Molecular Dynamics Simulations and Docking Studies of ACh




- Slides: 25

Molecular Dynamics Simulations and Docking Studies of ACh. BP and the Ligand Binding Domain of α 7 n. ACh. R Shiva Amiri JC 20 -04 -2005

1. Simulation studies of ACh. BP with Nicotine, Carbamylcholine, and HEPES as ligands > also one simulation of the ligand binding domain of α 7 n. ACh. R 2. Docking studies of α 7 n. ACh. R with Nicotine, Imidacloprid (an insecticide), and Acetylcholine (ACh)

n. ACh. R § § § a ligand gated ion channel (LGIC) found in central and peripheral nervous system endogenous ligand is acetylcholine (ACh) but reactive to many compounds such as nicotine, alcohol, and toxins mutations lead to various diseases such as epilepsy, myasthenic syndromes, etc. implicated in Alzheimer’s disease and Parkinson’s disease (not well understood) mediates nicotine addiction

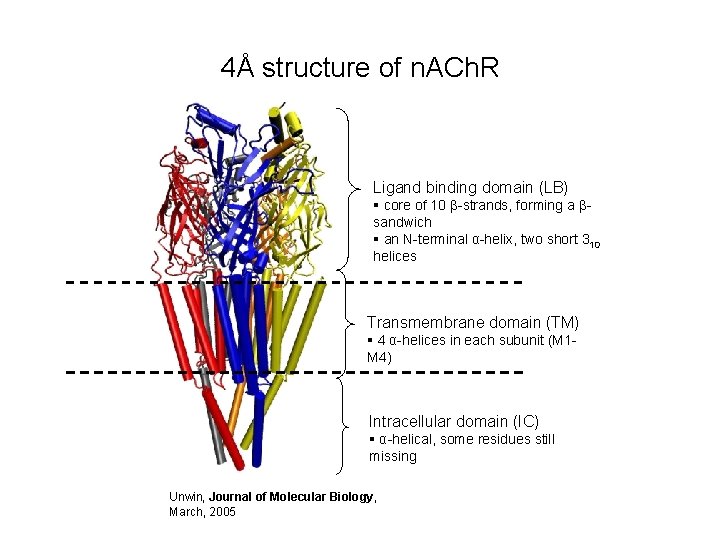
4Å structure of n. ACh. R Ligand binding domain (LB) § core of 10 β-strands, forming a βsandwich § an N-terminal α-helix, two short 310 helices Transmembrane domain (TM) § 4 α-helices in each subunit (M 1 M 4) Intracellular domain (IC) § α-helical, some residues still missing Unwin, Journal of Molecular Biology, March, 2005

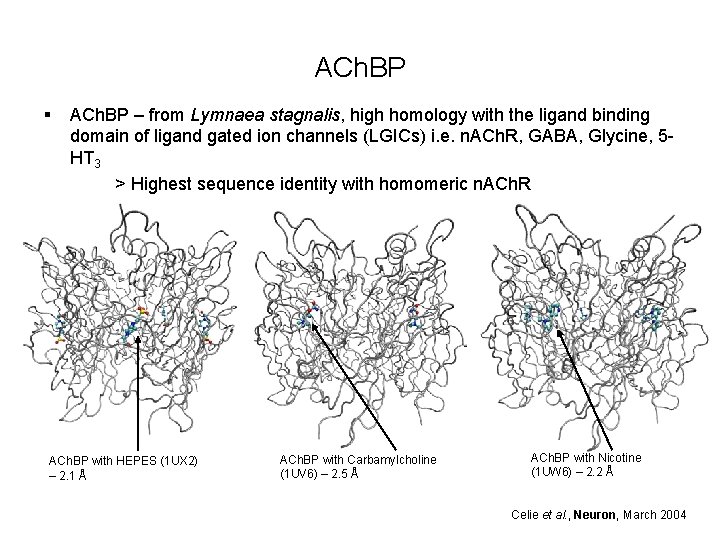
ACh. BP § ACh. BP – from Lymnaea stagnalis, high homology with the ligand binding domain of ligand gated ion channels (LGICs) i. e. n. ACh. R, GABA, Glycine, 5 HT 3 > Highest sequence identity with homomeric n. ACh. R ACh. BP with HEPES (1 UX 2) – 2. 1 Å ACh. BP with Carbamylcholine (1 UV 6) – 2. 5 Å ACh. BP with Nicotine (1 UW 6) – 2. 2 Å Celie et al. , Neuron, March 2004

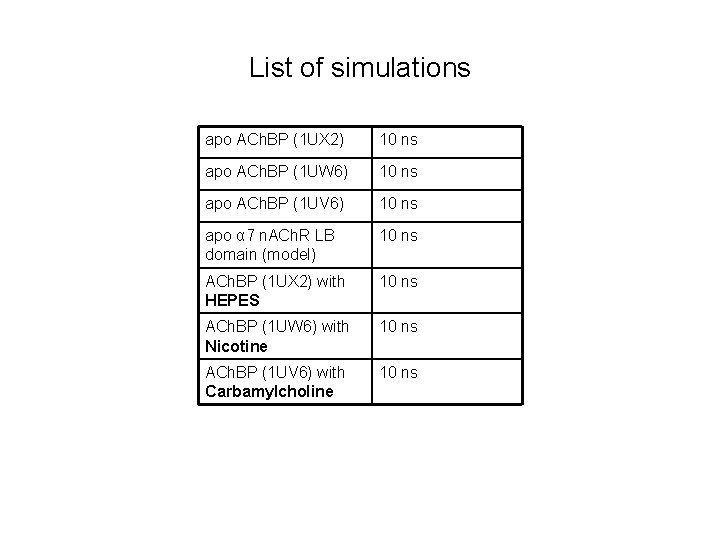
List of simulations apo ACh. BP (1 UX 2) 10 ns apo ACh. BP (1 UW 6) 10 ns apo ACh. BP (1 UV 6) 10 ns apo α 7 n. ACh. R LB domain (model) 10 ns ACh. BP (1 UX 2) with HEPES 10 ns ACh. BP (1 UW 6) with Nicotine 10 ns ACh. BP (1 UV 6) with Carbamylcholine 10 ns

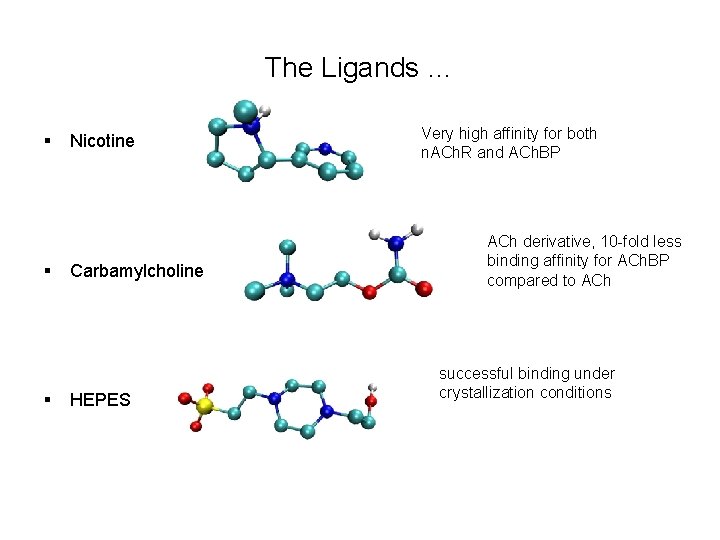
The Ligands … § § § Nicotine Carbamylcholine HEPES Very high affinity for both n. ACh. R and ACh. BP ACh derivative, 10 -fold less binding affinity for ACh. BP compared to ACh successful binding under crystallization conditions

Making the topologies… § Insight. II was used for protonating the ligands and Spartan was used to get the charges § Further details on making a topology on http: //indigo 1. biop. ox. ac. uk/wiki/index. php/Making_a_topology_file__a_quick_guide § For HEPES, I used PRODRG 2. 5 (beta), it gives GROMOS 96 topologies > have to check the topologies produced by this server…there are some bugs § A 1 ns simulation in water was run on each ligand after making its topology before including it with the protein

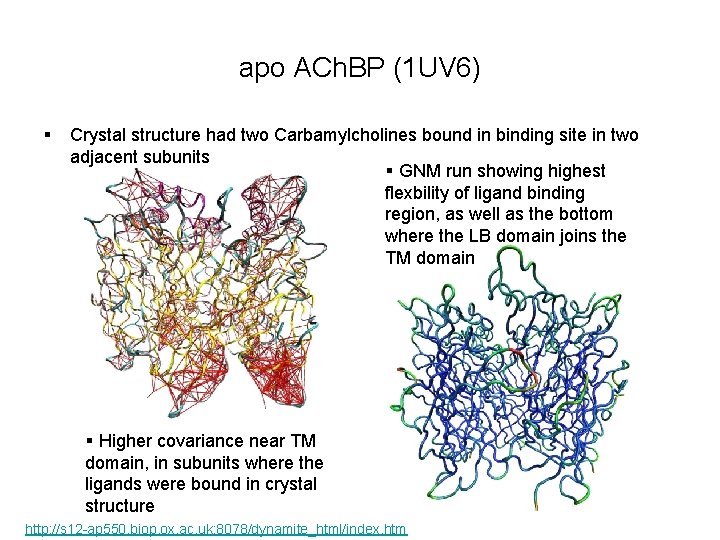
apo ACh. BP (1 UV 6) § Crystal structure had two Carbamylcholines bound in binding site in two adjacent subunits § GNM run showing highest flexbility of ligand binding region, as well as the bottom where the LB domain joins the TM domain § Higher covariance near TM domain, in subunits where the ligands were bound in crystal structure http: //s 12 -ap 550. biop. ox. ac. uk: 8078/dynamite_html/index. htm

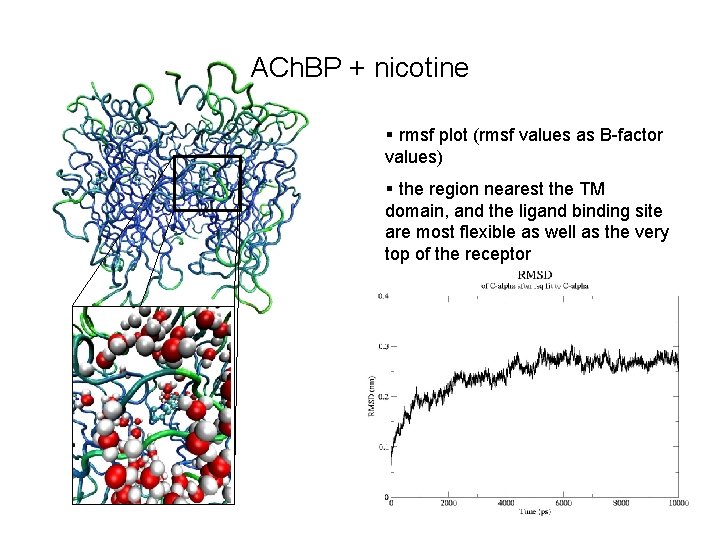
ACh. BP + nicotine § rmsf plot (rmsf values as B-factor values) § the region nearest the TM domain, and the ligand binding site are most flexible as well as the very top of the receptor

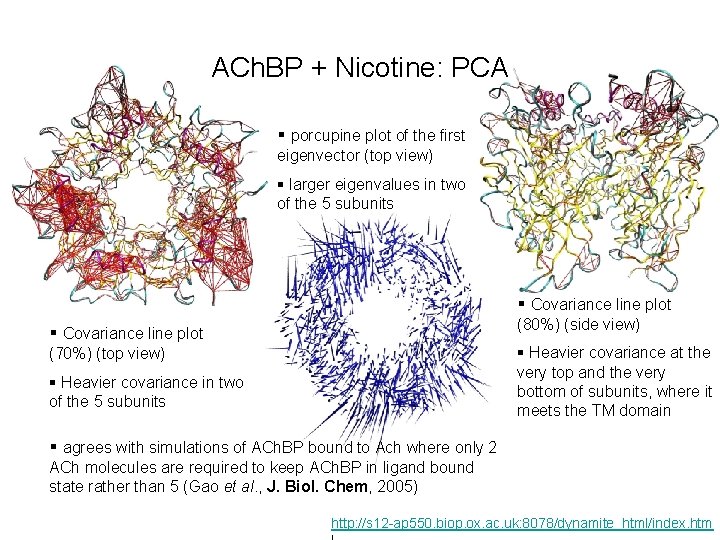
ACh. BP + Nicotine: PCA § porcupine plot of the first eigenvector (top view) § larger eigenvalues in two of the 5 subunits § Covariance line plot (80%) (side view) (70%) (top view) § Heavier covariance at the very top and the very bottom of subunits, where it meets the TM domain § Heavier covariance in two of the 5 subunits § agrees with simulations of ACh. BP bound to Ach where only 2 ACh molecules are required to keep ACh. BP in ligand bound state rather than 5 (Gao et al. , J. Biol. Chem, 2005) http: //s 12 -ap 550. biop. ox. ac. uk: 8078/dynamite_html/index. htm

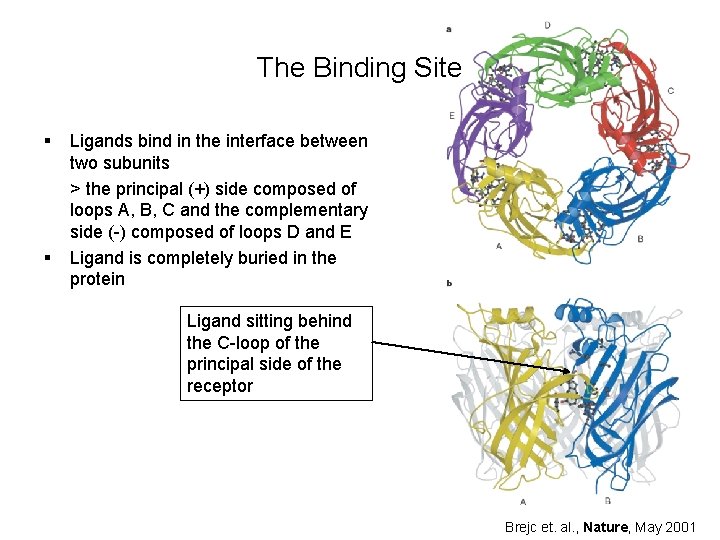
The Binding Site § § Ligands bind in the interface between two subunits > the principal (+) side composed of loops A, B, C and the complementary side (-) composed of loops D and E Ligand is completely buried in the protein Ligand sitting behind the C-loop of the principal side of the receptor Brejc et. al. , Nature, May 2001

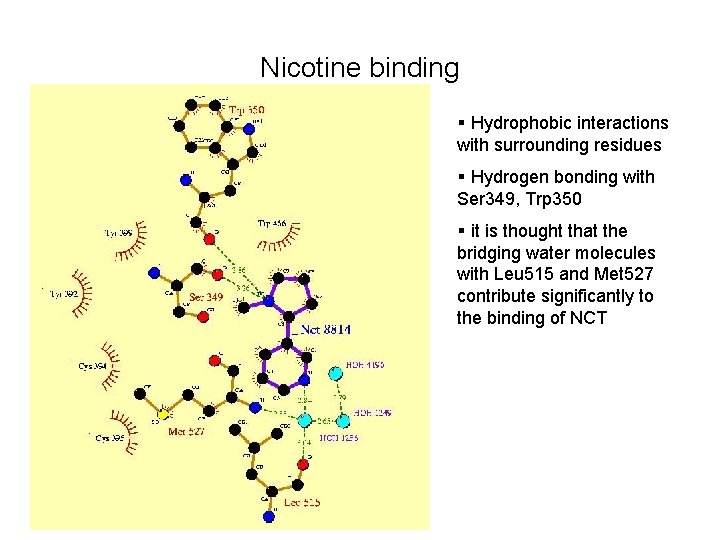
Nicotine binding § Hydrophobic interactions with surrounding residues § Hydrogen bonding with Ser 349, Trp 350 § it is thought that the bridging water molecules with Leu 515 and Met 527 contribute significantly to the binding of NCT

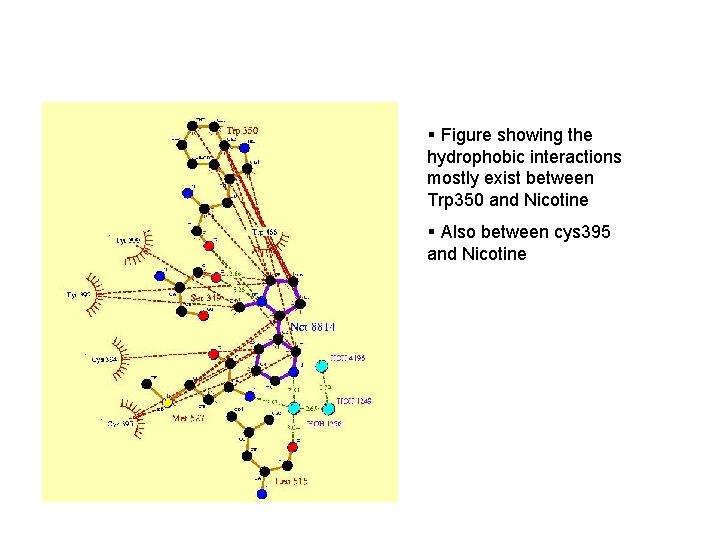
§ Figure showing the hydrophobic interactions mostly exist between Trp 350 and Nicotine § Also between cys 395 and Nicotine

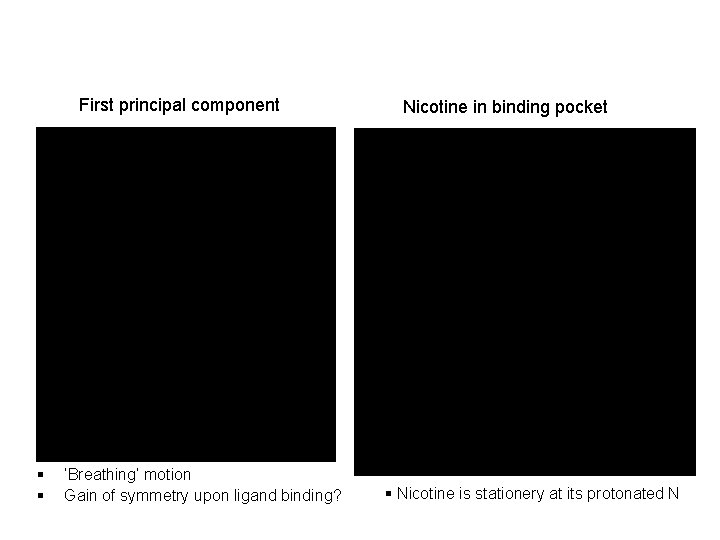
First principal component § § ‘Breathing’ motion Gain of symmetry upon ligand binding? Nicotine in binding pocket § Nicotine is stationery at its protonated N

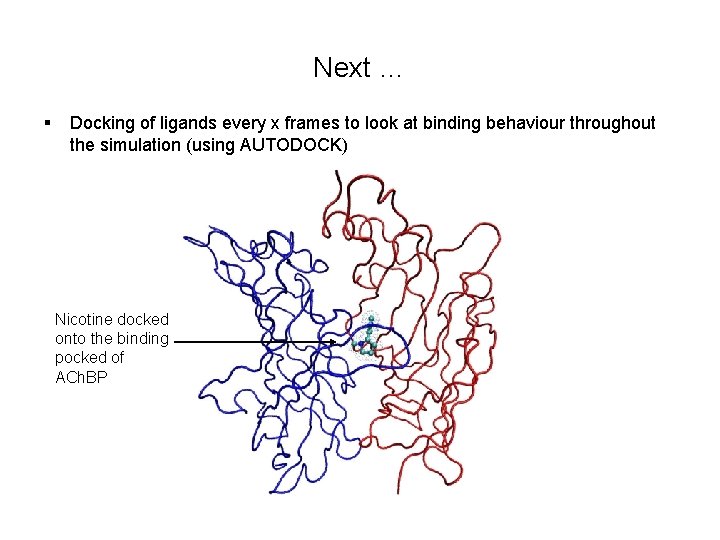
Next … § Docking of ligands every x frames to look at binding behaviour throughout the simulation (using AUTODOCK) Nicotine docked onto the binding pocked of ACh. BP

Docking studies of α 7 n. ACh. R Some α 7 background: § § § Homopentameric cationic channel Found in central nervous system Implicated in learning disabilities, Parkinson’s, Alzheimer’s, alcoholism, and nicotine addiction Docking: § § § The ligand binding domain is used for the docking studies with AUTODOCK Modelled on new ACh. BP HEPES bound structure (2. 1 Å) (Celie et al. , Neuron, March 2004) using MODELLER Nicotine (NCT), Acetylcholine (ACh), and Imidacloprid (IMI) used as ligands

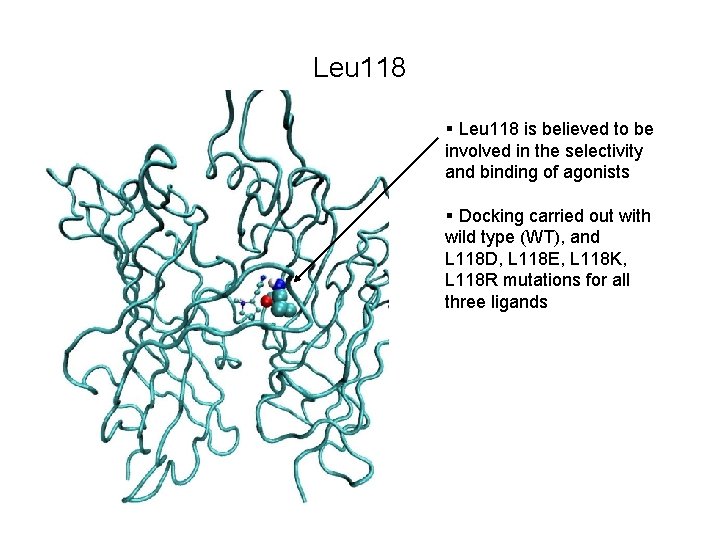
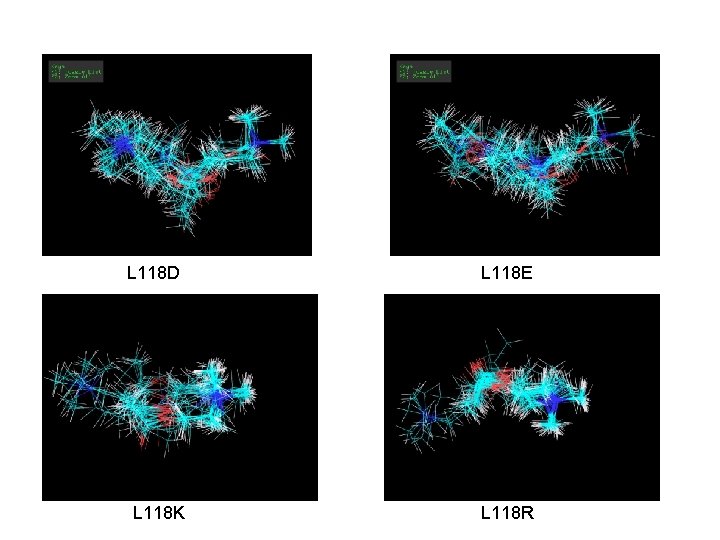
Leu 118 § Leu 118 is believed to be involved in the selectivity and binding of agonists § Docking carried out with wild type (WT), and L 118 D, L 118 E, L 118 K, L 118 R mutations for all three ligands

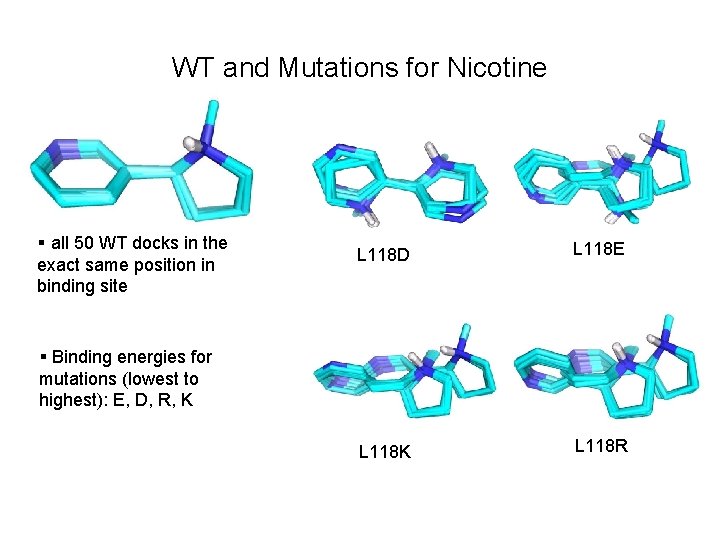
WT and Mutations for Nicotine § all 50 WT docks in the exact same position in binding site L 118 D L 118 E L 118 K L 118 R § Binding energies for mutations (lowest to highest): E, D, R, K

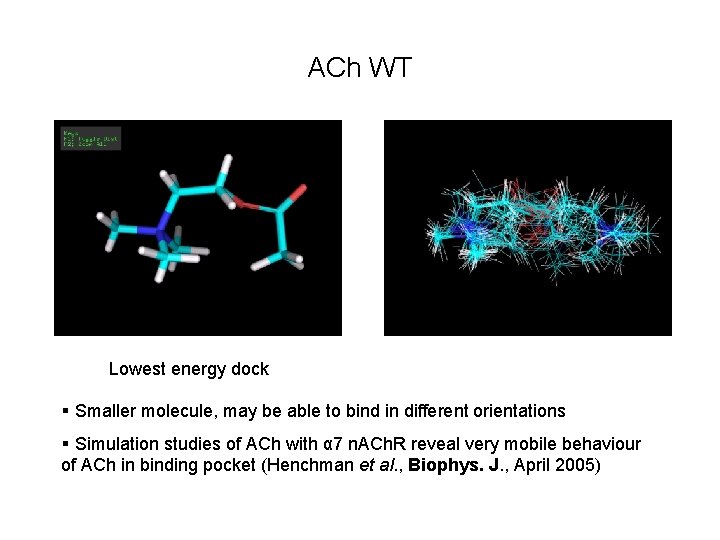
ACh WT Lowest energy dock § Smaller molecule, may be able to bind in different orientations § Simulation studies of ACh with α 7 n. ACh. R reveal very mobile behaviour of ACh in binding pocket (Henchman et al. , Biophys. J. , April 2005)

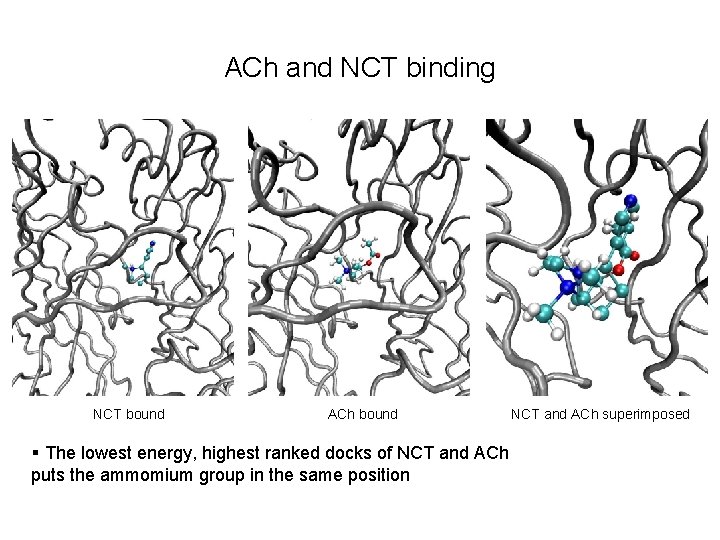
ACh and NCT binding NCT bound ACh bound § The lowest energy, highest ranked docks of NCT and ACh puts the ammomium group in the same position NCT and ACh superimposed

Further docking § § § Fighting with Imidacloprid docks… More ACh docking to look for a more clear pattern Using the 4Å Torpedo marmorata (Unwin, Journal of Molecular Biology, March, 2005) structure for docks to compare binding sites and modes of ligand binding

Summary + Future Directions Simulations § Simulations show highest covariance and flexibility near the TM domain, in ligand binding site, and at the very top of the receptor § Higher covariance in subunits with bound ligand, even in APO simulations § First eigenvector shows ‘breathing motion’ in agreement with Henchman’s data § Further analysis on individual subunits, binding site, ligand contacts and behaviour needs to be done Docking § Mutations cause incorrect binding orientations of nicotine § ACh … multiple binding modes? § IMI in progress § Heteropentameric EM structure will be used for further docking and comparison of different binding sites


L 118 D L 118 E L 118 K L 118 R