Light regulation of Plant Development Plants use light






- Slides: 79

Light regulation of Plant Development Plants use light as food and information Use information to control development

Light regulation of growth Plants sense 1. Light quantity 2. Light quality (colors) 3. Light duration 4. Direction it comes from Have photoreceptors that sense specific wavelengths

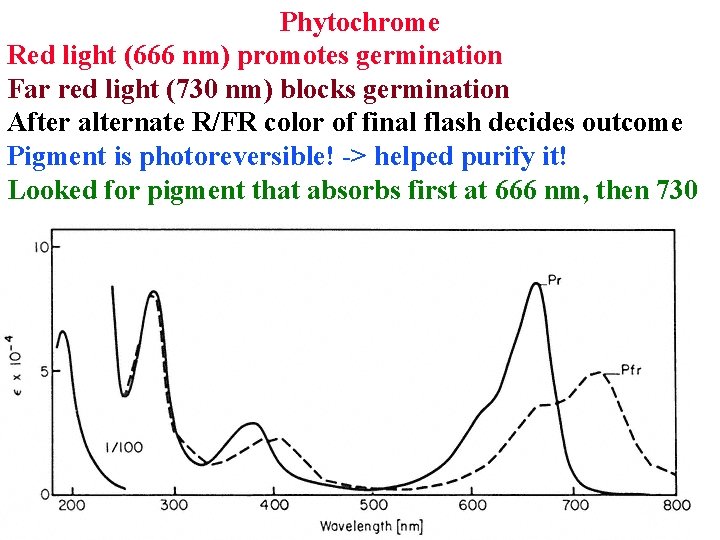
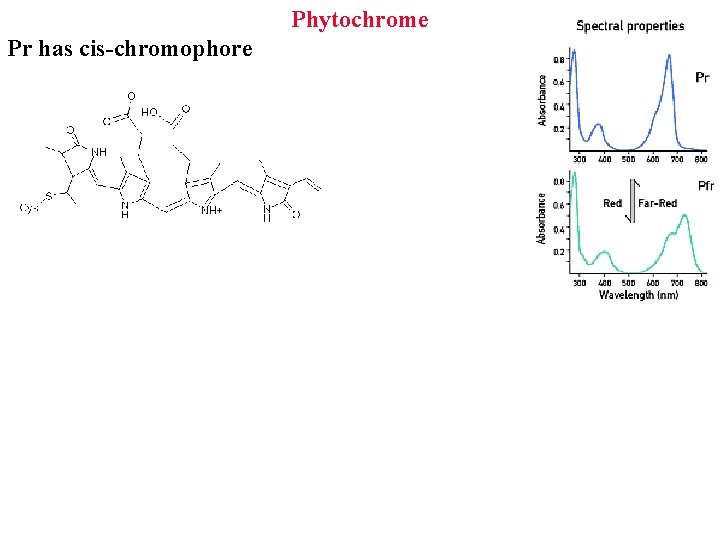
Phytochrome Red light (666 nm) promotes germination Far red light (730 nm) blocks germination After alternate R/FR color of final flash decides outcome Pigment is photoreversible! -> helped purify it! Looked for pigment that absorbs first at 666 nm, then 730

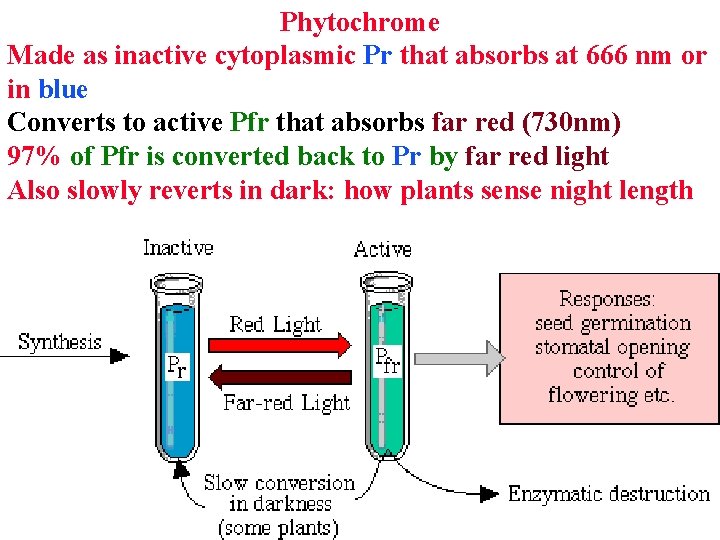
Phytochrome Made as inactive cytoplasmic Pr that absorbs at 666 nm or in blue Converts to active Pfr that absorbs far red (730 nm) 97% of Pfr is converted back to Pr by far red light Also slowly reverts in dark: how plants sense night length

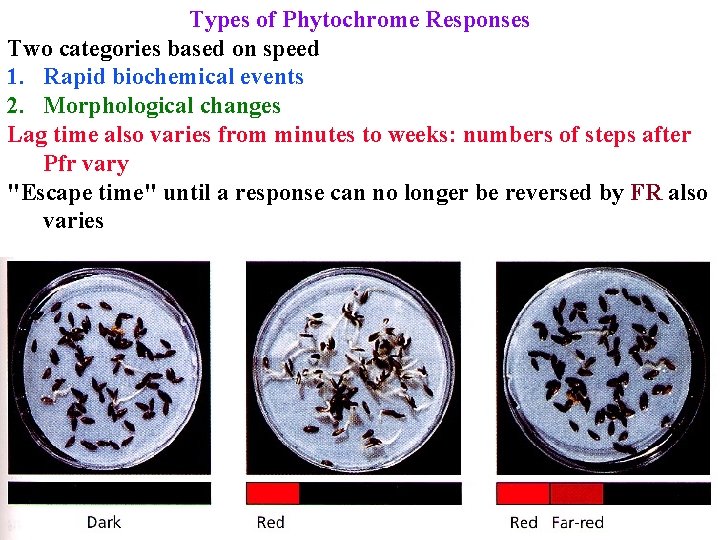
Types of Phytochrome Responses Two categories based on speed 1. Rapid biochemical events 2. Morphological changes Lag time also varies from minutes to weeks: numbers of steps after Pfr vary "Escape time" until a response can no longer be reversed by FR also varies

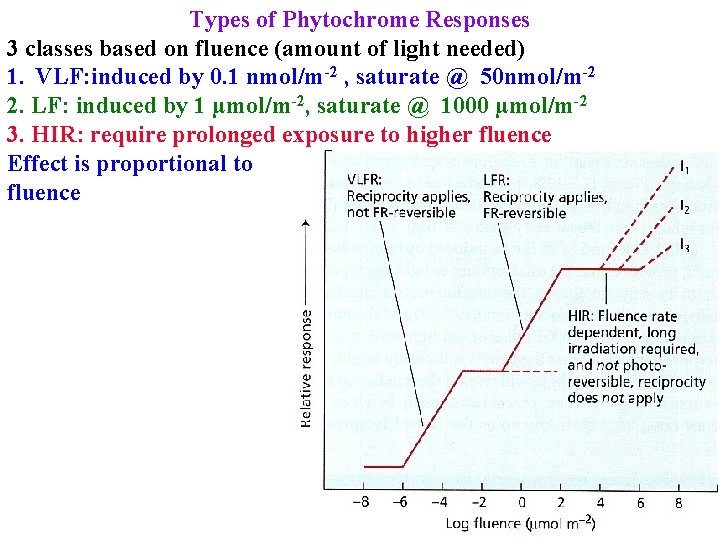
Types of Phytochrome Responses 3 classes based on fluence (amount of light needed) 1. VLF: induced by 0. 1 nmol/m-2 , saturate @ 50 nmol/m-2 2. LF: induced by 1 µmol/m-2, saturate @ 1000 µmol/m-2 3. HIR: require prolonged exposure to higher fluence Effect is proportional to fluence

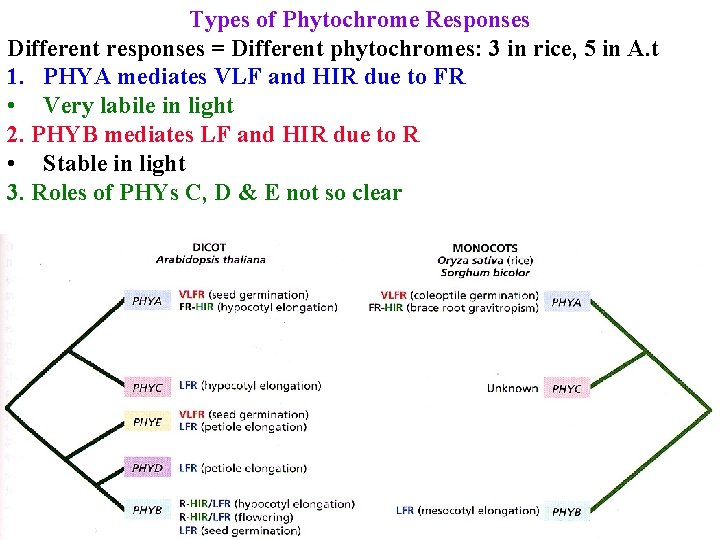
Types of Phytochrome Responses Different responses = Different phytochromes: 3 in rice, 5 in A. t 1. PHYA mediates VLF and HIR due to FR • Very labile in light 2. PHYB mediates LF and HIR due to R • Stable in light 3. Roles of PHYs C, D & E not so clear

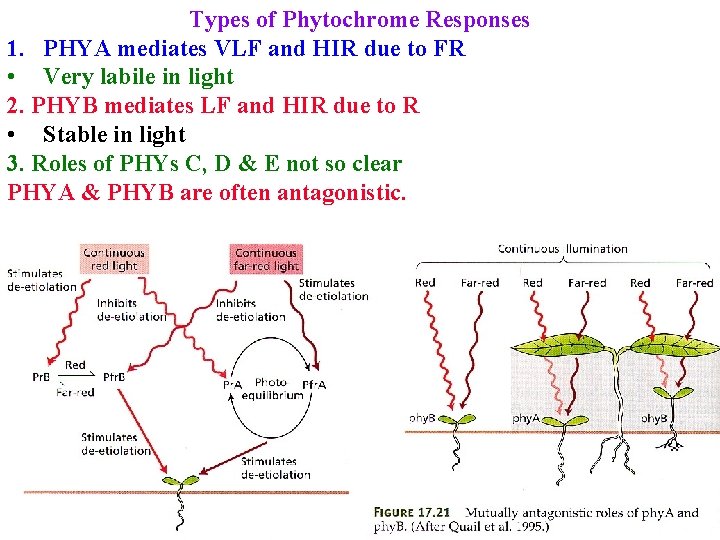
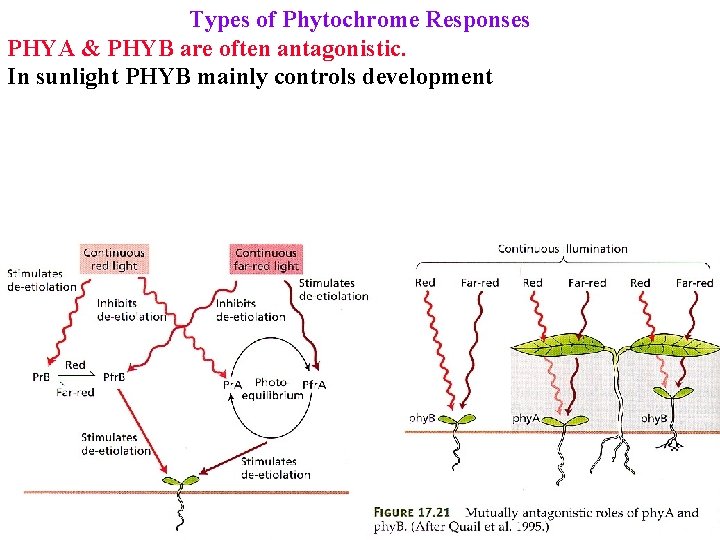
Types of Phytochrome Responses 1. PHYA mediates VLF and HIR due to FR • Very labile in light 2. PHYB mediates LF and HIR due to R • Stable in light 3. Roles of PHYs C, D & E not so clear PHYA & PHYB are often antagonistic.

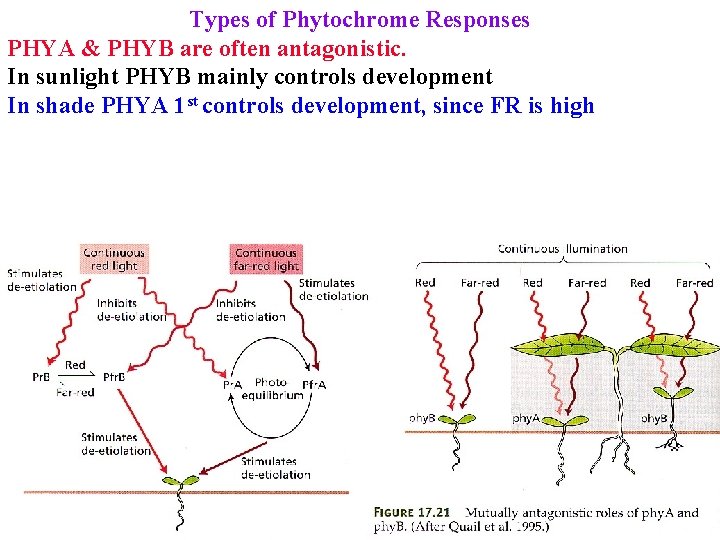
Types of Phytochrome Responses PHYA & PHYB are often antagonistic. In sunlight PHYB mainly controls development

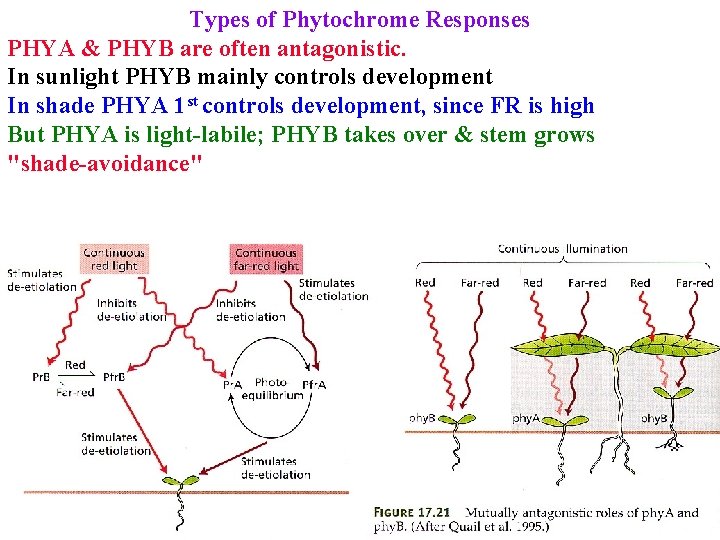
Types of Phytochrome Responses PHYA & PHYB are often antagonistic. In sunlight PHYB mainly controls development In shade PHYA 1 st controls development, since FR is high

Types of Phytochrome Responses PHYA & PHYB are often antagonistic. In sunlight PHYB mainly controls development In shade PHYA 1 st controls development, since FR is high But PHYA is light-labile; PHYB takes over & stem grows "shade-avoidance"

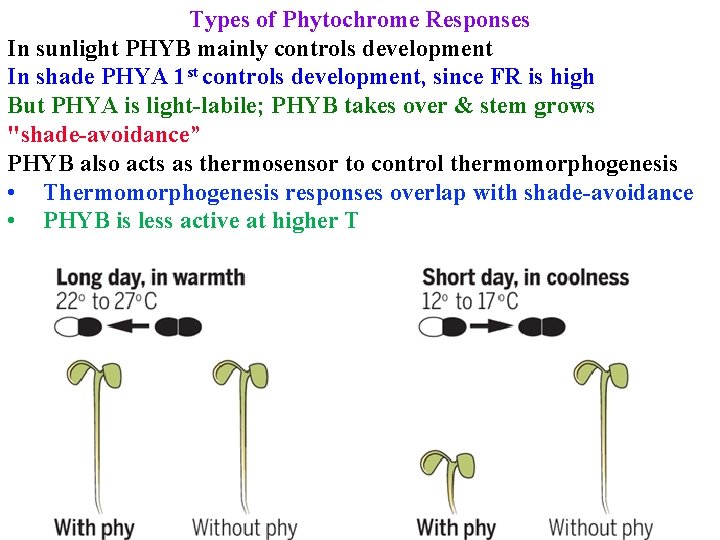
Types of Phytochrome Responses In sunlight PHYB mainly controls development In shade PHYA 1 st controls development, since FR is high But PHYA is light-labile; PHYB takes over & stem grows "shade-avoidance” PHYB also acts as thermosensor to control thermomorphogenesis • Thermomorphogenesis responses overlap with shade-avoidance • PHYB is less active at higher T

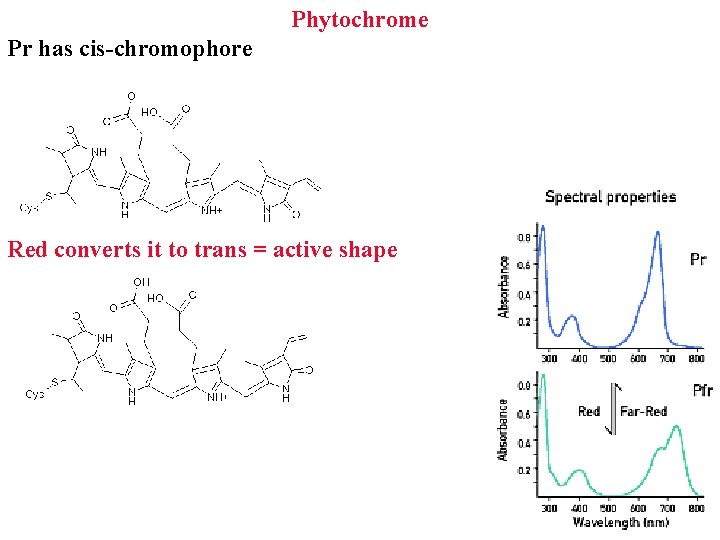
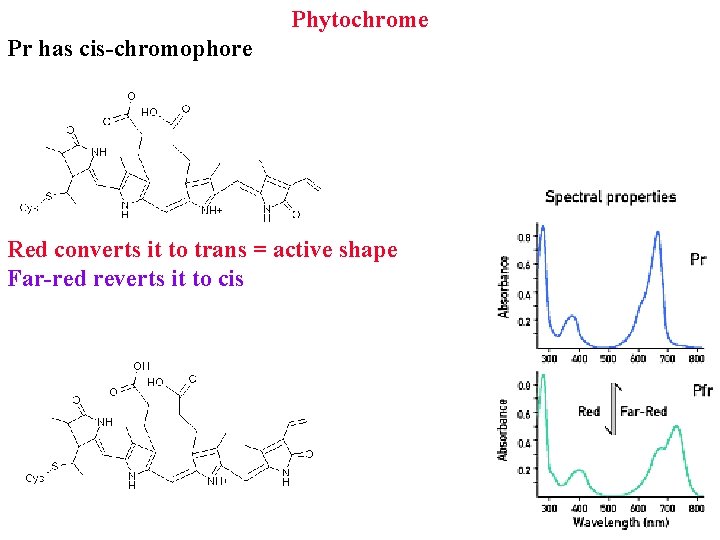
Phytochrome Pr has cis-chromophore

Phytochrome Pr has cis-chromophore Red converts it to trans = active shape

Phytochrome Pr has cis-chromophore Red converts it to trans = active shape Far-red reverts it to cis

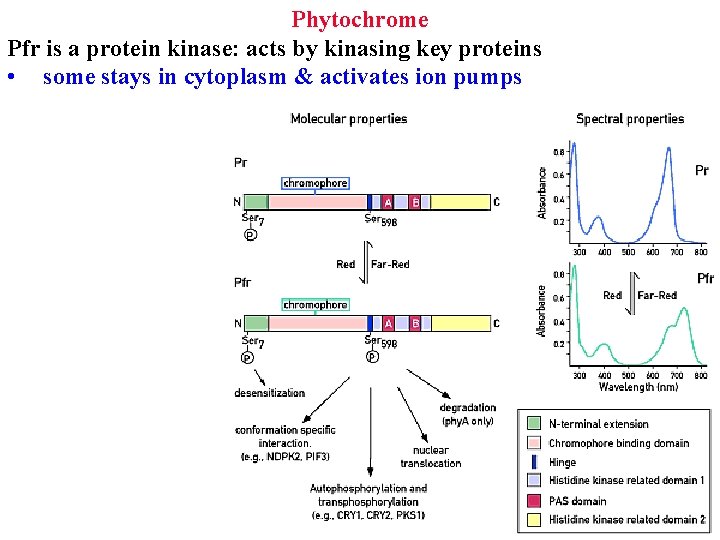
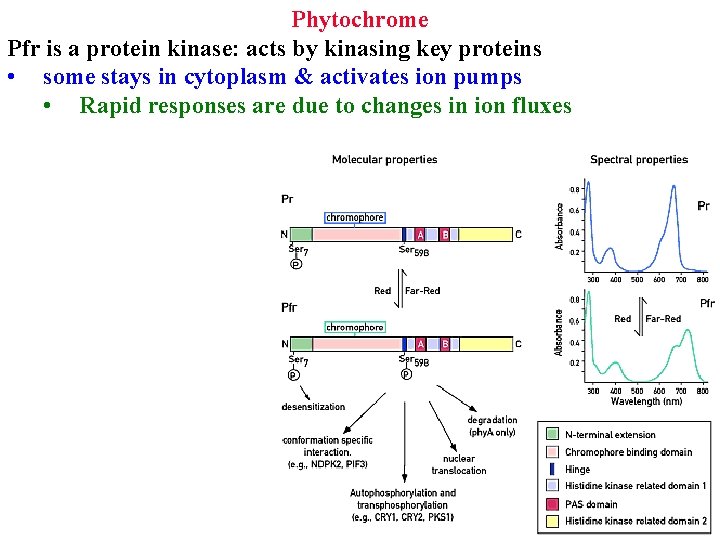
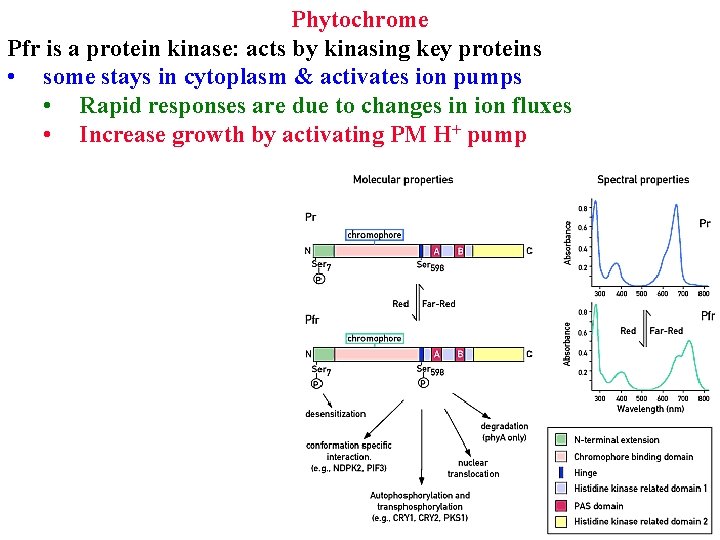
Phytochrome Pfr is a protein kinase: acts by kinasing key proteins • some stays in cytoplasm & activates ion pumps

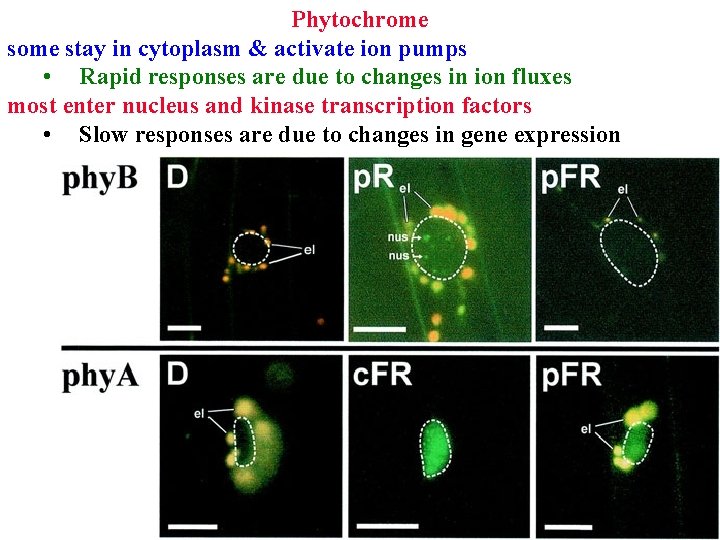
Phytochrome Pfr is a protein kinase: acts by kinasing key proteins • some stays in cytoplasm & activates ion pumps • Rapid responses are due to changes in ion fluxes

Phytochrome Pfr is a protein kinase: acts by kinasing key proteins • some stays in cytoplasm & activates ion pumps • Rapid responses are due to changes in ion fluxes • Increase growth by activating PM H+ pump

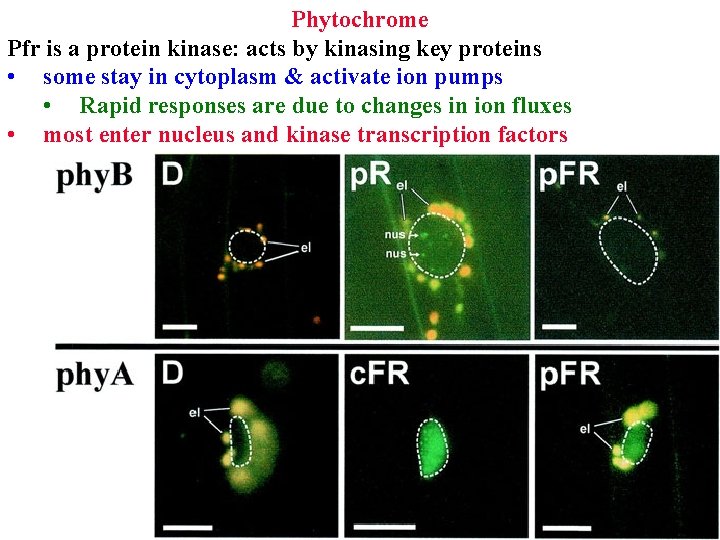
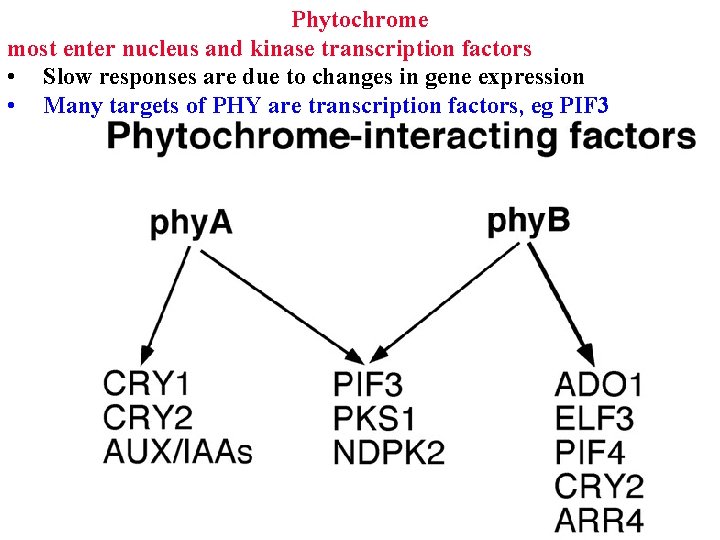
Phytochrome Pfr is a protein kinase: acts by kinasing key proteins • some stay in cytoplasm & activate ion pumps • Rapid responses are due to changes in ion fluxes • most enter nucleus and kinase transcription factors

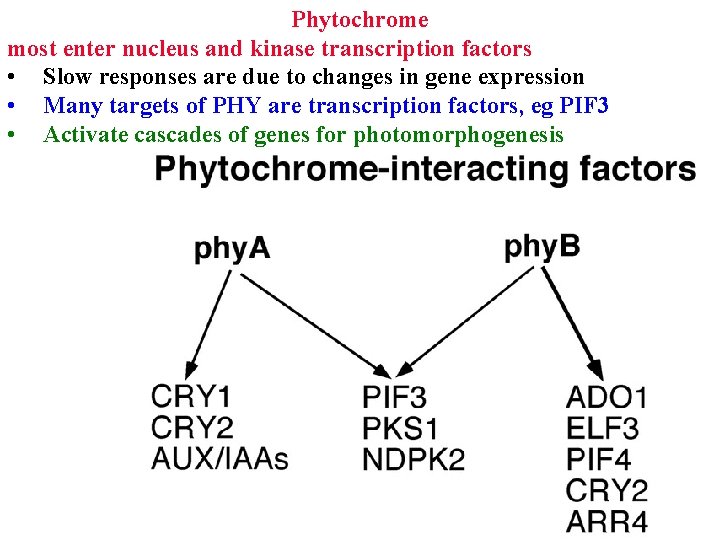
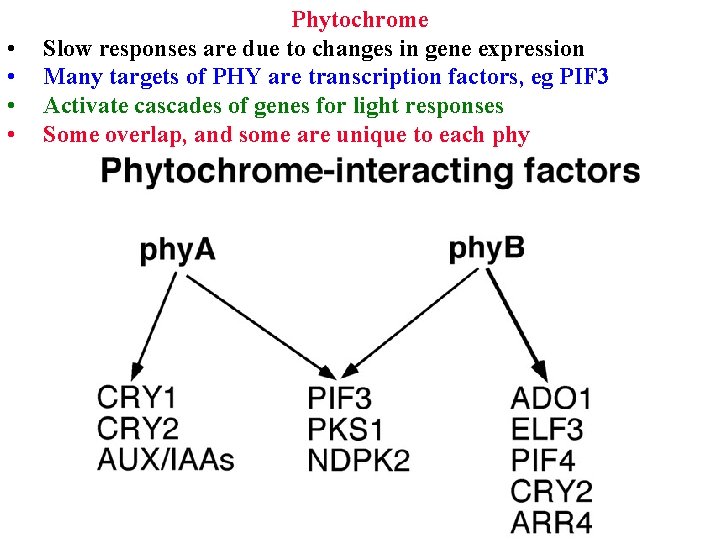
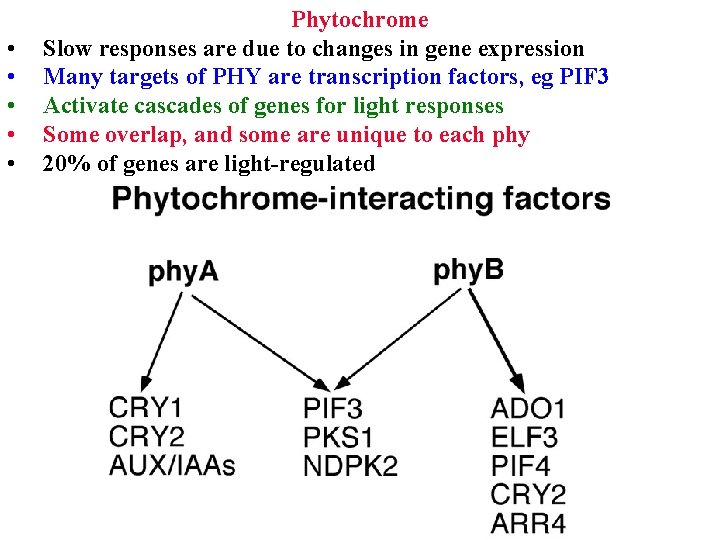
Phytochrome stay in cytoplasm & activate ion pumps • Rapid responses are due to changes in ion fluxes most enter nucleus and kinase transcription factors • Slow responses are due to changes in gene expression

Phytochrome most enter nucleus and kinase transcription factors • Slow responses are due to changes in gene expression • Many targets of PHY are transcription factors, eg PIF 3

Phytochrome most enter nucleus and kinase transcription factors • Slow responses are due to changes in gene expression • Many targets of PHY are transcription factors, eg PIF 3 • Activate cascades of genes for photomorphogenesis

• • Phytochrome Slow responses are due to changes in gene expression Many targets of PHY are transcription factors, eg PIF 3 Activate cascades of genes for light responses Some overlap, and some are unique to each phy

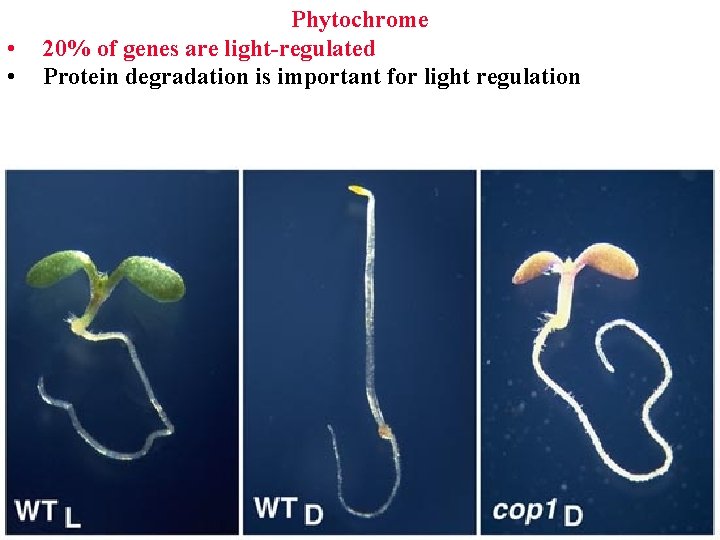
• • • Phytochrome Slow responses are due to changes in gene expression Many targets of PHY are transcription factors, eg PIF 3 Activate cascades of genes for light responses Some overlap, and some are unique to each phy 20% of genes are light-regulated

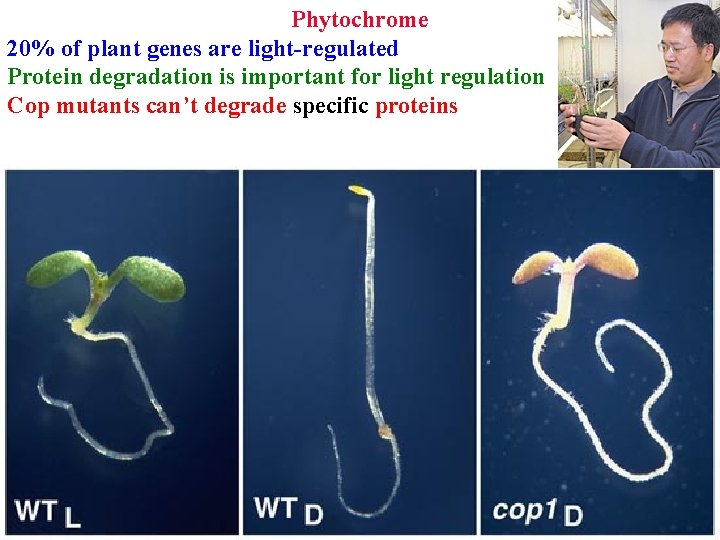
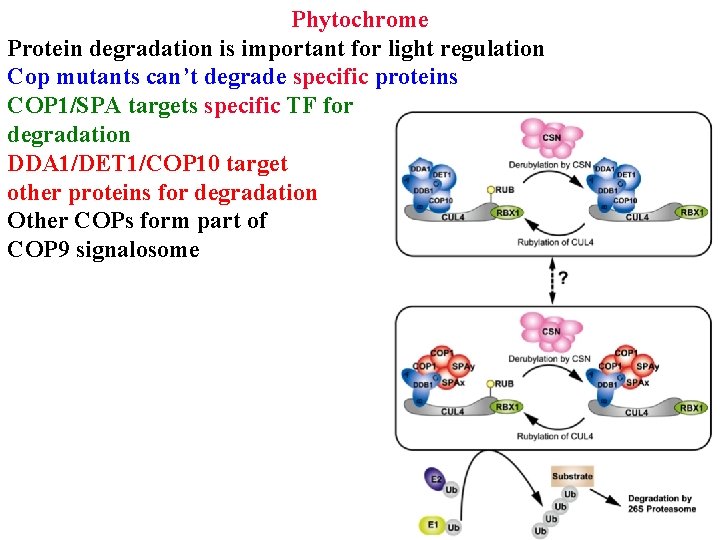
• • Phytochrome 20% of genes are light-regulated Protein degradation is important for light regulation

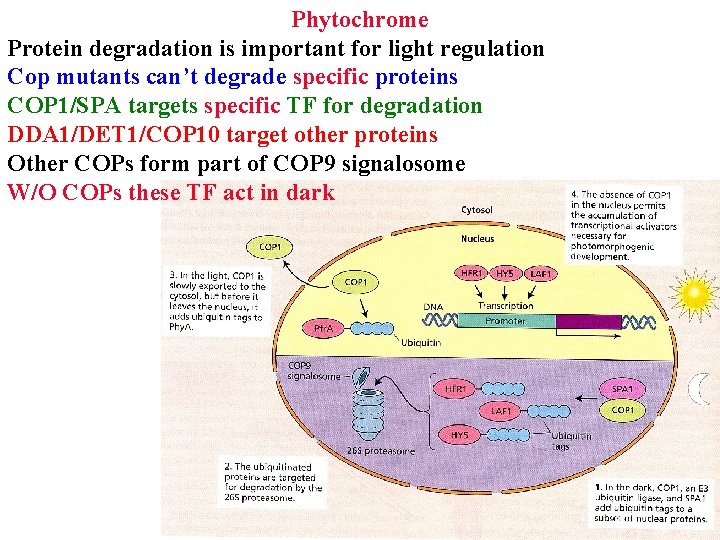
Phytochrome 20% of plant genes are light-regulated Protein degradation is important for light regulation Cop mutants can’t degrade specific proteins

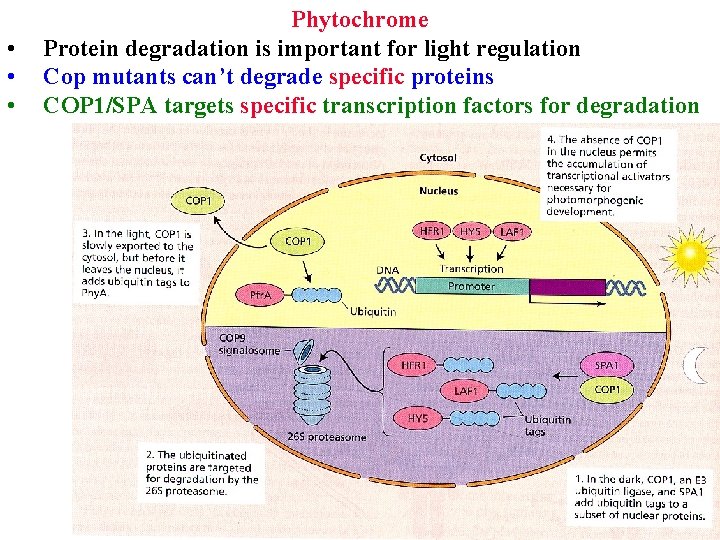
• • • Phytochrome Protein degradation is important for light regulation Cop mutants can’t degrade specific proteins COP 1/SPA targets specific transcription factors for degradation

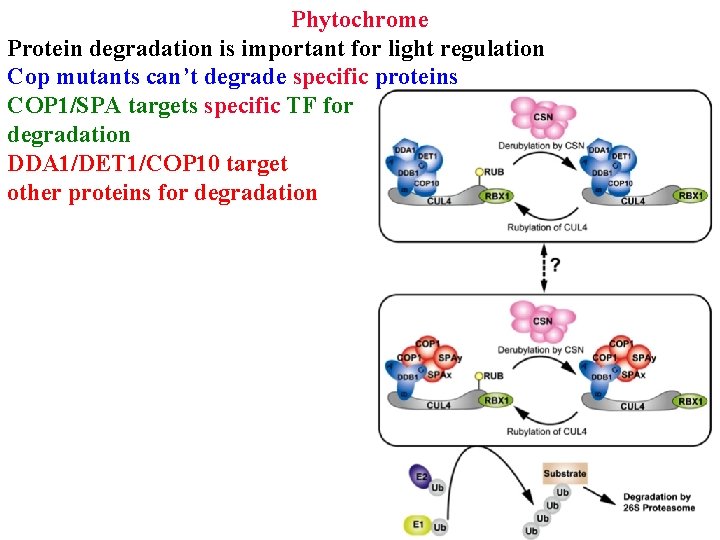
Phytochrome Protein degradation is important for light regulation Cop mutants can’t degrade specific proteins COP 1/SPA targets specific TF for degradation DDA 1/DET 1/COP 10 target other proteins for degradation

Phytochrome Protein degradation is important for light regulation Cop mutants can’t degrade specific proteins COP 1/SPA targets specific TF for degradation DDA 1/DET 1/COP 10 target other proteins for degradation Other COPs form part of COP 9 signalosome

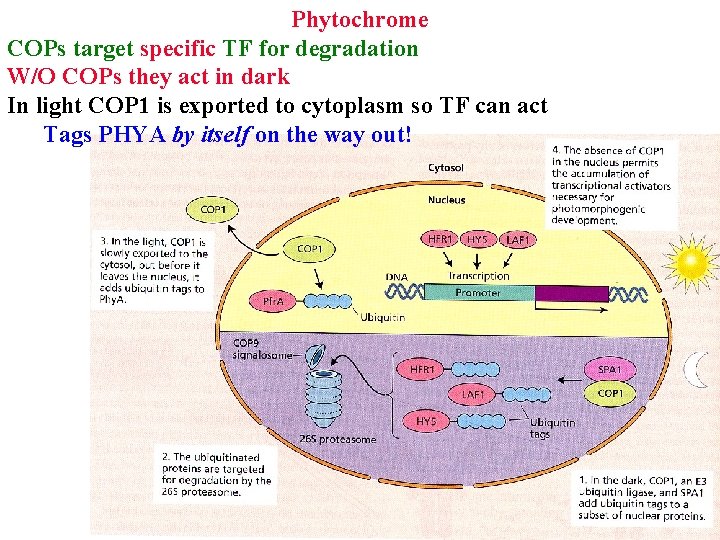
Phytochrome Protein degradation is important for light regulation Cop mutants can’t degrade specific proteins COP 1/SPA targets specific TF for degradation DDA 1/DET 1/COP 10 target other proteins Other COPs form part of COP 9 signalosome W/O COPs these TF act in dark

Phytochrome COPs target specific TF for degradation W/O COPs they act in dark In light COP 1 is exported to cytoplasm so TF can act Tags PHYA by itself on the way out!

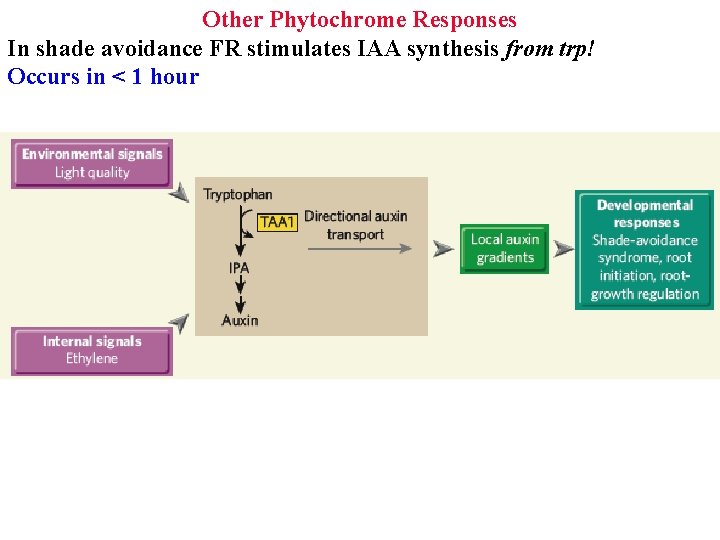
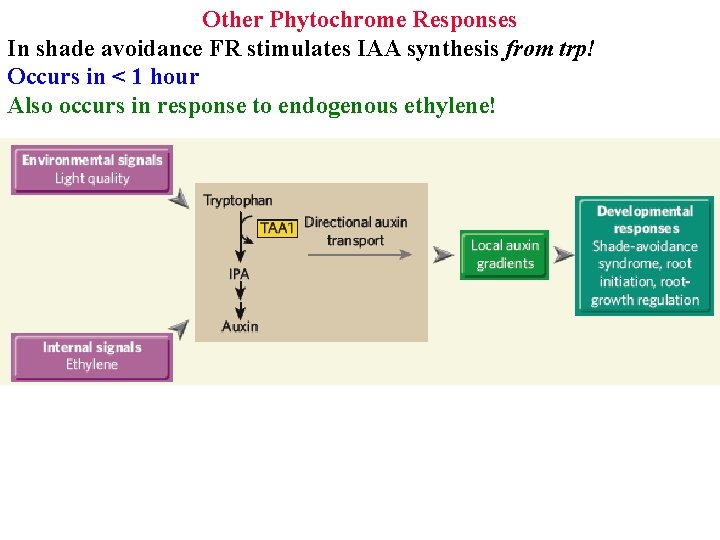
Other Phytochrome Responses In shade avoidance FR stimulates IAA synthesis from trp! Occurs in < 1 hour

Other Phytochrome Responses In shade avoidance FR stimulates IAA synthesis from trp! Occurs in < 1 hour Also occurs in response to endogenous ethylene!

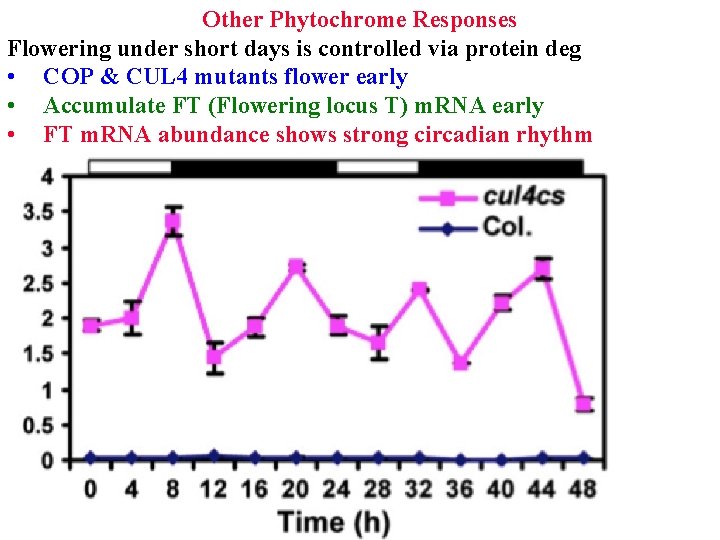
Other Phytochrome Responses Flowering under short days is controlled via protein deg • COP & CUL 4 mutants flower early

Other Phytochrome Responses Flowering under short days is controlled via protein deg • COP & CUL 4 mutants flower early • Accumulate FT (Flowering locus T) m. RNA early • FT m. RNA abundance shows strong circadian rhythm

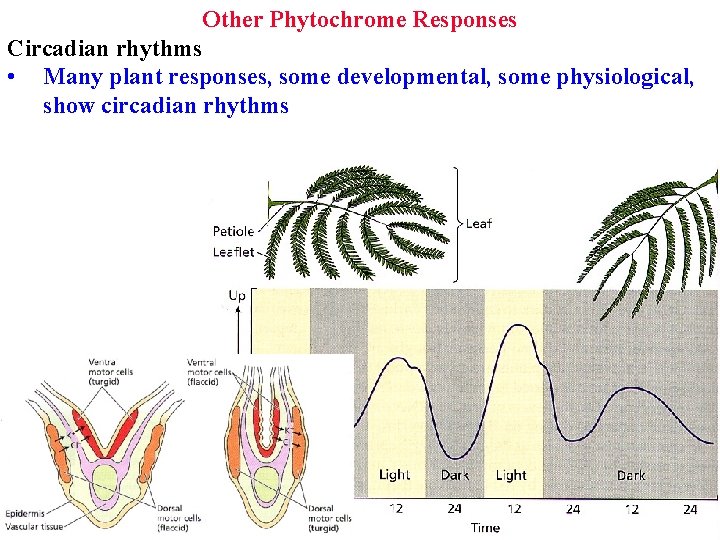
Other Phytochrome Responses Circadian rhythms • Many plant responses, some developmental, some physiological, show circadian rhythms

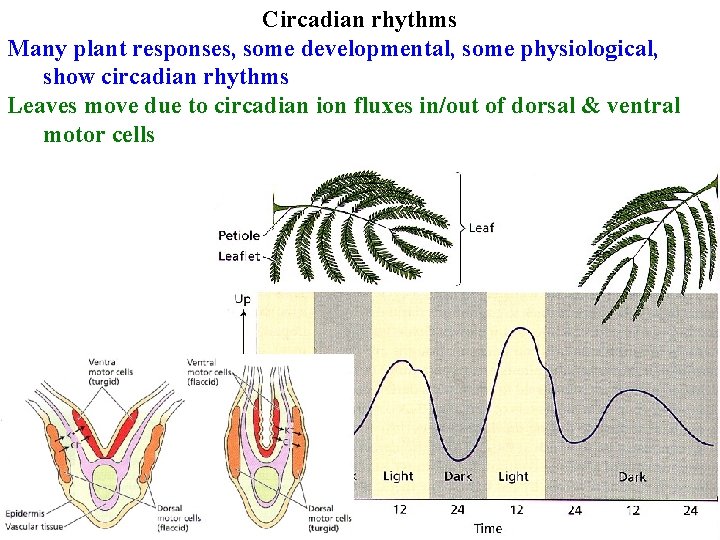
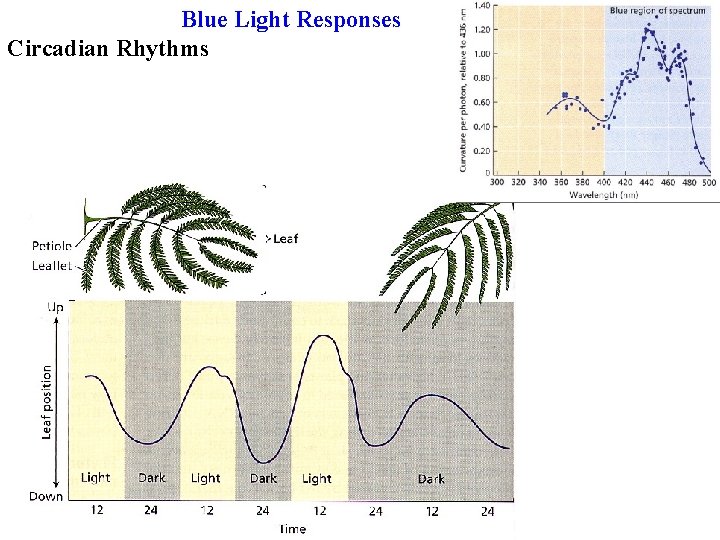
Circadian rhythms Many plant responses, some developmental, some physiological, show circadian rhythms Leaves move due to circadian ion fluxes in/out of dorsal & ventral motor cells

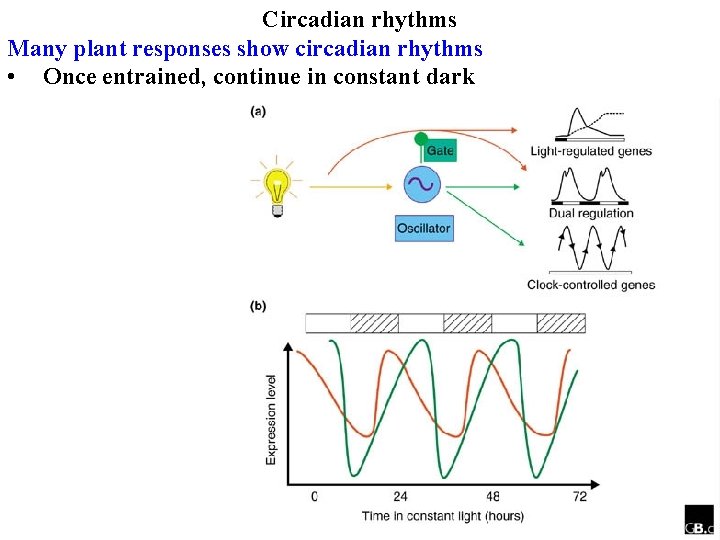
Circadian rhythms Many plant responses show circadian rhythms • Once entrained, continue in constant dark

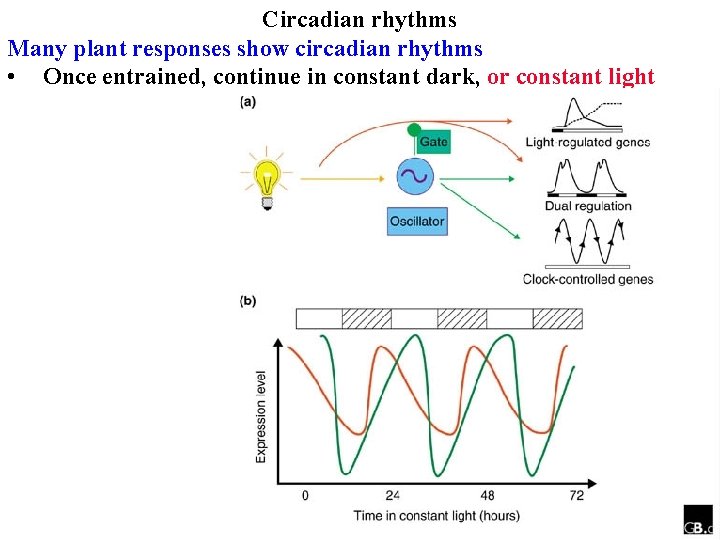
Circadian rhythms Many plant responses show circadian rhythms • Once entrained, continue in constant dark, or constant light

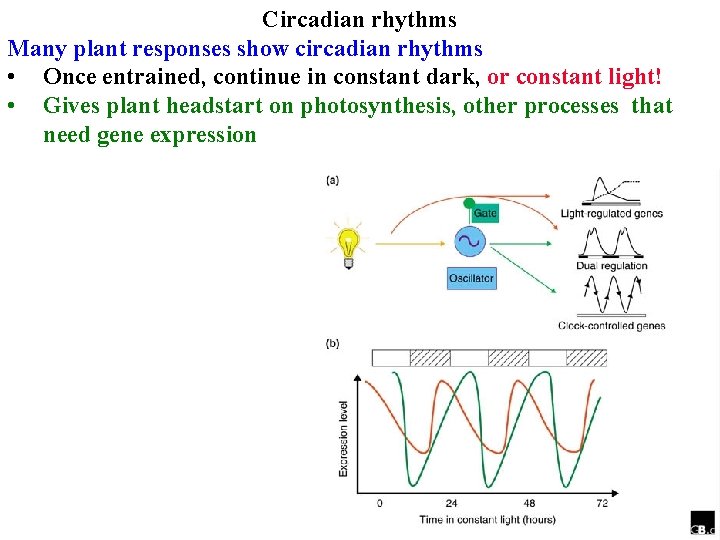
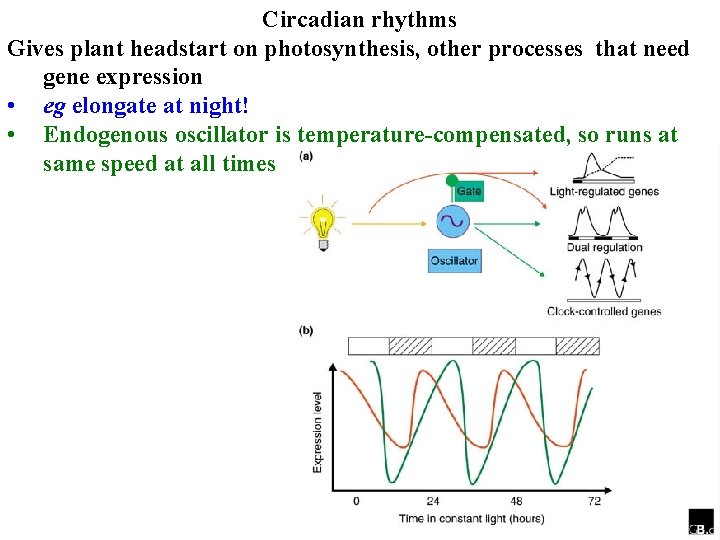
Circadian rhythms Many plant responses show circadian rhythms • Once entrained, continue in constant dark, or constant light! • Gives plant headstart on photosynthesis, other processes that need gene expression

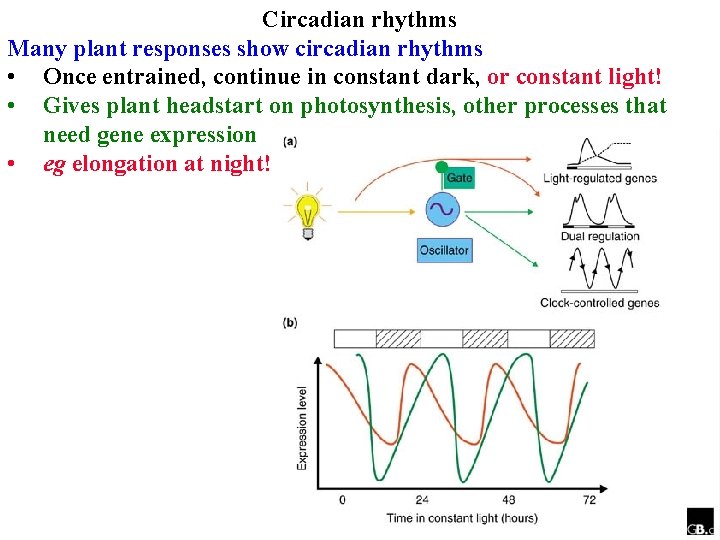
Circadian rhythms Many plant responses show circadian rhythms • Once entrained, continue in constant dark, or constant light! • Gives plant headstart on photosynthesis, other processes that need gene expression • eg elongation at night!

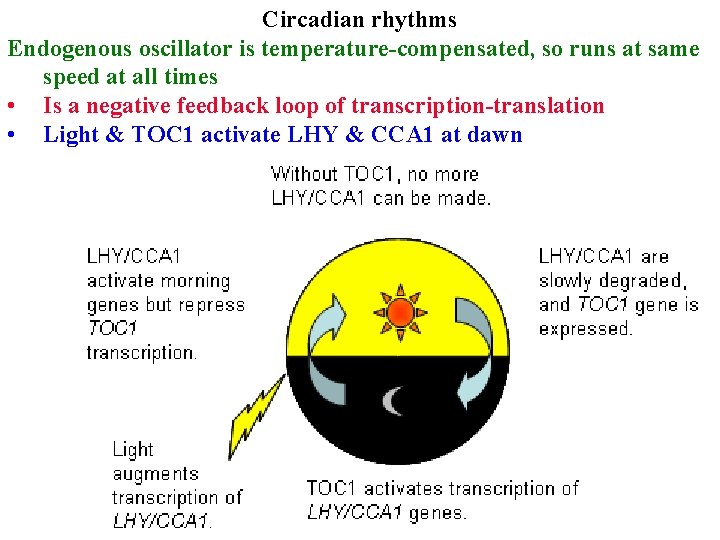
Circadian rhythms Gives plant headstart on photosynthesis, other processes that need gene expression • eg elongate at night! • Endogenous oscillator is temperature-compensated, so runs at same speed at all times

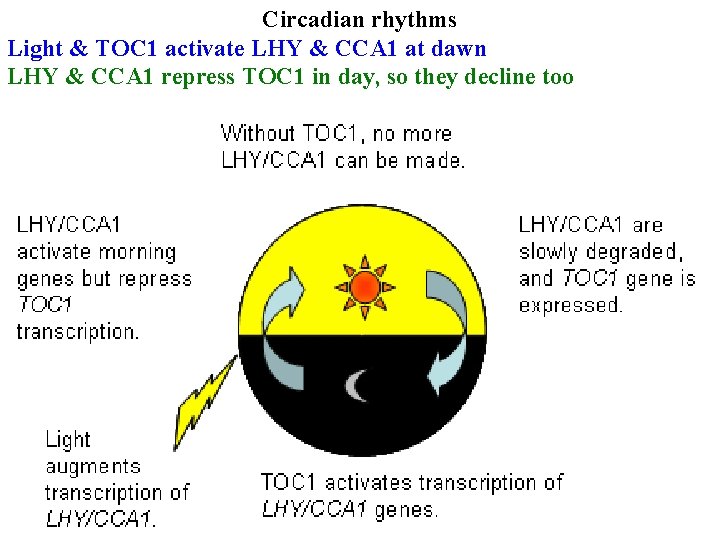
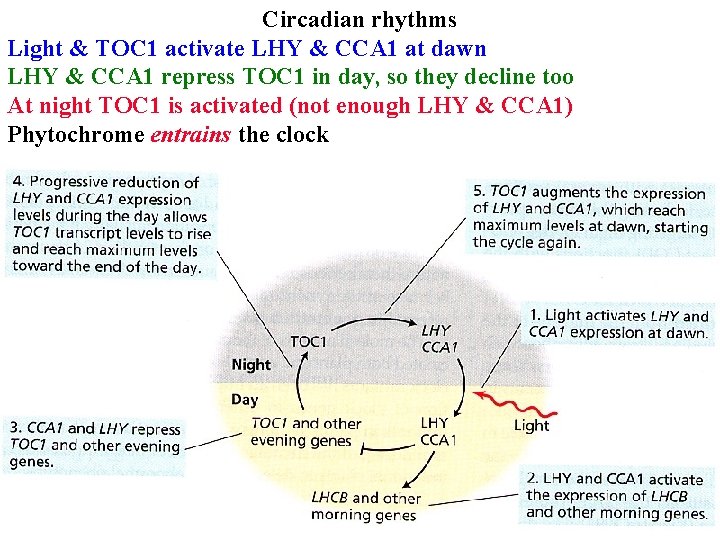
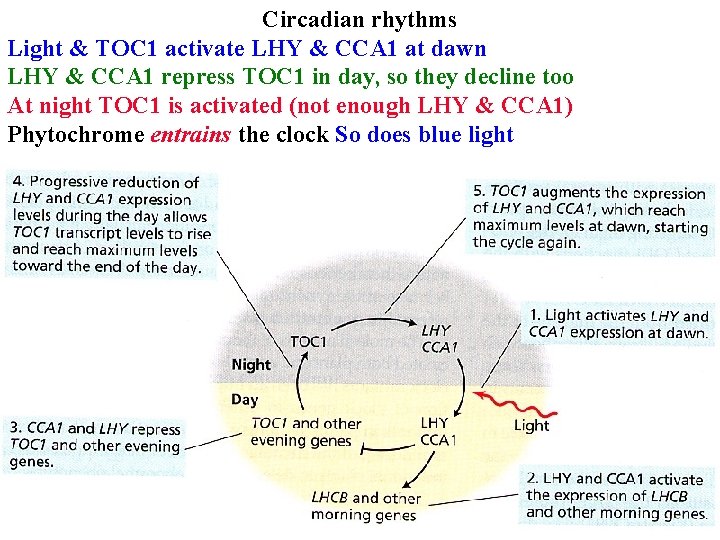
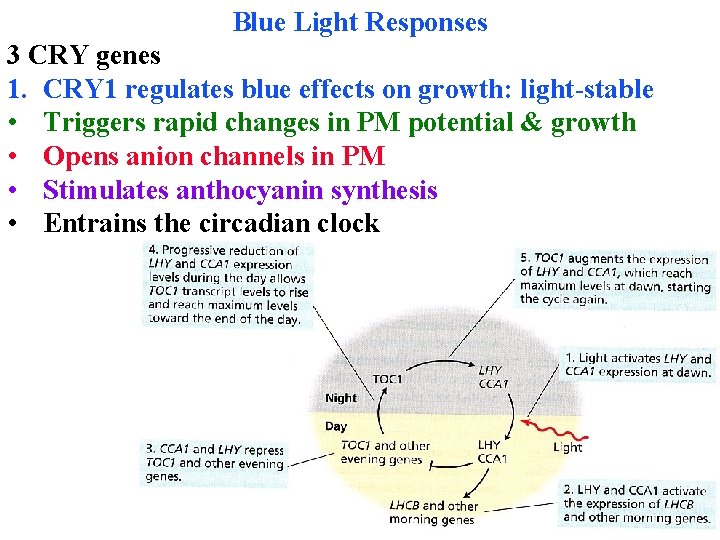
Circadian rhythms Endogenous oscillator is temperature-compensated, so runs at same speed at all times • Is a negative feedback loop of transcription-translation • Light & TOC 1 activate LHY & CCA 1 at dawn

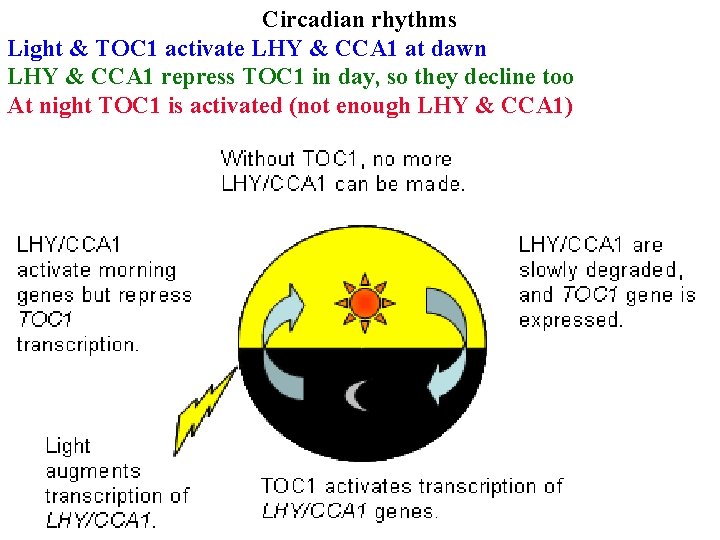
Circadian rhythms Light & TOC 1 activate LHY & CCA 1 at dawn LHY & CCA 1 repress TOC 1 in day, so they decline too

Circadian rhythms Light & TOC 1 activate LHY & CCA 1 at dawn LHY & CCA 1 repress TOC 1 in day, so they decline too At night TOC 1 is activated (not enough LHY & CCA 1)

Circadian rhythms Light & TOC 1 activate LHY & CCA 1 at dawn LHY & CCA 1 repress TOC 1 in day, so they decline too At night TOC 1 is activated (not enough LHY & CCA 1) Phytochrome entrains the clock

Circadian rhythms Light & TOC 1 activate LHY & CCA 1 at dawn LHY & CCA 1 repress TOC 1 in day, so they decline too At night TOC 1 is activated (not enough LHY & CCA 1) Phytochrome entrains the clock So does blue light

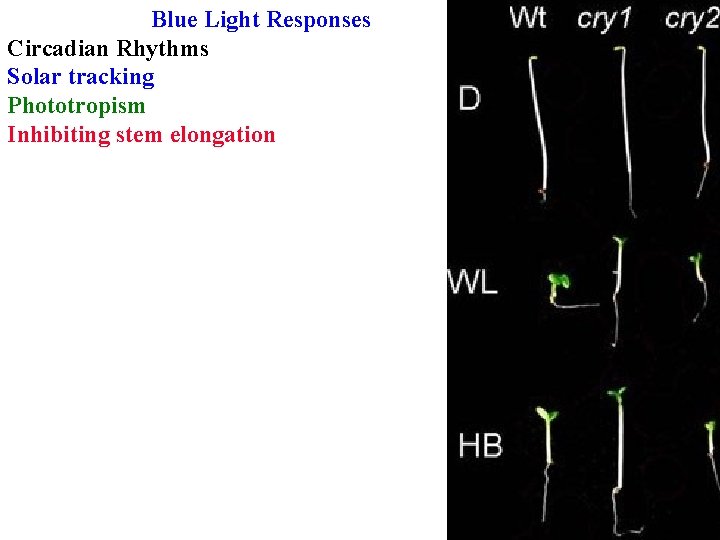
Blue Light Responses Circadian Rhythms

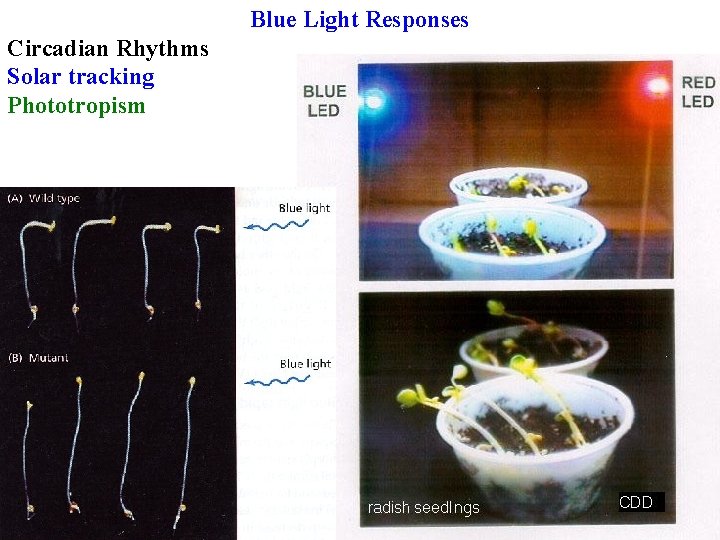
Blue Light Responses Circadian Rhythms Solar tracking

Blue Light Responses Circadian Rhythms Solar tracking Phototropism

Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation

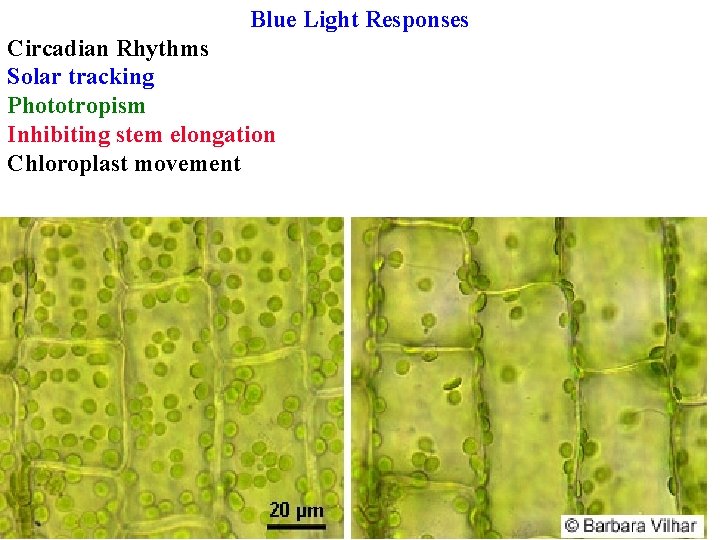
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement

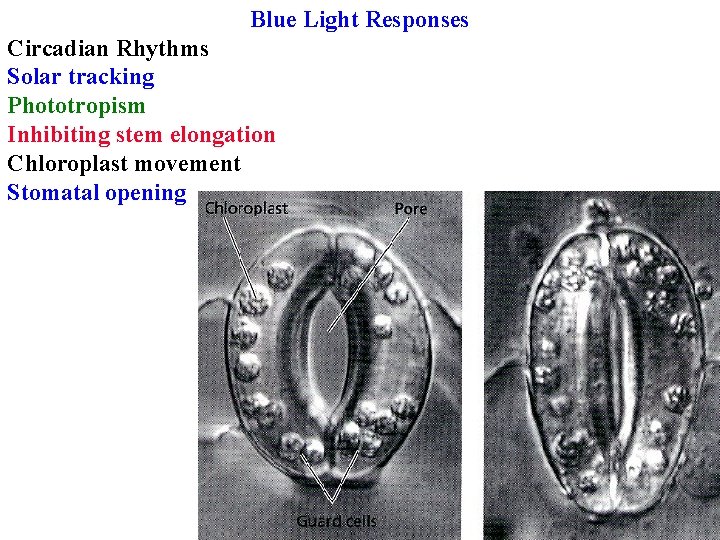
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement Stomatal opening

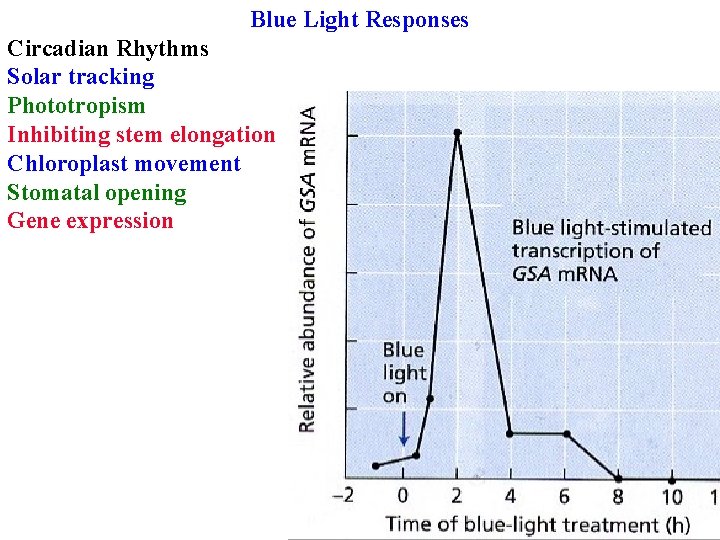
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement Stomatal opening Gene expression

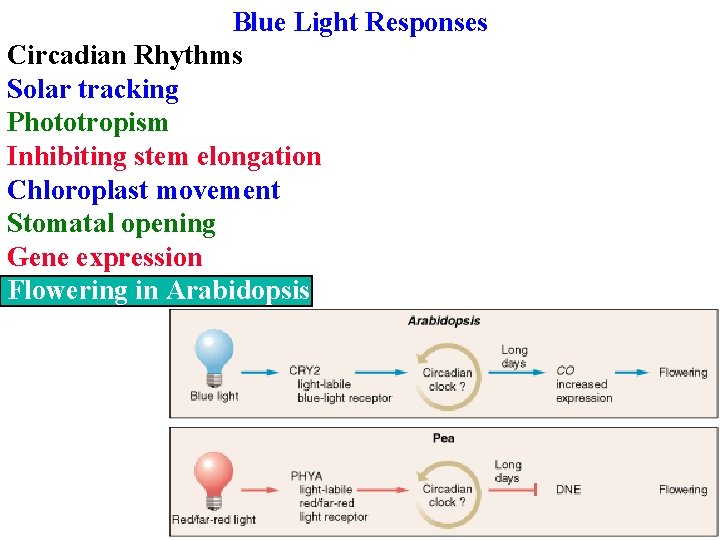
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement Stomatal opening Gene expression Flowering in Arabidopsis

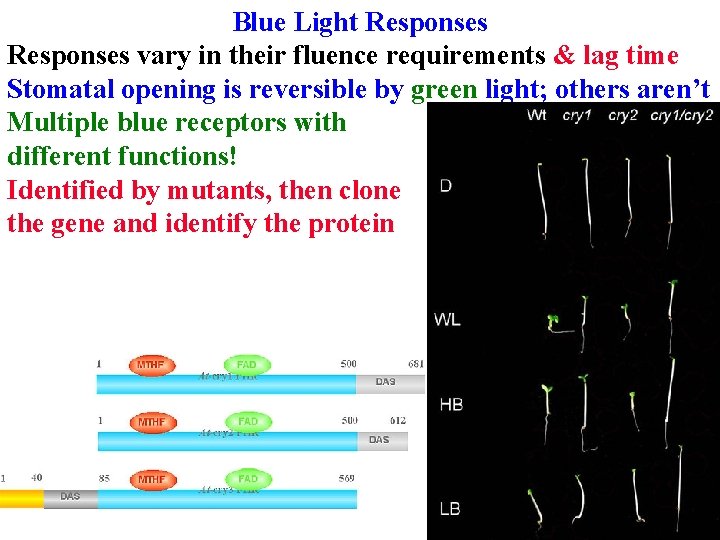
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement Stomatal opening Gene expression Flowering in Arabidopsis Responses vary in their fluence requirements

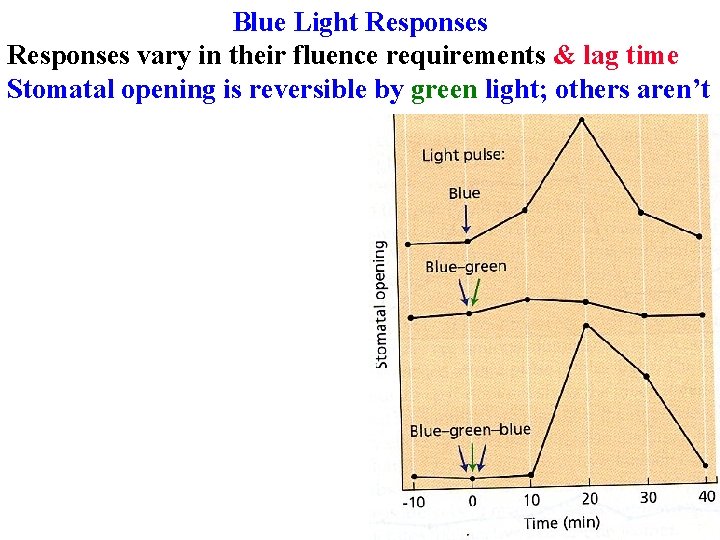
Blue Light Responses Circadian Rhythms Solar tracking Phototropism Inhibiting stem elongation Chloroplast movement Stomatal opening Gene expression Flowering in Arabidopsis Responses vary in their fluence requirements & lag times

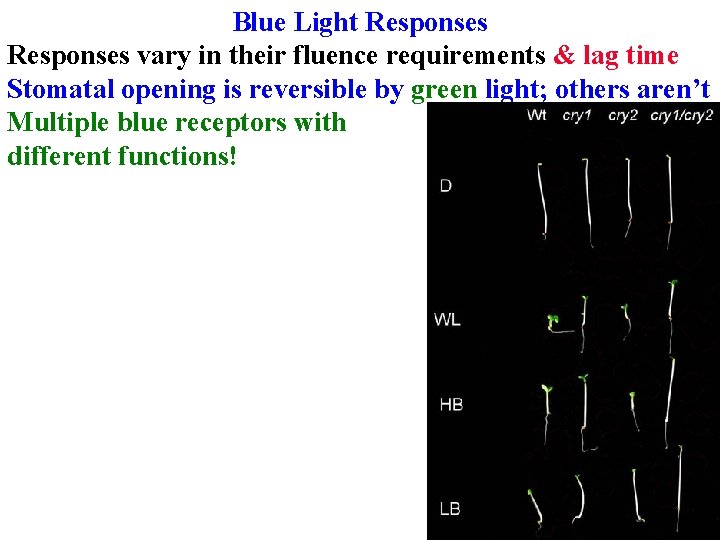
Blue Light Responses vary in their fluence requirements & lag time Stomatal opening is reversible by green light; others aren’t

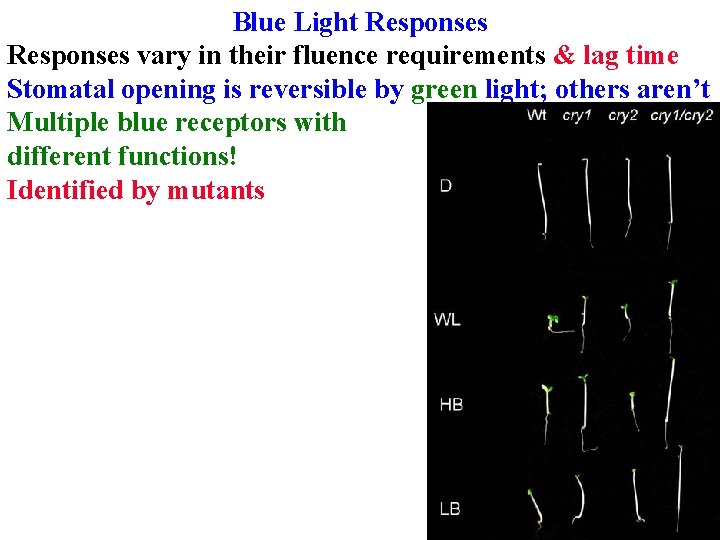
Blue Light Responses vary in their fluence requirements & lag time Stomatal opening is reversible by green light; others aren’t Multiple blue receptors with different functions!

Blue Light Responses vary in their fluence requirements & lag time Stomatal opening is reversible by green light; others aren’t Multiple blue receptors with different functions! Identified by mutants

Blue Light Responses vary in their fluence requirements & lag time Stomatal opening is reversible by green light; others aren’t Multiple blue receptors with different functions! Identified by mutants, then clone the gene and identify the protein

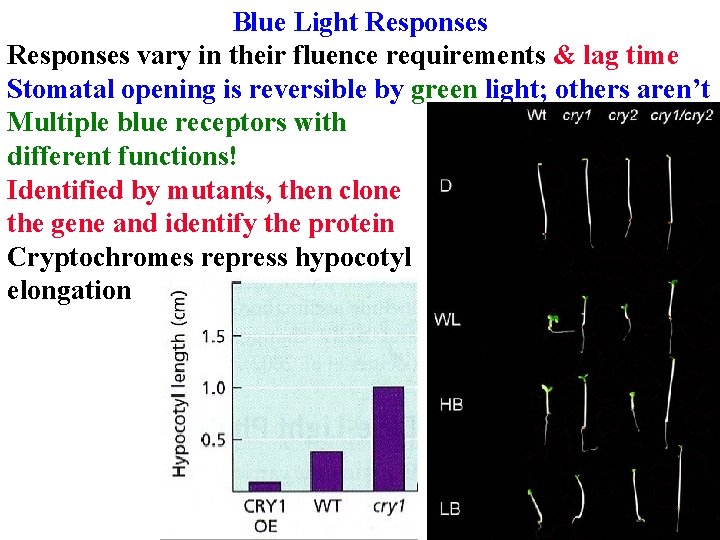
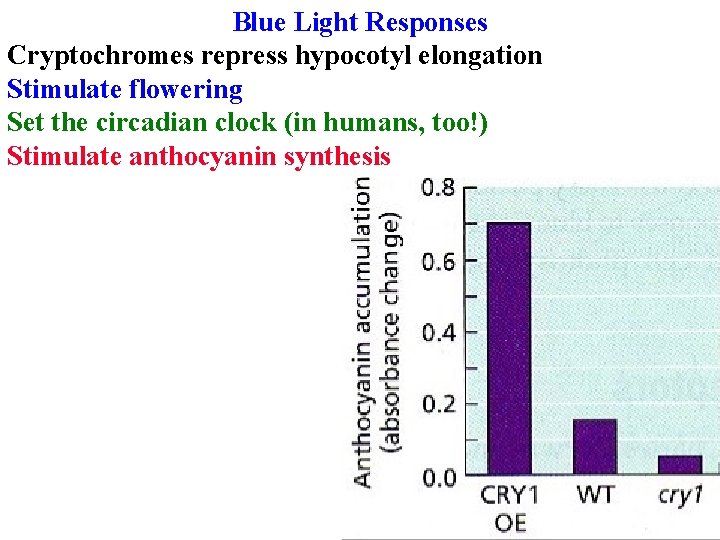
Blue Light Responses vary in their fluence requirements & lag time Stomatal opening is reversible by green light; others aren’t Multiple blue receptors with different functions! Identified by mutants, then clone the gene and identify the protein Cryptochromes repress hypocotyl elongation

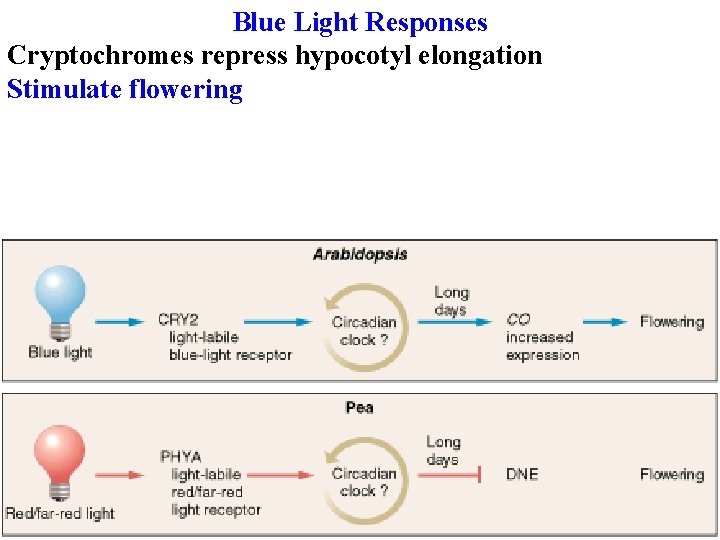
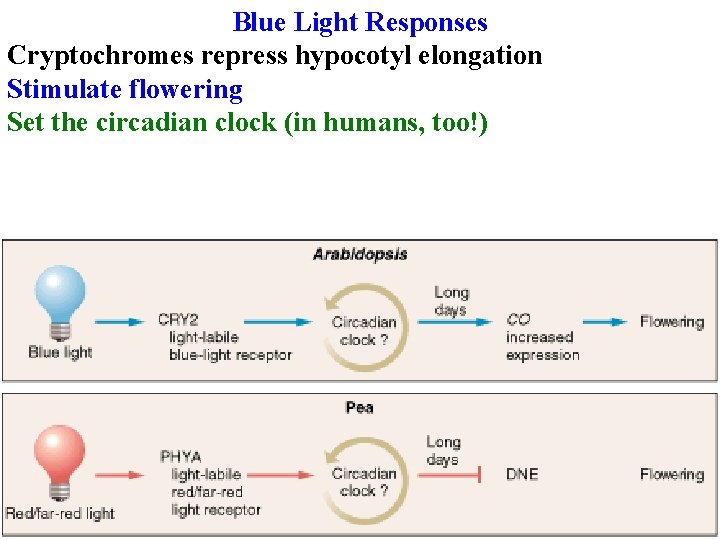
Blue Light Responses Cryptochromes repress hypocotyl elongation Stimulate flowering

Blue Light Responses Cryptochromes repress hypocotyl elongation Stimulate flowering Set the circadian clock (in humans, too!)

Blue Light Responses Cryptochromes repress hypocotyl elongation Stimulate flowering Set the circadian clock (in humans, too!) Stimulate anthocyanin synthesis

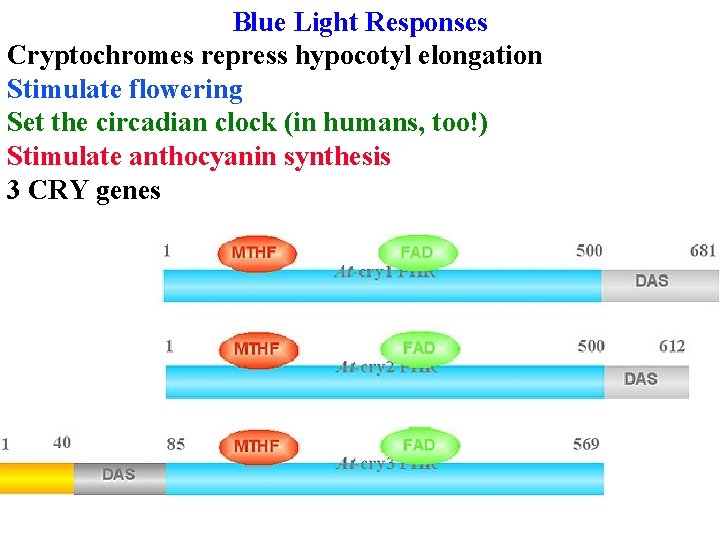
Blue Light Responses Cryptochromes repress hypocotyl elongation Stimulate flowering Set the circadian clock (in humans, too!) Stimulate anthocyanin synthesis 3 CRY genes

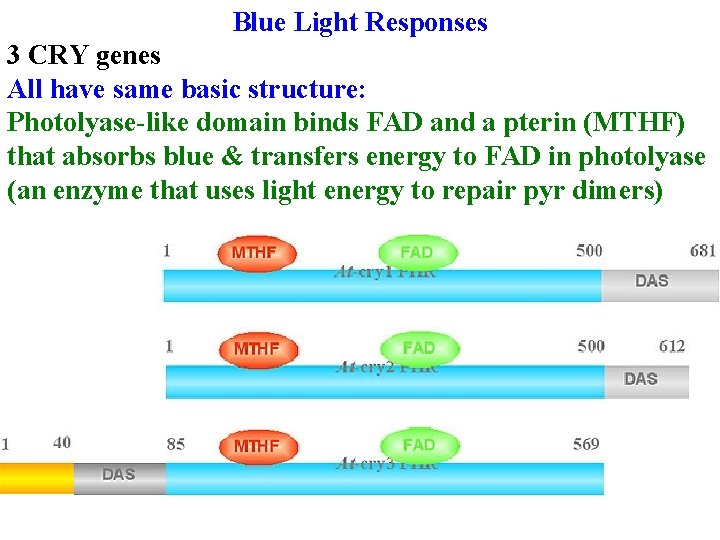
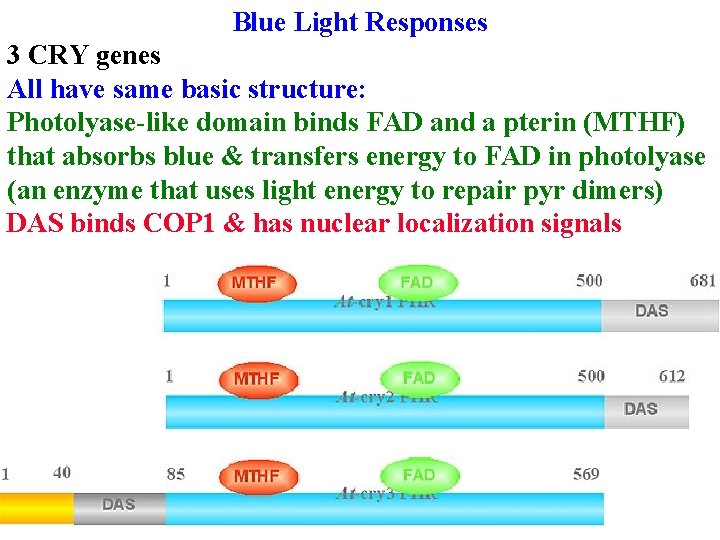
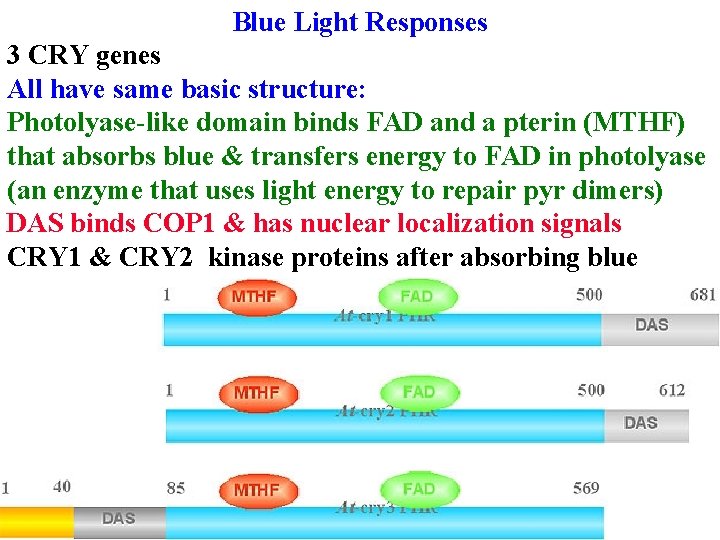
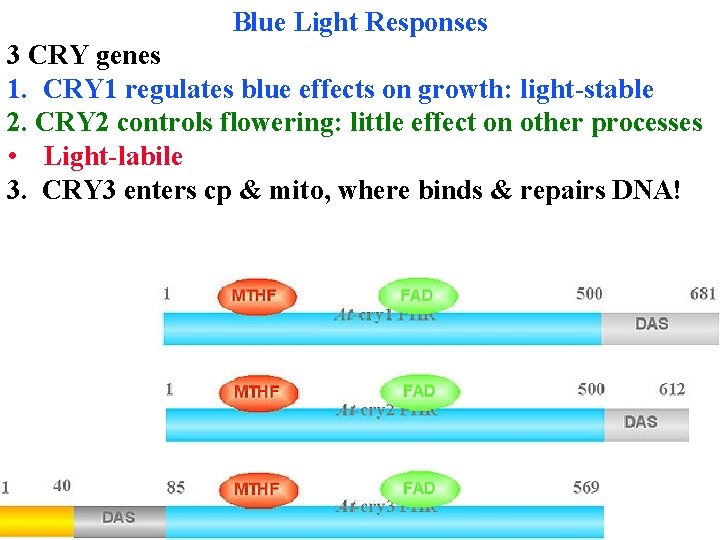
Blue Light Responses 3 CRY genes All have same basic structure: Photolyase-like domain binds FAD and a pterin (MTHF) that absorbs blue & transfers energy to FAD in photolyase (an enzyme that uses light energy to repair pyr dimers)

Blue Light Responses 3 CRY genes All have same basic structure: Photolyase-like domain binds FAD and a pterin (MTHF) that absorbs blue & transfers energy to FAD in photolyase (an enzyme that uses light energy to repair pyr dimers) DAS binds COP 1 & has nuclear localization signals

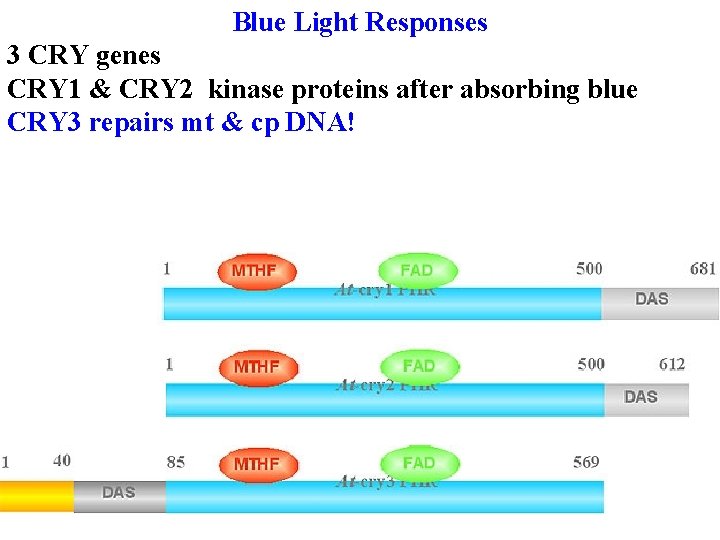
Blue Light Responses 3 CRY genes All have same basic structure: Photolyase-like domain binds FAD and a pterin (MTHF) that absorbs blue & transfers energy to FAD in photolyase (an enzyme that uses light energy to repair pyr dimers) DAS binds COP 1 & has nuclear localization signals CRY 1 & CRY 2 kinase proteins after absorbing blue

Blue Light Responses 3 CRY genes CRY 1 & CRY 2 kinase proteins after absorbing blue CRY 3 repairs mt & cp DNA!

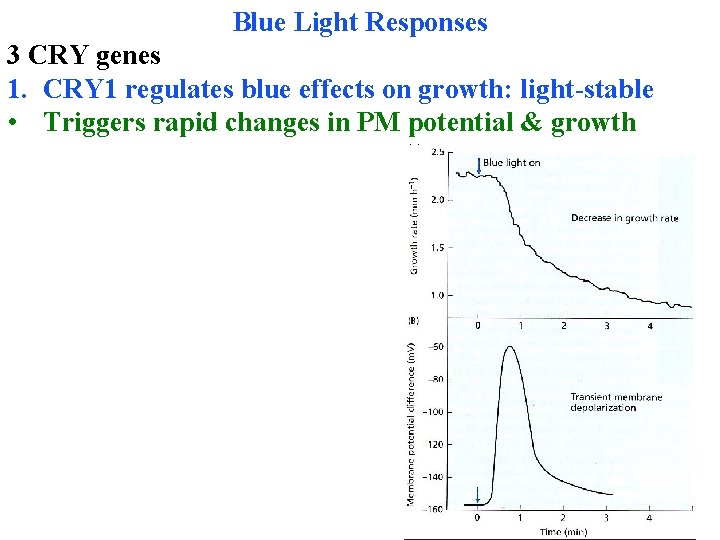
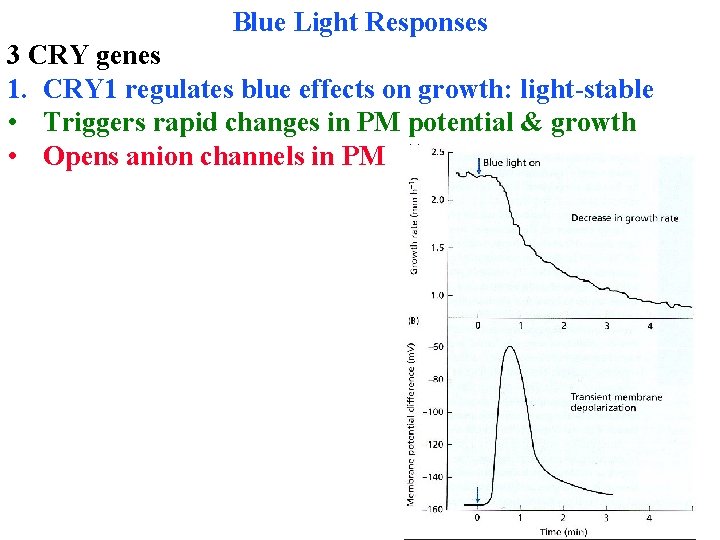
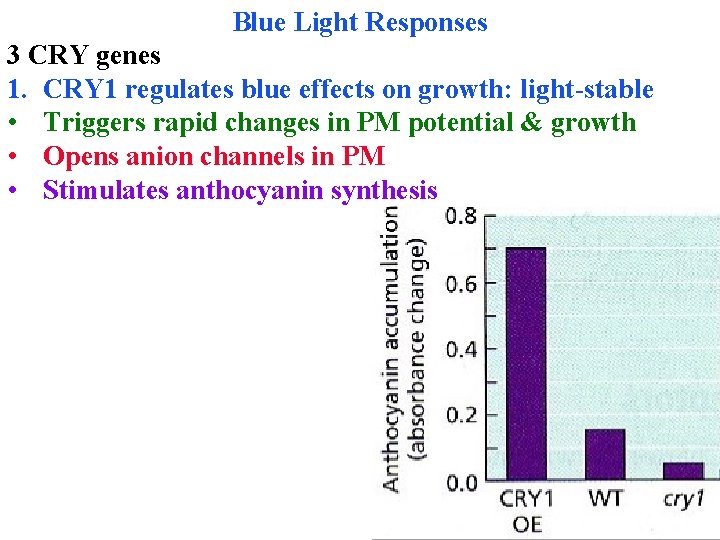
Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth

Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth • Opens anion channels in PM

Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth • Opens anion channels in PM • Stimulates anthocyanin synthesis

Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth • Opens anion channels in PM • Stimulates anthocyanin synthesis • Entrains the circadian clock

Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth • Opens anion channels in PM • Stimulates anthocyanin synthesis • Entrains the circadian clock • Also accumulates in nucleus & interacts with PHY & COP 1 to regulate photomorphogenesis, probably by kinasing substrates

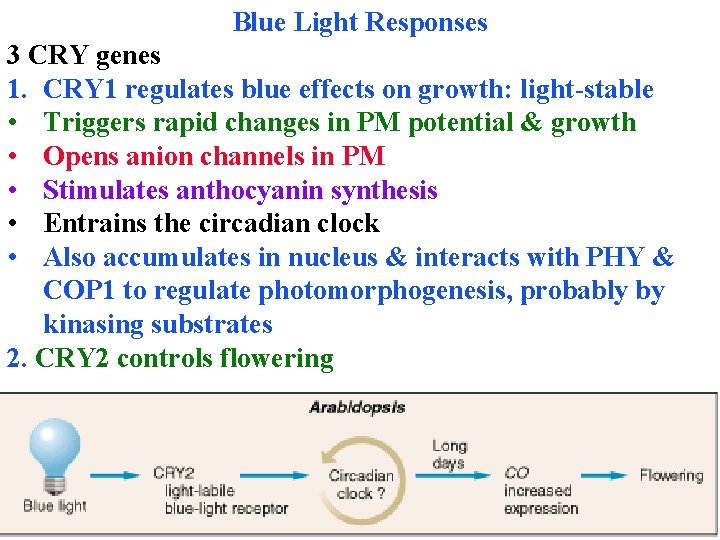
Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable • Triggers rapid changes in PM potential & growth • Opens anion channels in PM • Stimulates anthocyanin synthesis • Entrains the circadian clock • Also accumulates in nucleus & interacts with PHY & COP 1 to regulate photomorphogenesis, probably by kinasing substrates 2. CRY 2 controls flowering

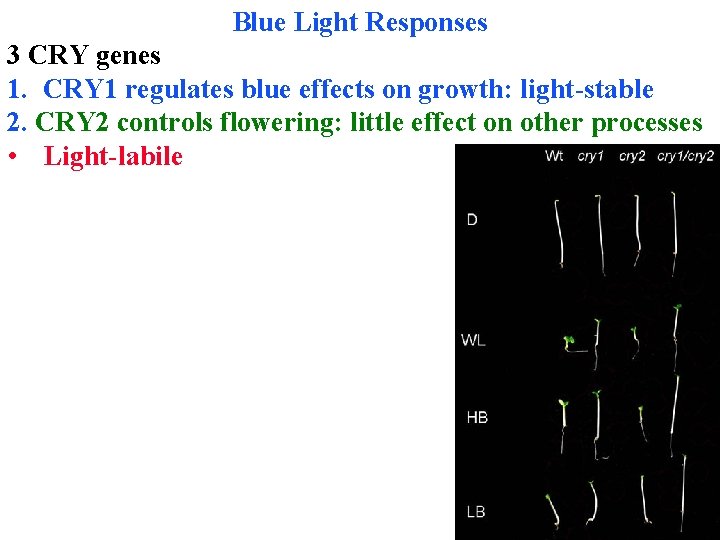
Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable 2. CRY 2 controls flowering: little effect on other processes • Light-labile

Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth: light-stable 2. CRY 2 controls flowering: little effect on other processes • Light-labile 3. CRY 3 enters cp & mito, where binds & repairs DNA!

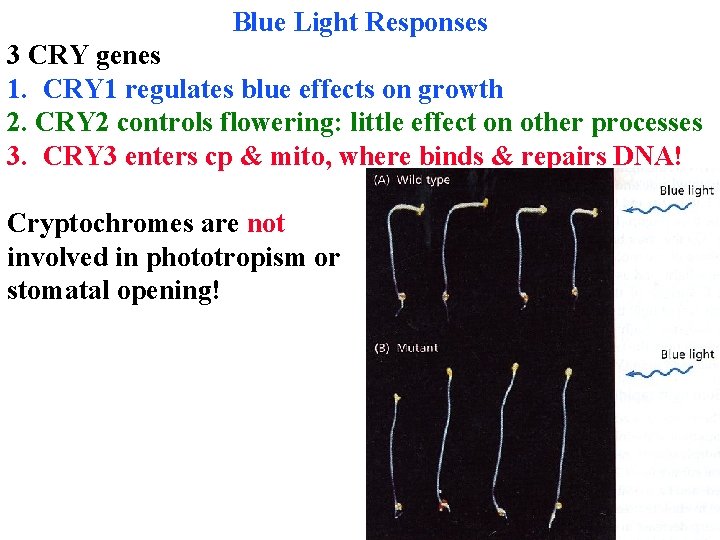
Blue Light Responses 3 CRY genes 1. CRY 1 regulates blue effects on growth 2. CRY 2 controls flowering: little effect on other processes 3. CRY 3 enters cp & mito, where binds & repairs DNA! Cryptochromes are not involved in phototropism or stomatal opening!
 Nonvascular plants
Nonvascular plants Non vascular plants
Non vascular plants Characteristics of non-flowering plants
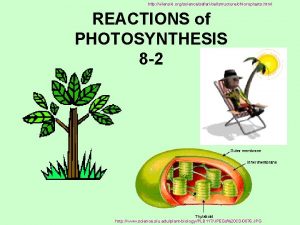
Characteristics of non-flowering plants Photosynthesis equation
Photosynthesis equation Light light light chapter 23
Light light light chapter 23 Into the light chapter 22
Into the light chapter 22 Chapter 22
Chapter 22 Unit 2 plant taxonomy how plants are named
Unit 2 plant taxonomy how plants are named Scope of plant breeding
Scope of plant breeding Plant breeding for disease resistance
Plant breeding for disease resistance Plant introduction in plant breeding
Plant introduction in plant breeding Tronsmo plant pathology and plant diseases download
Tronsmo plant pathology and plant diseases download Tronsmo plant pathology and plant diseases download
Tronsmo plant pathology and plant diseases download Albugo eye
Albugo eye Animal cells
Animal cells Natural source of nitrogen
Natural source of nitrogen Plants gather energy with light-absorbing pigments called *
Plants gather energy with light-absorbing pigments called * Plant tissue culture application
Plant tissue culture application Rice growth phases and stages
Rice growth phases and stages Cytokinin function
Cytokinin function Chapter 35 plant structure growth and development
Chapter 35 plant structure growth and development Apical meristem
Apical meristem Vascular ray
Vascular ray Yesterday
Yesterday Traditionally light cured gels relied on
Traditionally light cured gels relied on Plane mirror used
Plane mirror used Othello put out the light
Othello put out the light Leucoplast double membrane
Leucoplast double membrane Or the bending of light and the bouncing off of light
Or the bending of light and the bouncing off of light Materials that completely block light
Materials that completely block light Homometric regulation of cardiac output
Homometric regulation of cardiac output Ddb board regulation no.1 series of 2014
Ddb board regulation no.1 series of 2014 Line regulation
Line regulation Jrotc hair regulations
Jrotc hair regulations Loop of henle
Loop of henle Homeostasis mechanisms for regulation of body temperature
Homeostasis mechanisms for regulation of body temperature Body temperature maintenance
Body temperature maintenance Zones of regulation
Zones of regulation Ar 623 3
Ar 623 3 Army bah regulation 608 99
Army bah regulation 608 99 Section 4 gene regulation and mutations
Section 4 gene regulation and mutations Rule and regulation of table tennis
Rule and regulation of table tennis Gene expression in prokaryotes and eukaryotes
Gene expression in prokaryotes and eukaryotes Gastric emptying is promoted by
Gastric emptying is promoted by Regulation of blood pressure
Regulation of blood pressure Section 72 of regulation o.reg.941
Section 72 of regulation o.reg.941 Prescriptive vs performance based regulation
Prescriptive vs performance based regulation Malaysia elv regulation
Malaysia elv regulation Population regulation in the serengeti answer key
Population regulation in the serengeti answer key Army evaluation regulation
Army evaluation regulation Dd form 2560 advance pay certification/authorization
Dd form 2560 advance pay certification/authorization Baroreceptors vs chemoreceptors
Baroreceptors vs chemoreceptors Guided self regulation
Guided self regulation Coregulation parenting
Coregulation parenting Temperature regulation
Temperature regulation Recyclability and self regulation in nature
Recyclability and self regulation in nature Model of professional nursing practice regulation
Model of professional nursing practice regulation Mandatory removal date army regulation
Mandatory removal date army regulation Army ncoer non rated codes
Army ncoer non rated codes Regulation of blood pressure negative feedback
Regulation of blood pressure negative feedback Mcit bir regulation 9-98
Mcit bir regulation 9-98 14cfr91
14cfr91 Outcomes focused regulation
Outcomes focused regulation Top down regulation
Top down regulation Regulation of glycolysis and gluconeogenesis
Regulation of glycolysis and gluconeogenesis Functions of glycogen
Functions of glycogen 6h6 tolerance
6h6 tolerance 12-5 gene regulation
12-5 gene regulation Positive vs negative controls
Positive vs negative controls Line regulation formula
Line regulation formula Deviance regulation theory
Deviance regulation theory Nco authority
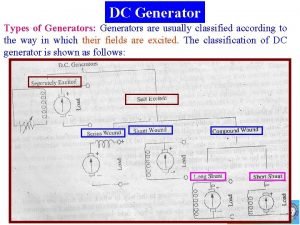
Nco authority Compound generator diagram
Compound generator diagram Cimah regulation
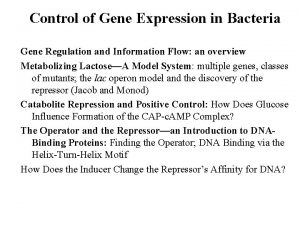
Cimah regulation Regulation of gene expression in bacteria
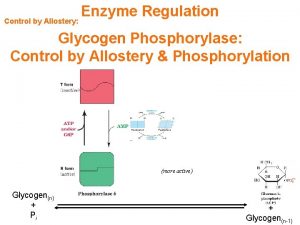
Regulation of gene expression in bacteria Glycogen regulation
Glycogen regulation Collision regulation
Collision regulation Indonesia elv regulation
Indonesia elv regulation Regulation of recruitment and placement activities
Regulation of recruitment and placement activities In a linear regulator the control transistor is conducting
In a linear regulator the control transistor is conducting