Kmer Finder Ole Lund Center for Genomic Epidemiology











- Slides: 19

Kmer. Finder Ole Lund Center for Genomic Epidemiology

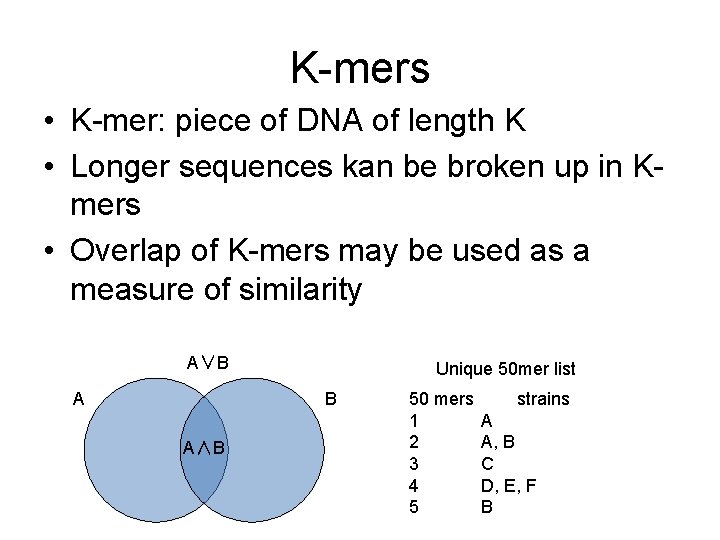
K-mers • K-mer: piece of DNA of length K • Longer sequences kan be broken up in Kmers • Overlap of K-mers may be used as a measure of similarity A∨B A Unique 50 mer list B A∧B 50 mers 1 2 3 4 5 strains A A, B C D, E, F B

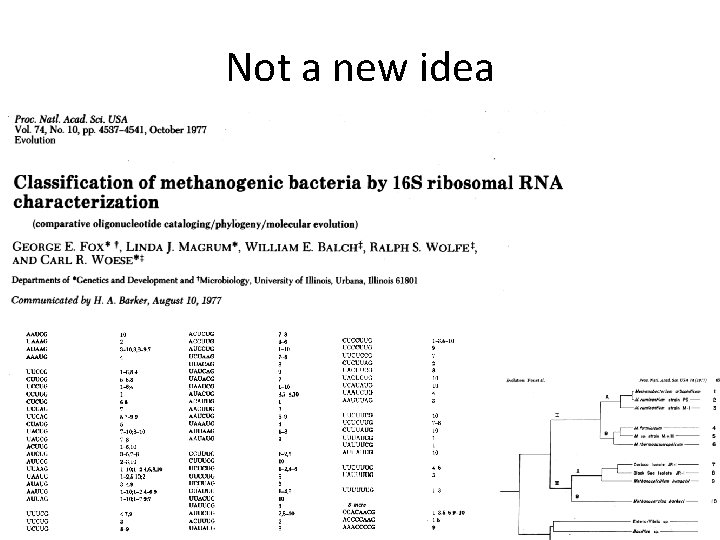
Not a new idea

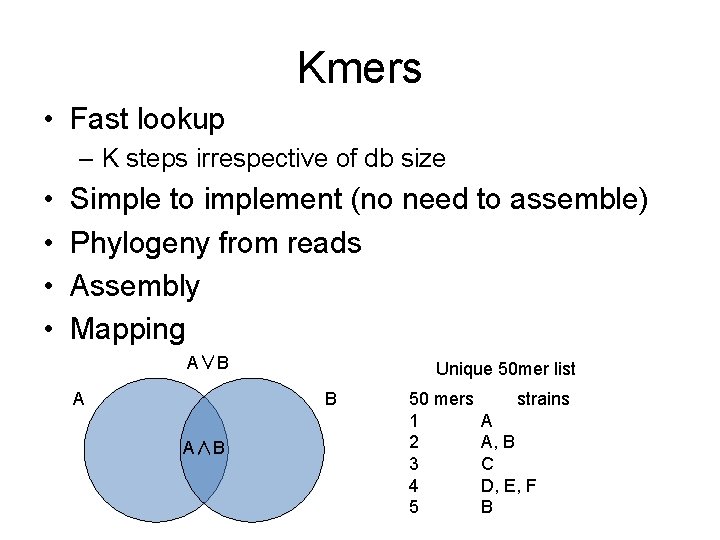
Kmers • Fast lookup – K steps irrespective of db size • • Simple to implement (no need to assemble) Phylogeny from reads Assembly Mapping A∨B A Unique 50 mer list B A∧B 50 mers 1 2 3 4 5 strains A A, B C D, E, F B

Genome hash hack Genome: >AP 011135 Acetobacter pasteurianus IFO 3283 -07 ACTGCAGGCGTCGAGTTCATTGAAGAGAAAGGTGGTGGGGCTGGAGTTAGGTTGAGGAAA TGAATTGTGTAAAAGTCGATCCCAATAAAAGTTTCACTGCCCCTACCGGCTTTCCGGTAT GCCCATCACCCGCGCACGGAGGGCCTCAAACCGTGCAATCTGGGCAGGGCTTGCG Python: myhashtable = {} Myhashtable["ACTGCAGGCGTCGAGT"] = "Acetobacter pasteurianus” myhashtable["CTGCAGGCGTCGAGTT"] = "Acetobacter pasteurianus” print myhashtable["CTGCAGGCGTCGAGTT"] >Acetobacter pasteurianus Perl: $myhashtable = {}; $myhashtable{'ACTGCAGGCGTCGAGT'} = 'Acetobacter pasteurianus'; $myhashtable{'CTGCAGGCGTCGAGTT'} = 'Acetobacter pasteurianus'; print $myhashtable{'ACTGCAGGCGTCGAGT'}, "n” >Acetobacter pasteurianus

Limitation • The computer may not have memory to store all the kmers

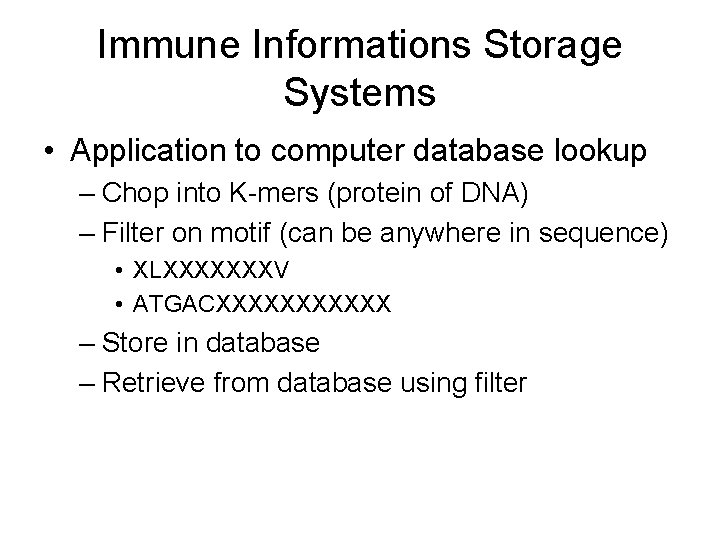
Immune Informations Storage Systems • Store information about encountered pathogens in database • Look up new infections in database of previously encountered infections

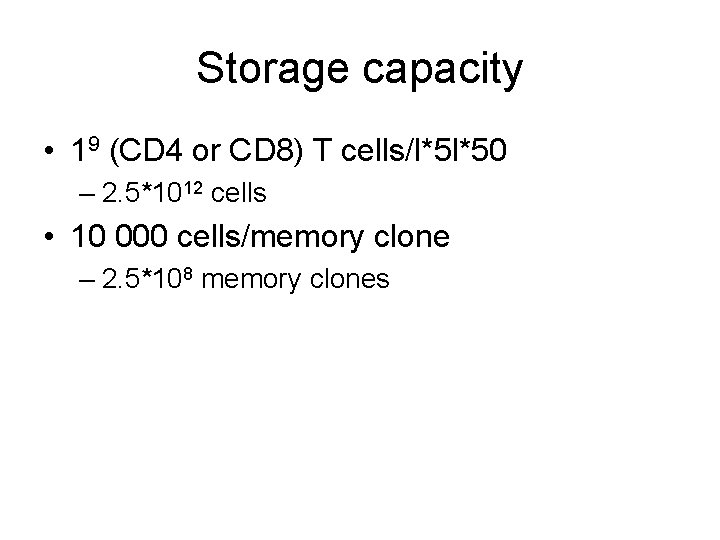
Storage capacity • 19 (CD 4 or CD 8) T cells/l*5 l*50 – 2. 5*1012 cells • 10 000 cells/memory clone – 2. 5*108 memory clones

Storage need – Pathogens/ peptides • 200 bacteria *2000 proteins *400 aa • 1. 6*108 residues/peptide starting positions • + variants + viruses, parasites etc. .

Down sampling • A two amino acid motif – XLXXXXXXXV – 1: 400 peptides bind (assuming all aa are …) – Reduce needed storage capacity by a factor 400! – Better to decide before what 1/400 to store so storing and retrieving filters can be matched rather than randomly choosing – NB: The immune system do not store the peptide but a “TCR imprint” of it

Immune Informations Storage Systems • Application to computer database lookup – Chop into K-mers (protein of DNA) – Filter on motif (can be anywhere in sequence) • XLXXXXXXXV • ATGACXXXXXX – Store in database – Retrieve from database using filter

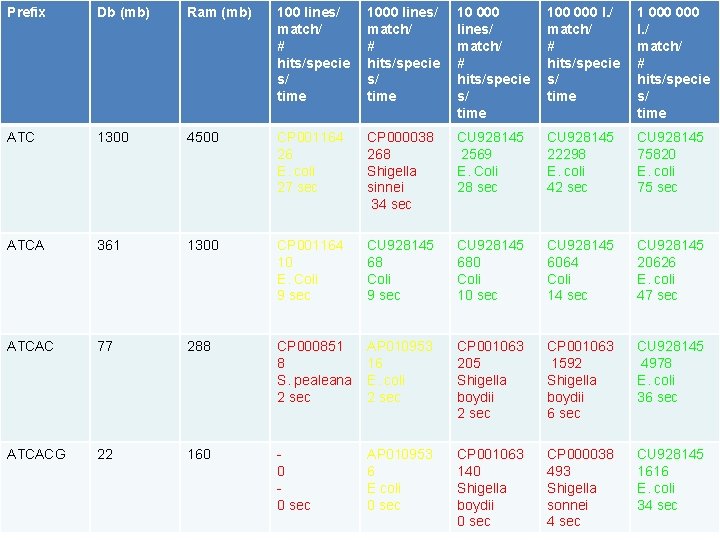
Immune inspired database • Store 16 mers of DNA from 5. 8 Gb DNA from 1647 bacterial genomes • Filter on prefix ATG, ATGA, etc. • Search with 100 -1 000 lines of fastq files (~2 500 – 25 000) nucleotides • Report best match

Prefix Db (mb) Ram (mb) 100 lines/ match/ # hits/specie s/ time 10 000 lines/ match/ # hits/specie s/ time 100 000 l. / match/ # hits/specie s/ time 1 000 l. / match/ # hits/specie s/ time ATC 1300 4500 CP 001164 26 E. coli 27 sec CP 000038 268 Shigella sinnei 34 sec CU 928145 2569 E. Coli 28 sec CU 928145 22298 E. coli 42 sec CU 928145 75820 E. coli 75 sec ATCA 361 1300 CP 001164 10 E. Coli 9 sec CU 928145 680 Coli 10 sec CU 928145 6064 Coli 14 sec CU 928145 20626 E. coli 47 sec ATCAC 77 288 CP 000851 8 S. pealeana 2 sec AP 010953 16 E. coli 2 sec CP 001063 205 Shigella boydii 2 sec CP 001063 1592 Shigella boydii 6 sec CU 928145 4978 E. coli 36 sec ATCACG 22 160 0 0 sec AP 010953 6 E coli 0 sec CP 001063 140 Shigella boydii 0 sec CP 000038 493 Shigella sonnei 4 sec CU 928145 1616 E. coli 34 sec

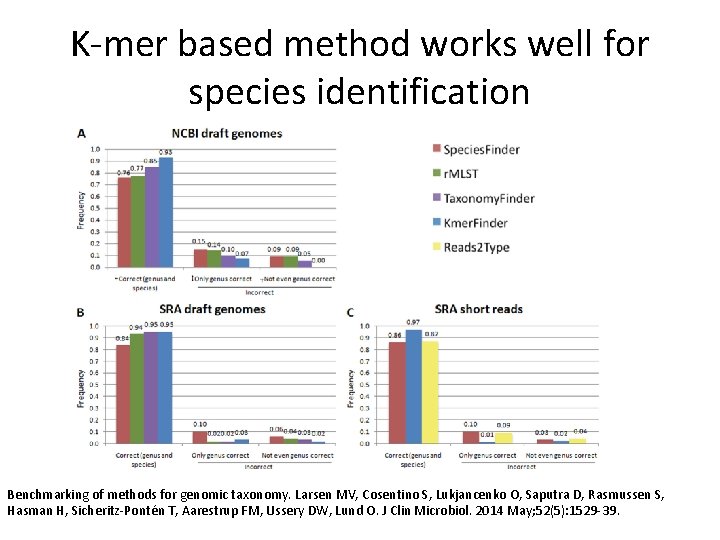
K-mer based method works well for species identification Benchmarking of methods for genomic taxonomy. Larsen MV, Cosentino S, Lukjancenko O, Saputra D, Rasmussen S, Hasman H, Sicheritz-Pontén T, Aarestrup FM, Ussery DW, Lund O. J Clin Microbiol. 2014 May; 52(5): 1529 -39.

Limitations for metagenomic use • Close homologues to best hit also score high • This is not unusual – the same happens when you run blast • Different solutions – Kmer can be shared between hits • Baysian statistics – Unique regions • Like specific pcr primers – The winner takes it all • A kmer belong to the best scoring hit that contains it

kmerdb https: //bitbucket. org/genomicepidemiology/kmerdb

Perfect hash • A hash with no clashes • A, T, C, G is assigned to 0, 1 , 2, 3 • The value of a 3 -mer GAC may be calculated as • 3*43+0*42+2*40 • Presence/absence of all 416 = 4. 3*109 sixteen mers may be stored in a 1/2 G Byte bit vector

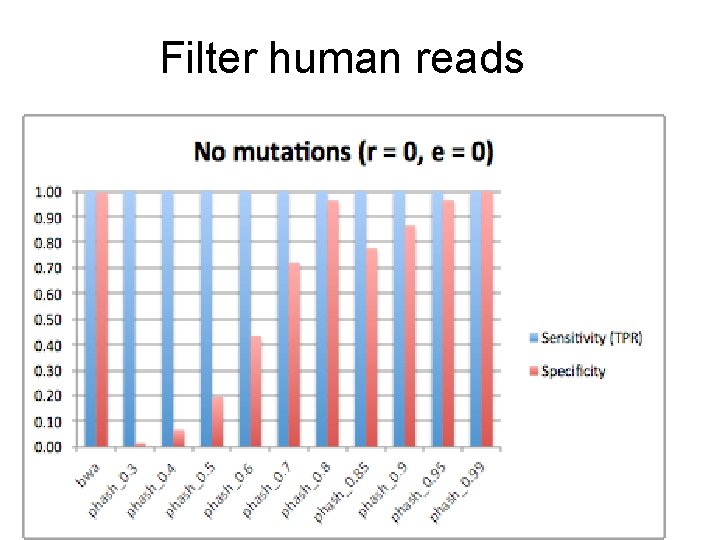
Filter human reads

 Kmer finder
Kmer finder Kmer analysis
Kmer analysis Genomic signal processing
Genomic signal processing Genomic equivalence definition
Genomic equivalence definition Genomic instability
Genomic instability Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic england
Genomic england Genomic
Genomic Genomic equivalence definition
Genomic equivalence definition Genomic england
Genomic england Genomic imprinting definition
Genomic imprinting definition Anneke seller
Anneke seller Formula for attack rate
Formula for attack rate How dr. wafaa elsadr epidemiology professor
How dr. wafaa elsadr epidemiology professor Distribution in epidemiology
Distribution in epidemiology Defination of epidemiology
Defination of epidemiology Spurious association
Spurious association Difference between descriptive and analytical epidemiology
Difference between descriptive and analytical epidemiology John snow epidemiology
John snow epidemiology Define epidemiology
Define epidemiology