 5. 1 4. 5 8. 6 0 0. 1 1. 4 0. 6 1. 6 2. 8 Rates in Substitutions/site/Billion Years as measured on Mouse Vs Human (80 Million years) Ks Synonymous Mutations, Ka Non-Neutral. Cédric Notredame (21/11/2020)

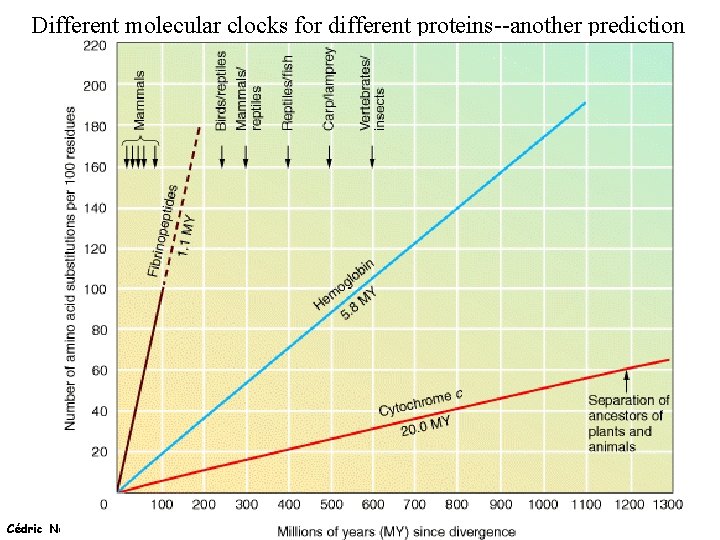
Different molecular clocks for different proteins--another prediction Cédric Notredame (21/11/2020)

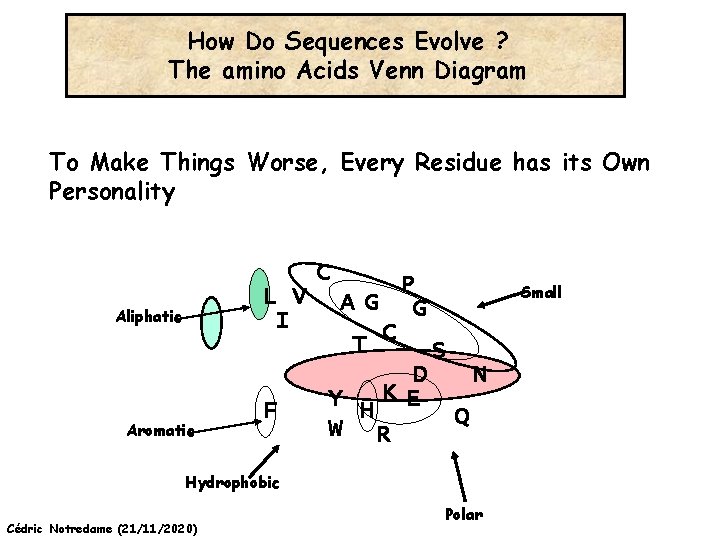
How Do Sequences Evolve ? The amino Acids Venn Diagram To Make Things Worse, Every Residue has its Own Personality C L V I Aliphatic Aromatic F P AG G T C S D N KE Y H Q W R Hydrophobic Cédric Notredame (21/11/2020) Polar Small

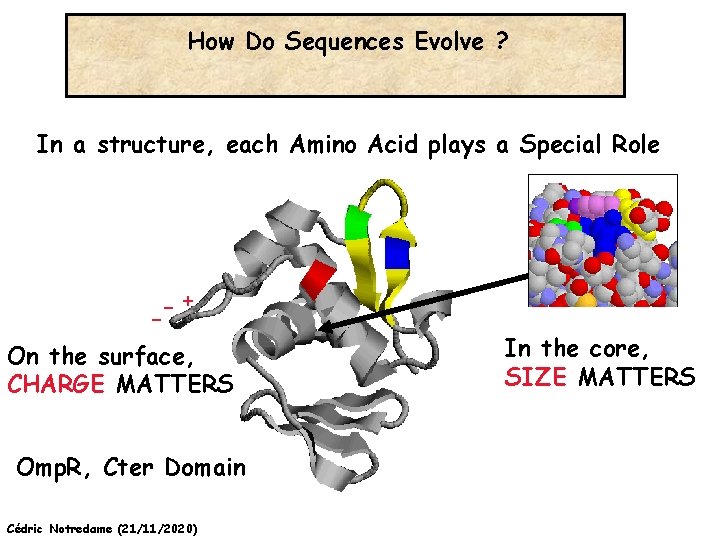
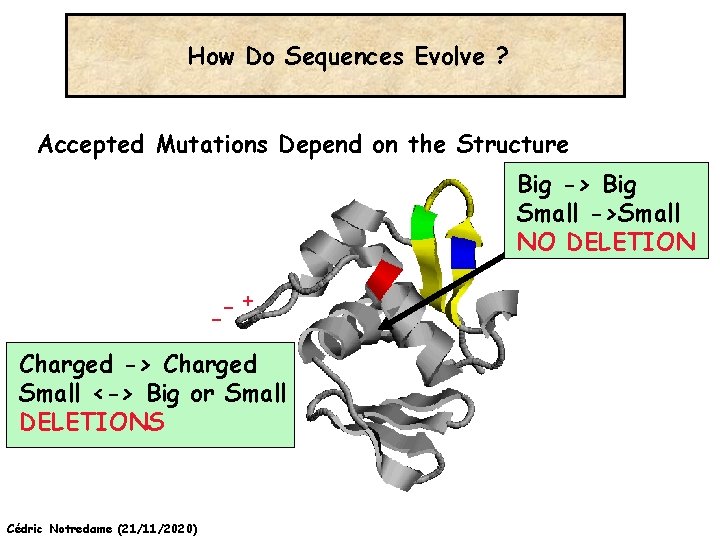
How Do Sequences Evolve ? In a structure, each Amino Acid plays a Special Role -+ On the surface, CHARGE MATTERS Omp. R, Cter Domain Cédric Notredame (21/11/2020) In the core, SIZE MATTERS

How Do Sequences Evolve ? Accepted Mutations Depend on the Structure Big -> Big Small ->Small NO DELETION + Charged -> Charged Small <-> Big or Small DELETIONS Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? Substitution Matrices Cédric Notredame (21/11/2020)

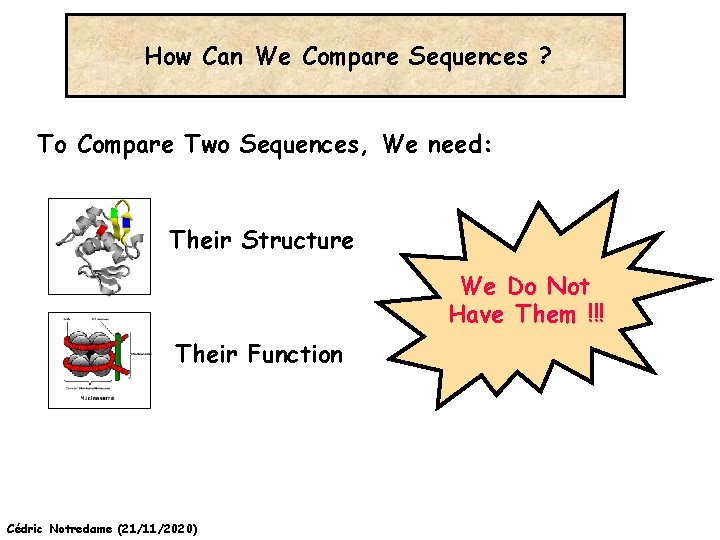
How Can We Compare Sequences ? To Compare Two Sequences, We need: Their Structure We Do Not Have Them !!! Their Function Cédric Notredame (21/11/2020)

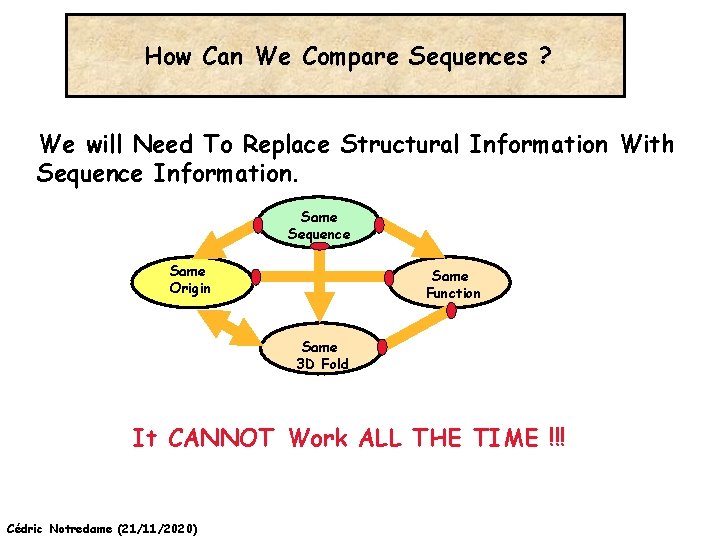
How Can We Compare Sequences ? We will Need To Replace Structural Information With Sequence Information. Same Sequence Same Origin Same Function Same 3 D Fold It CANNOT Work ALL THE TIME !!! Cédric Notredame (21/11/2020)

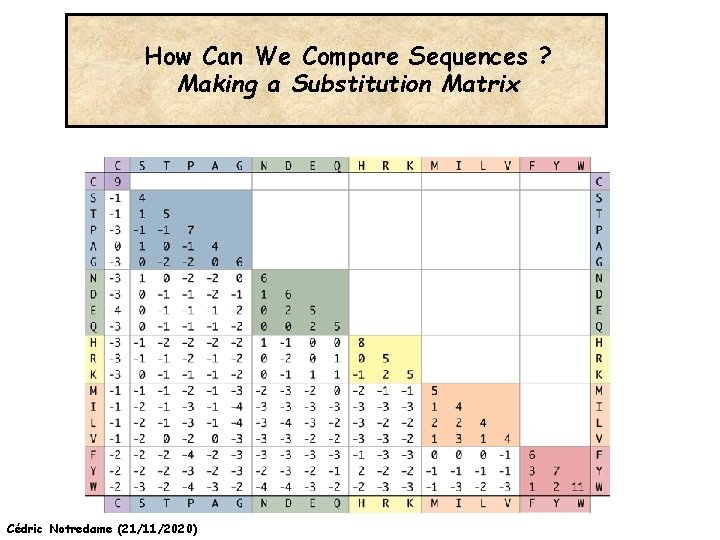
How Can We Compare Sequences ? To Compare Sequences, We need to Compare Residues We Need to Know How Much it COSTS to SUBSTITUTE an Alanine into an Isoleucine a Tryptophan into a Glycine … The table that contains the costs for all the possible substitutions is called the SUBSTITUTION MATRIX How to derive that matrix? Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? Using Knowledge Could Work C Aliphatic L V I A G T Aromatic F Y W H Small P G CC D K E R S N Q Hydrophobic Polar But we do not know enough about Evolution and Structure. Using Data works better. Cédric Notredame (21/11/2020)

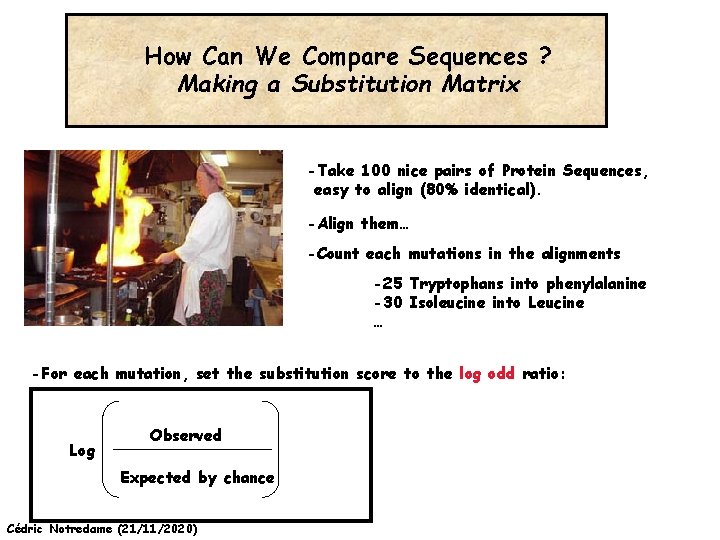
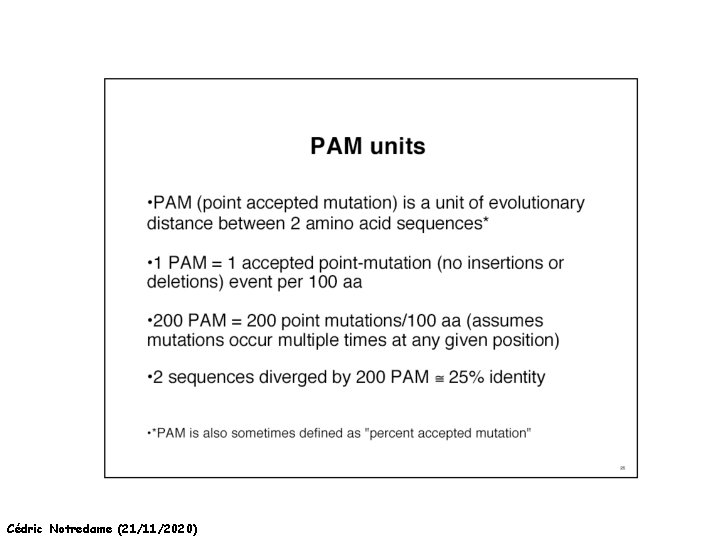
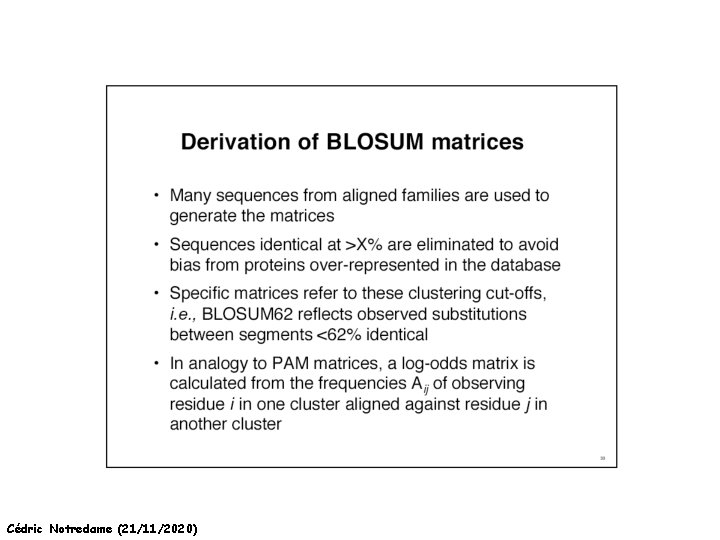
How Can We Compare Sequences ? Making a Substitution Matrix -Take 100 nice pairs of Protein Sequences, easy to align (80% identical). -Align them… -Count each mutations in the alignments -25 Tryptophans into phenylalanine -30 Isoleucine into Leucine … -For each mutation, set the substitution score to the log odd ratio: Log Observed Expected by chance Cédric Notredame (21/11/2020)

You’re kidding! … I was struck by a lightning twice too!! Cédric Notredame (21/11/2020) Garry Larson, The Far Side

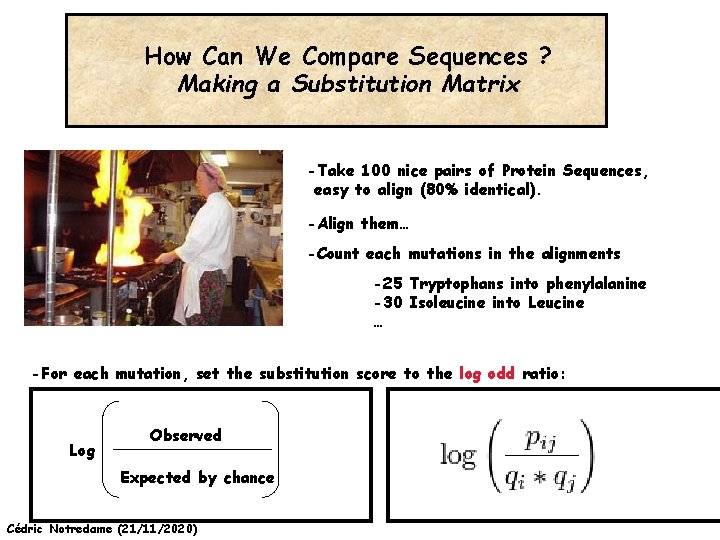
How Can We Compare Sequences ? Making a Substitution Matrix -Take 100 nice pairs of Protein Sequences, easy to align (80% identical). -Align them… -Count each mutations in the alignments -25 Tryptophans into phenylalanine -30 Isoleucine into Leucine … -For each mutation, set the substitution score to the log odd ratio: Log Observed Expected by chance Cédric Notredame (21/11/2020)

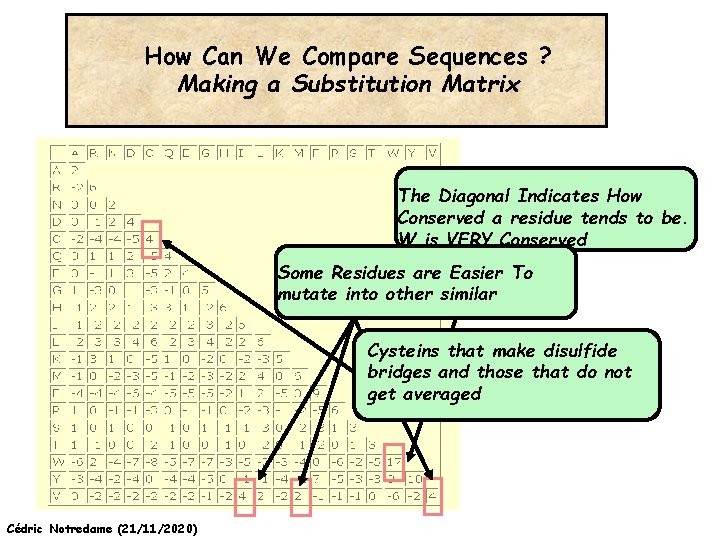
How Can We Compare Sequences ? Making a Substitution Matrix The Diagonal Indicates How Conserved a residue tends to be. W is VERY Conserved Some Residues are Easier To mutate into other similar Cysteins that make disulfide bridges and those that do not get averaged Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? Making a Substitution Matrix Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? Using Substitution Matrix Given two Sequences and a substitution Matrix, We must Compute the CHEAPEST Alignment Insertion Deletion ADKPRRP---LS-YMLWLN ADKPKRPKPRLSAYMLWLN Mutation Cédric Notredame (21/11/2020)

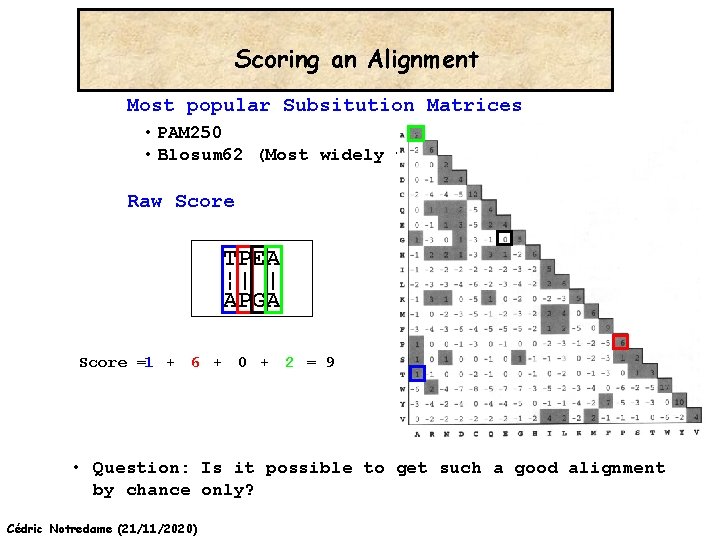
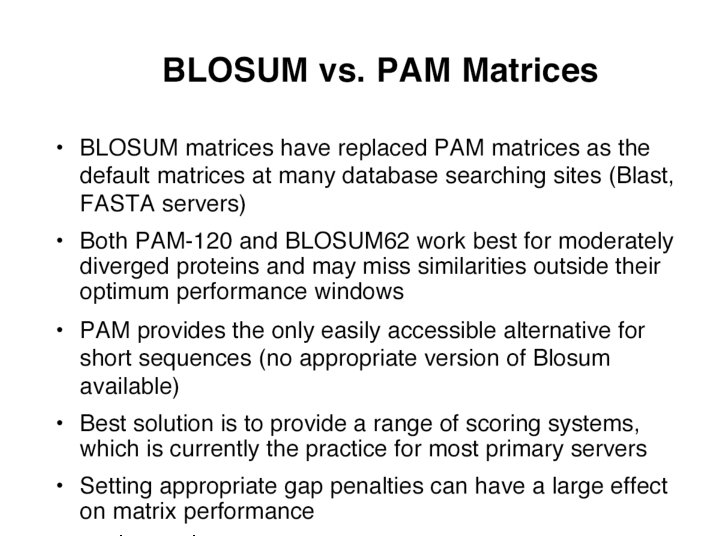
Scoring an Alignment Most popular Subsitution Matrices • PAM 250 • Blosum 62 (Most widely used) Raw Score TPEA ¦| | APGA Score =1 + 6 + 0 + 2 = 9 • Question: Is it possible to get such a good alignment by chance only? Cédric Notredame (21/11/2020)

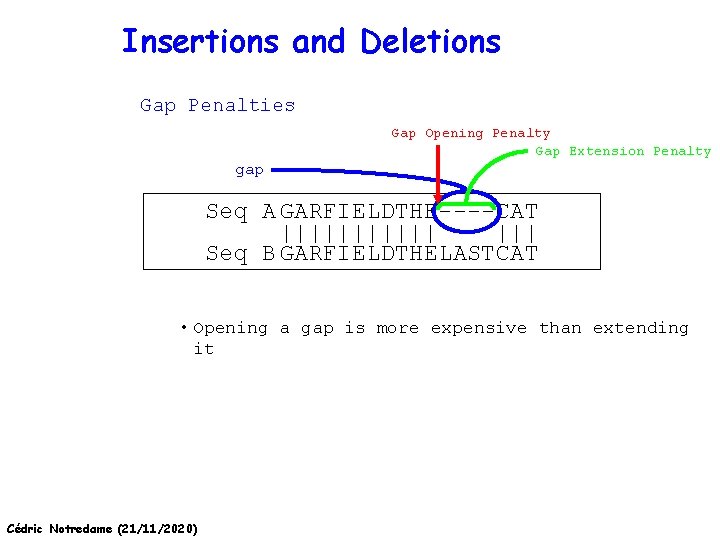
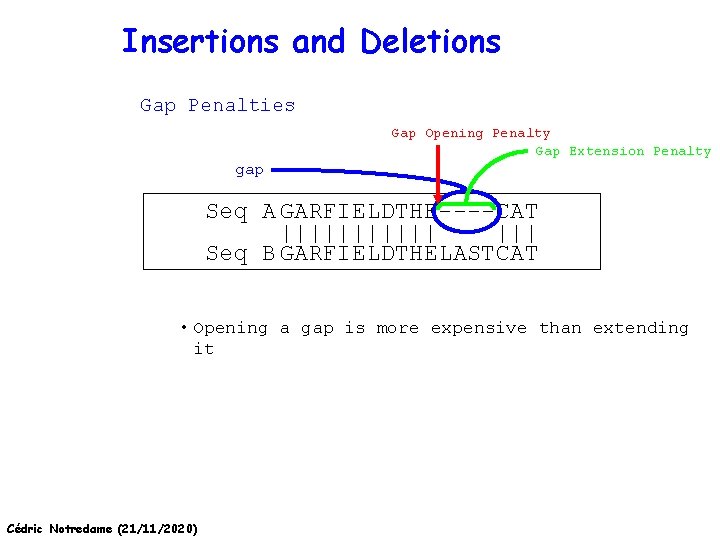
Insertions and Deletions Gap Penalties Gap Opening Penalty Gap Extension Penalty gap Seq A GARFIELDTHE----CAT |||||| ||| Seq B GARFIELDTHELASTCAT • Opening a gap is more expensive than extending it Cédric Notredame (21/11/2020)

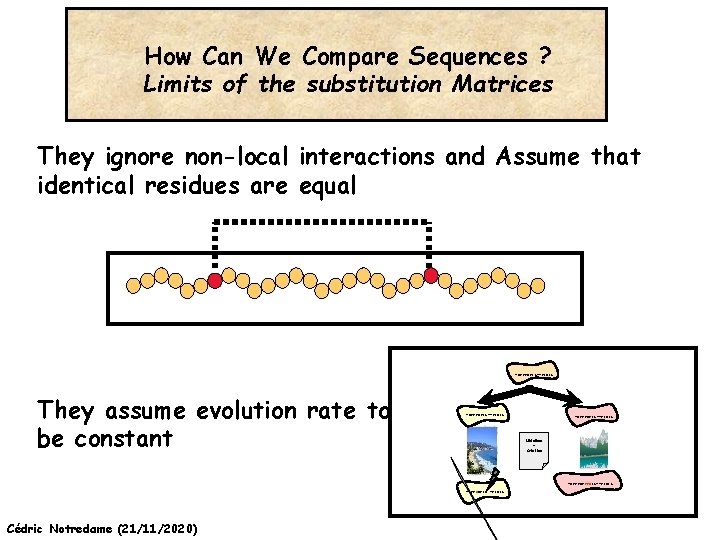
How Can We Compare Sequences ? Limits of the substitution Matrices They ignore non-local interactions and Assume that identical residues are equal ADKPKRPLSAYMLWLN They assume evolution rate to be constant ADKPKRPLSAYMLWLN Mutations + Selection ADKPKRPKPRLSAYMLWLN ADKPRRPLS-YMLWLN Cédric Notredame (21/11/2020)

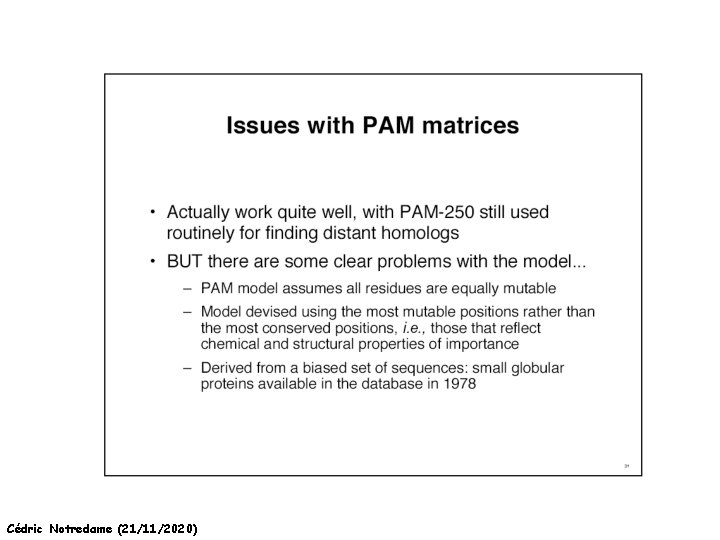
How Can We Compare Sequences ? Limits of the substitution Matrices Substitution Matrices Cannot Work !!! Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? Limits of the substitution Matrices I know… But at least, could I get some idea of when they are likely to do all right Cédric Notredame (21/11/2020)

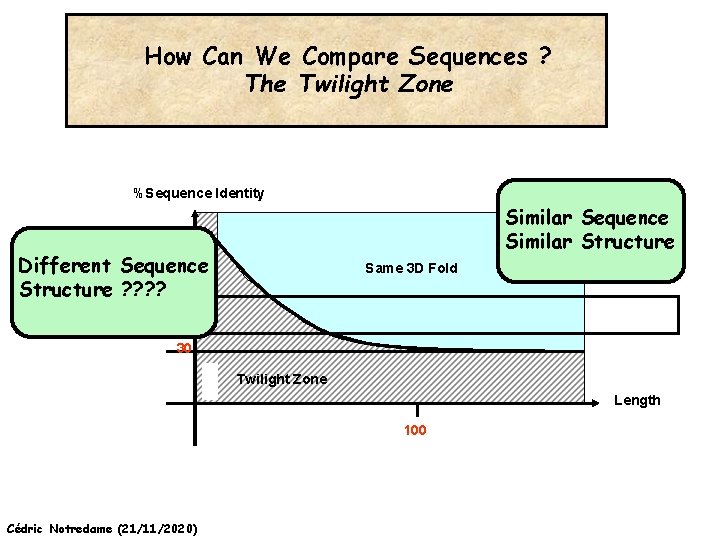
How Can We Compare Sequences ? The Twilight Zone %Sequence Identity Similar Sequence Similar Structure Different Sequence Structure ? ? Same 3 D Fold 30% 30 Twilight Zone Length 100 Cédric Notredame (21/11/2020)

How Can We Compare Sequences ? The Twilight Zone Substitution Matrices Work Reasonably Well on Sequences that have more than 30 % identity over more than 100 residues Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

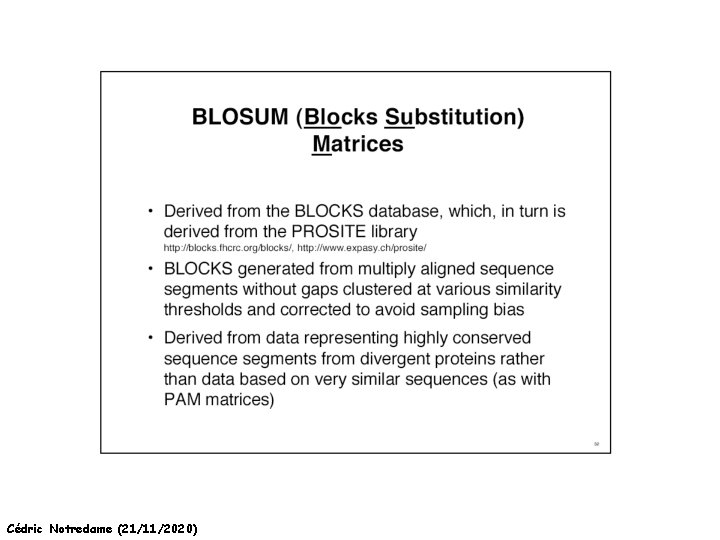
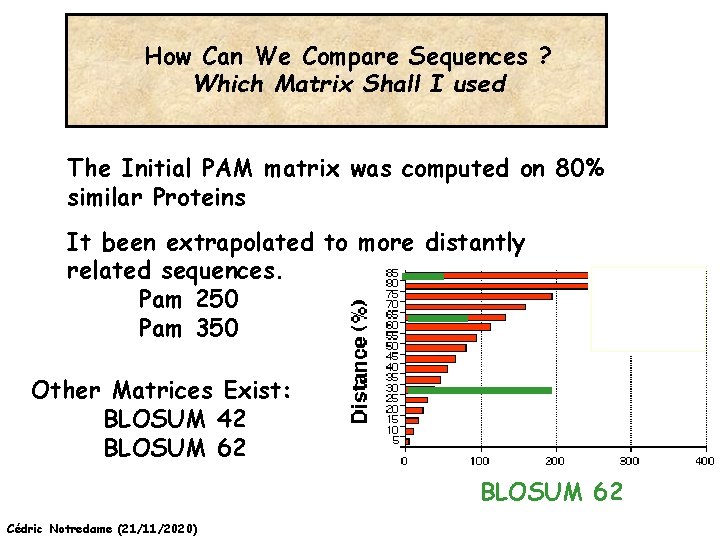
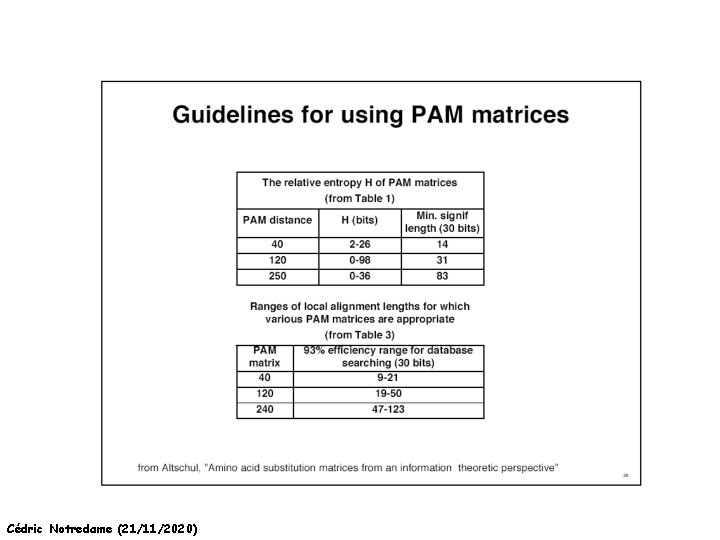
How Can We Compare Sequences ? Which Matrix Shall I used The Initial PAM matrix was computed on 80% similar Proteins It been extrapolated to more distantly related sequences. Pam 250 Pam 350 Other Matrices Exist: BLOSUM 42 BLOSUM 62 Cédric Notredame (21/11/2020)

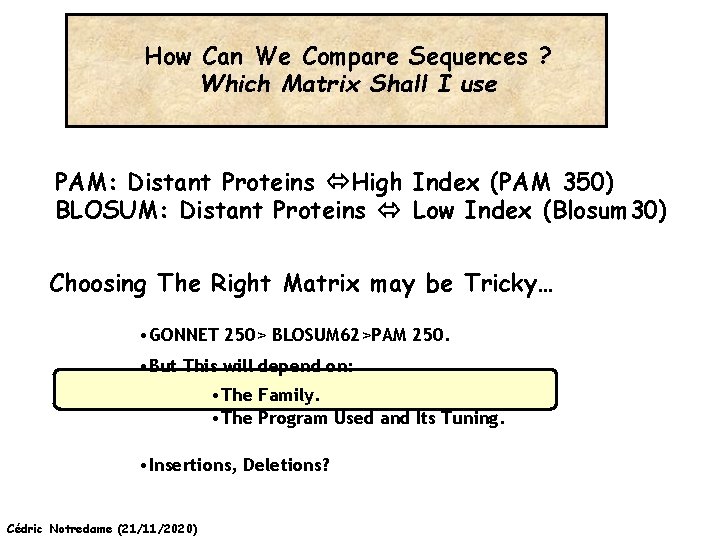
How Can We Compare Sequences ? Which Matrix Shall I use PAM: Distant Proteins High Index (PAM 350) BLOSUM: Distant Proteins Low Index (Blosum 30) Choosing The Right Matrix may be Tricky… • GONNET 250> BLOSUM 62>PAM 250. • But This will depend on: • The Family. • The Program Used and Its Tuning. • Insertions, Deletions? Cédric Notredame (21/11/2020)

HOW Can we Align Two Sequences ? Dot Matrices Global Alignments Local Alignment Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

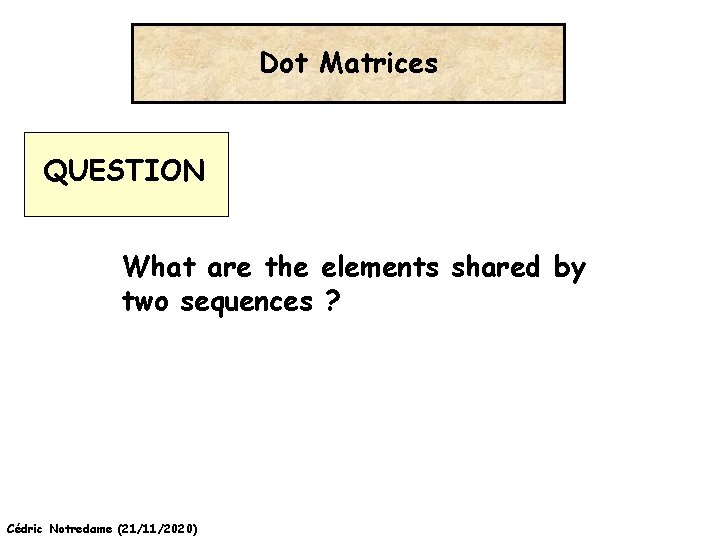
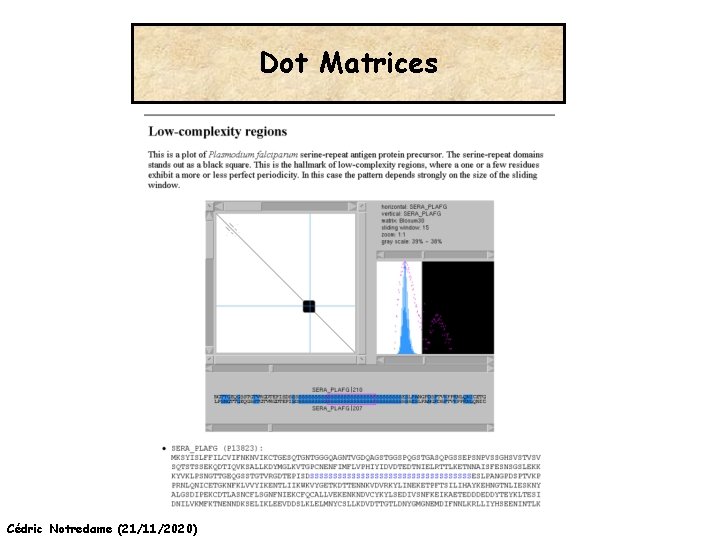
Dot Matrices QUESTION What are the elements shared by two sequences ? Cédric Notredame (21/11/2020)

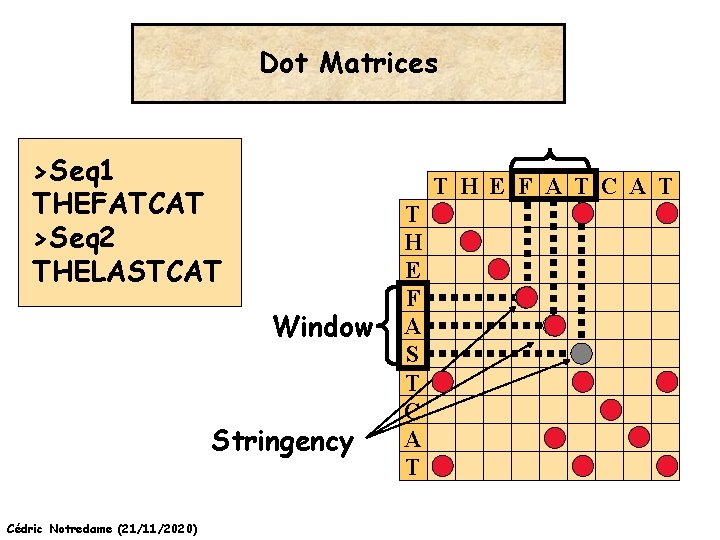
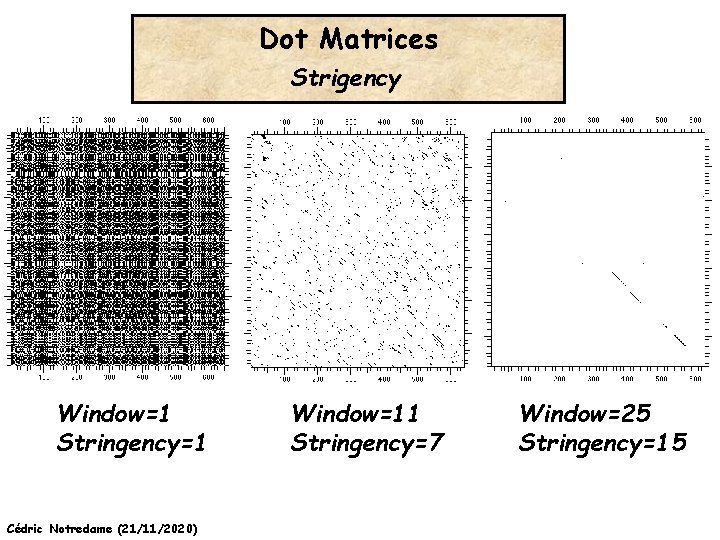
Dot Matrices >Seq 1 THEFATCAT >Seq 2 THELASTCAT T H E F A T C A T Window Stringency Cédric Notredame (21/11/2020) T H E F A S T C A T

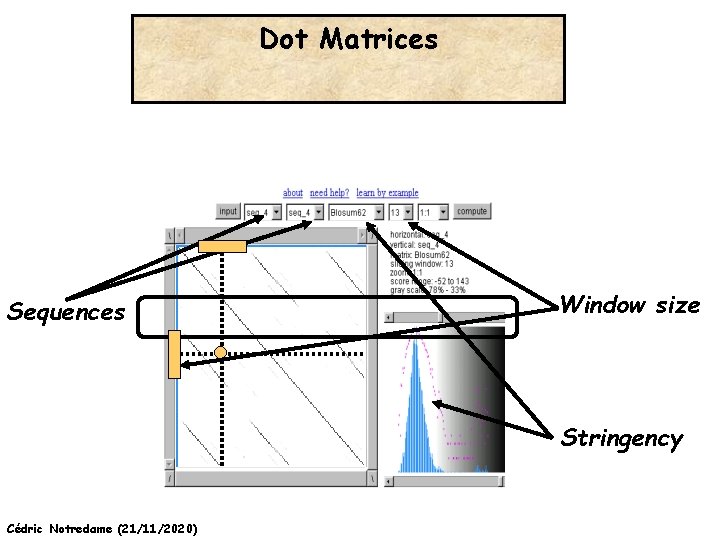
Dot Matrices Sequences Window size Stringency Cédric Notredame (21/11/2020)

Dot Matrices Strigency Window=1 Stringency=1 Cédric Notredame (21/11/2020) Window=11 Stringency=7 Window=25 Stringency=15

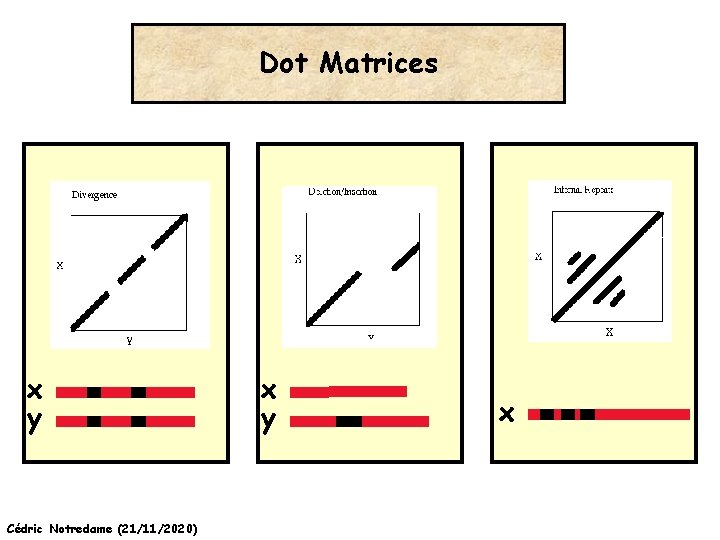
Dot Matrices x y Cédric Notredame (21/11/2020) x y x

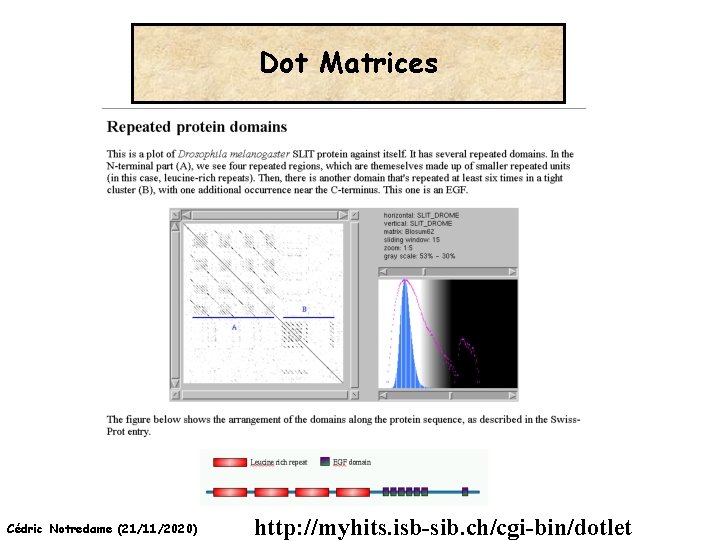
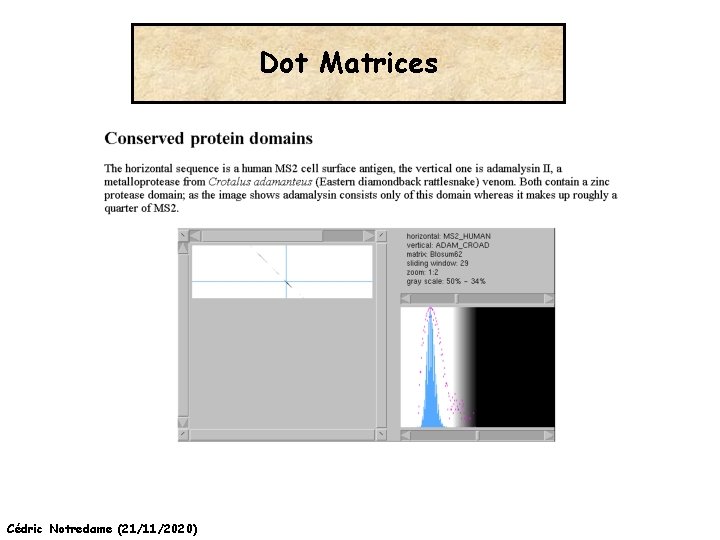
Dot Matrices Cédric Notredame (21/11/2020) http: //myhits. isb-sib. ch/cgi-bin/dotlet

Dot Matrices Cédric Notredame (21/11/2020)

Dot Matrices Cédric Notredame (21/11/2020)

Dot Matrices Cédric Notredame (21/11/2020)

Dot Matrices Limits -Visual aid -Best Way to EXPLORE the Sequence Organisation -Does NOT provide us with an ALIGNMENT wheat ? ? ? --DPNKPKRAMTSFVFFMSEFRSEFKQKHSKLKSIVEMVKAAGER | | |||| ||| | |||| KKDSNAPKRAMTSFMFFSSDFRS----KHSDL-SIVEMSKAAGAA Cédric Notredame (21/11/2020)

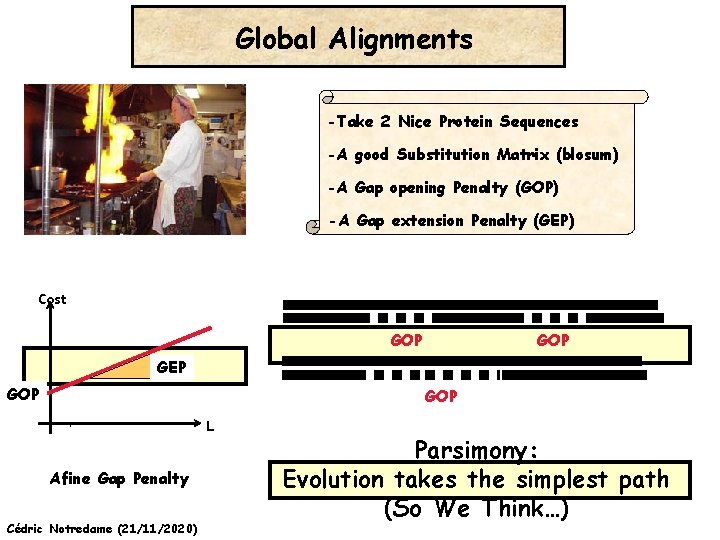
Global Alignments -Take 2 Nice Protein Sequences -A good Substitution Matrix (blosum) -A Gap opening Penalty (GOP) -A Gap extension Penalty (GEP) Cost GOP GEP GOP L Afine Gap Penalty Cédric Notredame (21/11/2020) Parsimony: Evolution takes the simplest path (So We Think…)

Insertions and Deletions Gap Penalties Gap Opening Penalty Gap Extension Penalty gap Seq A GARFIELDTHE----CAT |||||| ||| Seq B GARFIELDTHELASTCAT • Opening a gap is more expensive than extending it Cédric Notredame (21/11/2020)

Global Alignments -Take 2 Nice Protein Sequences -A good Substitution Matrix (blosum) -A Gap opening Penalty (GOP) -A Gap extension Penalty (GEP) -DYNAMIC PROGRAMMING >Seq 1 THEFATCAT >Seq 2 THEFASTCAT Cédric Notredame (21/11/2020) THEFA-TCAT THEFASTCAT DYNAMIC PROGRAMMING

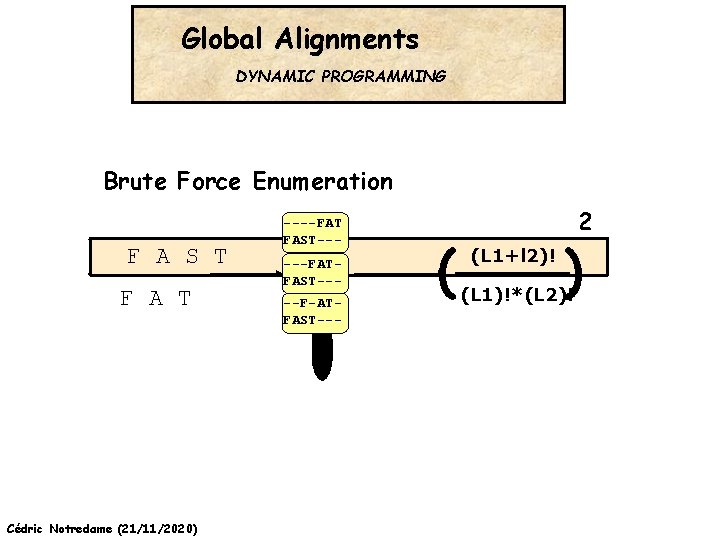
Global Alignments DYNAMIC PROGRAMMING Brute Force Enumeration F A S T F A T Cédric Notredame (21/11/2020) ----FAT FAST-----FATFAST----F-ATFAST--- ( 2 (L 1+l 2)! ) (L 1)!*(L 2)!

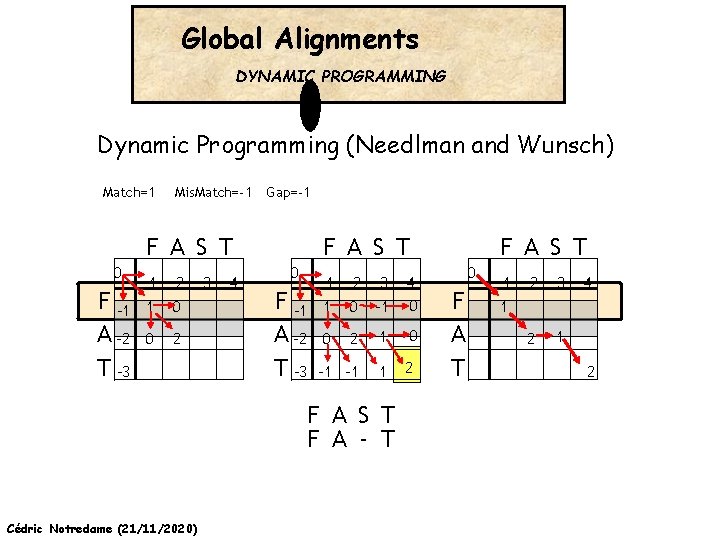
Global Alignments DYNAMIC PROGRAMMING Dynamic Programming (Needlman and Wunsch) Match=1 Mis. Match=-1 Gap=-1 F A S T 0 0 F A -2 T -3 -1 -2 -3 -4 -1 1 0 0 2 -1 -2 -3 -4 -1 1 0 -1 0 2 1 0 -1 -1 1 2 0 F A S T F A - T Cédric Notredame (21/11/2020) F A S T 0 F A T -1 -2 -3 -4 1 2

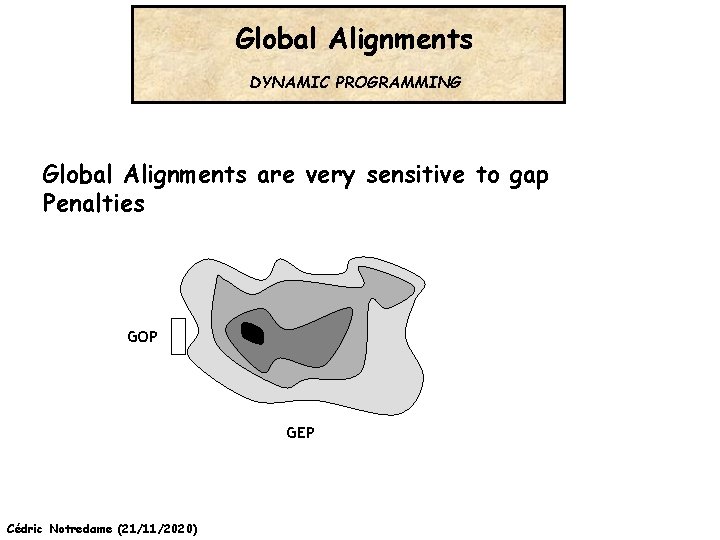
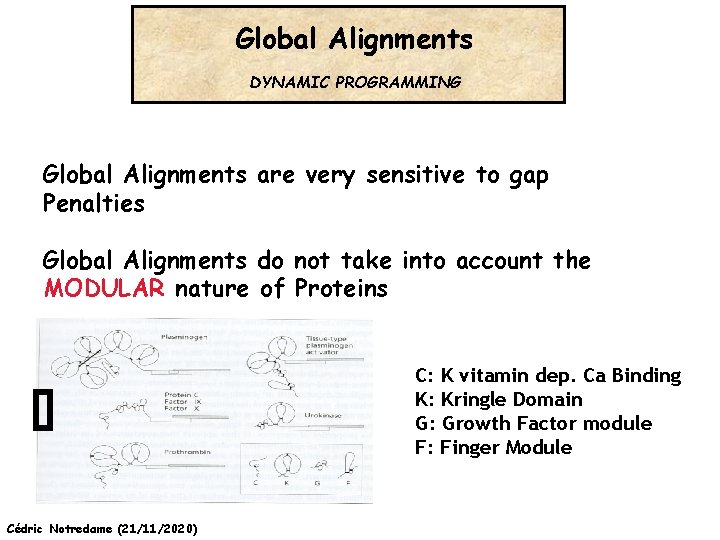
Global Alignments DYNAMIC PROGRAMMING Global Alignments are very sensitive to gap Penalties GOP GEP Cédric Notredame (21/11/2020)

Global Alignments DYNAMIC PROGRAMMING Global Alignments are very sensitive to gap Penalties Global Alignments do not take into account the MODULAR nature of Proteins C: K vitamin dep. Ca Binding K: Kringle Domain G: Growth Factor module F: Finger Module Cédric Notredame (21/11/2020)

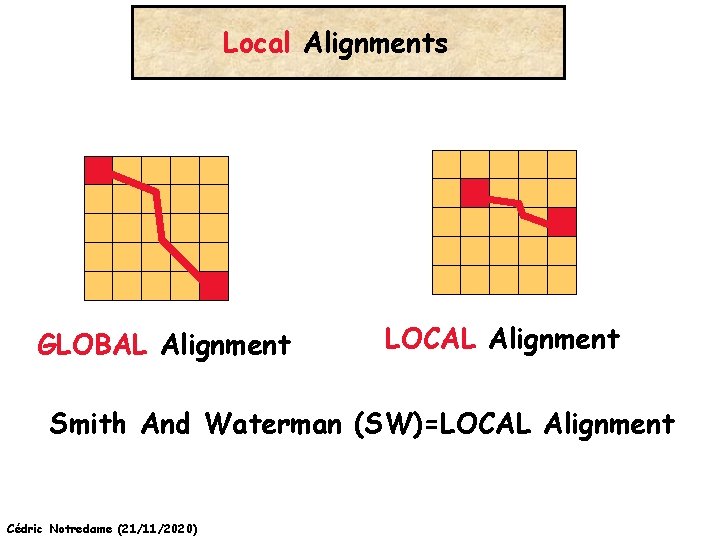
Local Alignments GLOBAL Alignment LOCAL Alignment Smith And Waterman (SW)=LOCAL Alignment Cédric Notredame (21/11/2020)

Local Alignments We now have a Pair. Wise Comparison Algorithm, We are ready to search Databases Cédric Notredame (21/11/2020)

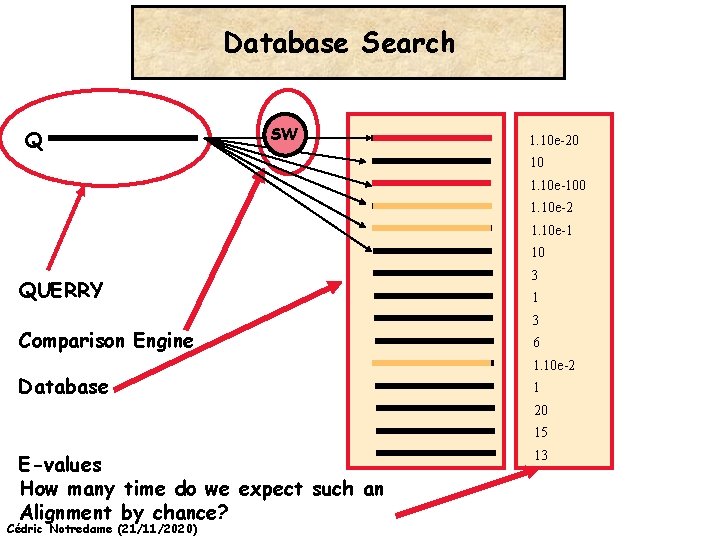
Database Search Q SW 1. 10 e-20 10 1. 10 e-100 1. 10 e-2 1. 10 e-1 10 QUERRY Comparison Engine Database 3 1 3 6 1. 10 e-2 1 20 15 E-values How many time do we expect such an Alignment by chance? Cédric Notredame (21/11/2020) 13

Cédric Notredame (21/11/2020)

CONCLUSION Cédric Notredame (21/11/2020)

Sequence Comparison -Thanks to evolution, We CAN compare Sequences -There is a relation between Sequence and Structure. -Substitution matrices only work well with similar Sequences (More than 30% id). The Easiest way to Compare Two Sequences is a dotplot. Cédric Notredame (21/11/2020)

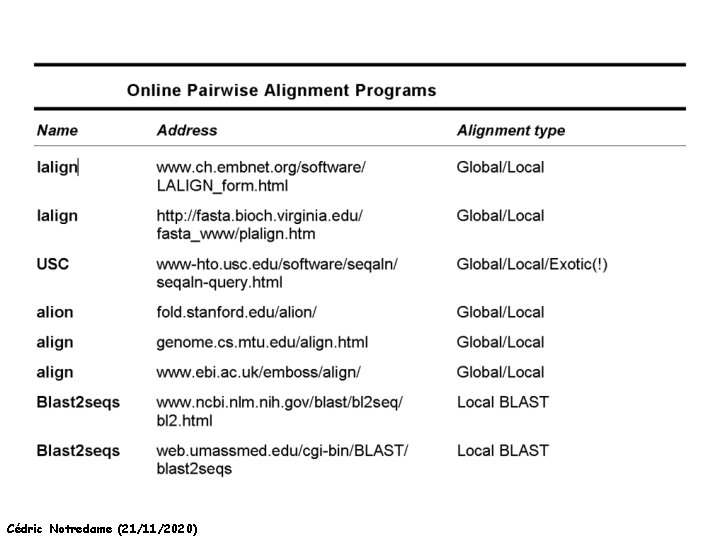
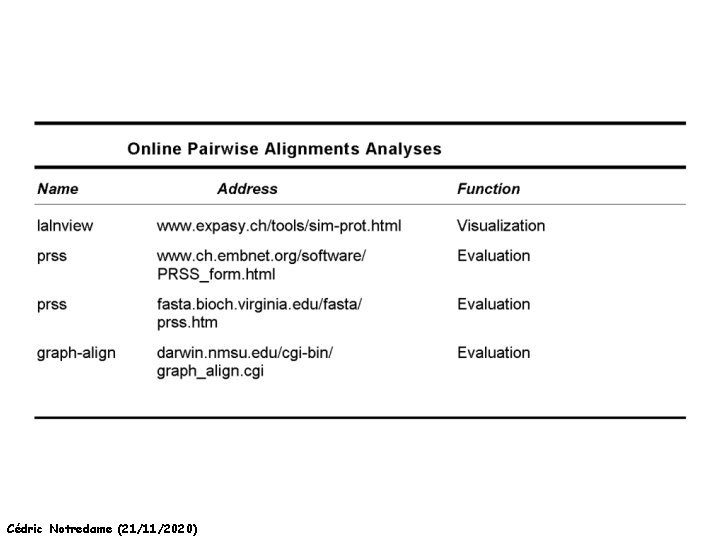
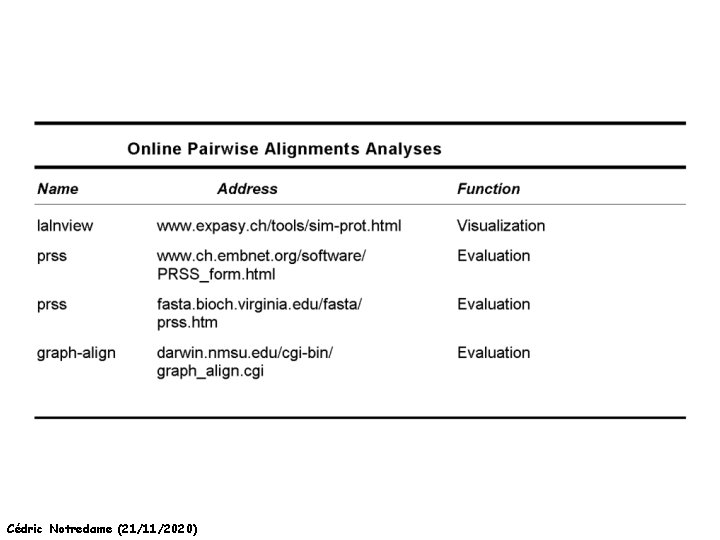
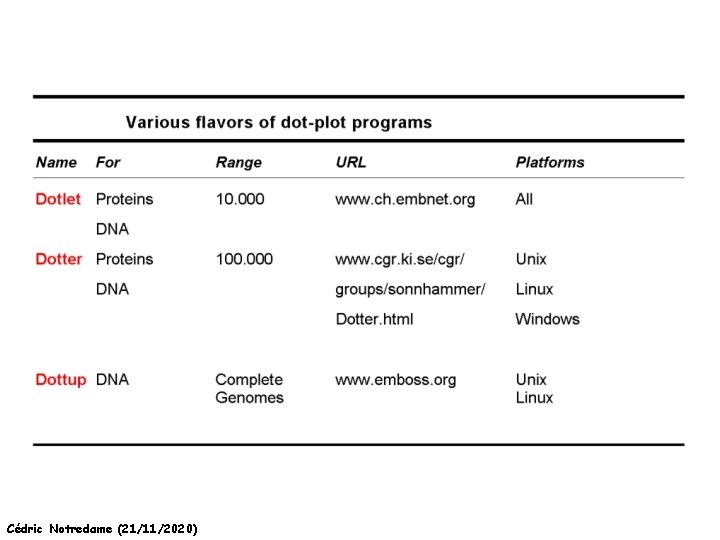
A few Addresses Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)

Cédric Notredame (21/11/2020)