Introduction To Next Generation Sequencing NGS Data Analysis







- Slides: 39

Introduction To Next Generation Sequencing (NGS) Data Analysis Jenny Wu UCI Genomics High Throughput Facility

Outline • Goals : Practical guide to NGS data processing • Bioinformatics in NGS data analysis – Basics: terminology, data file formats, general workflow – Data Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • Example: RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

Why Next Generation Sequencing One can sequence hundreds of millions of short sequences (35 bp‐ 120 bp) in a single run in a short period of time with low per base cost. • Illumina/Solexa GA II / Hi. Seq 2000, 2500 • Life Technologies/Applied Biosystems SOLi. D • Roche/454 FLX, Titanium Reviews: Michael Metzker (2010) Nature Reviews Genetics 11: 31 Quail et al (2012) BMC Genomics Jul 24; 13: 341.

Why Bioinformatics Informatics (wall. hms. harvard. edu)

Bioinformatics Challenges in NGS Data Analysis • VERY large text files (tens of millions of lines long) – Can’t do ‘business as usual’ with familiar tools – Impossible memory usage and execution time – Manage, analyze, store, transfer and archive huge files • Need for powerful computers and expertise – Informatics groups must manage compute clusters – New algorithms and software required and often time they are open source Unix/Linux based. – Collaboration of IT, bioinformaticians and biologists

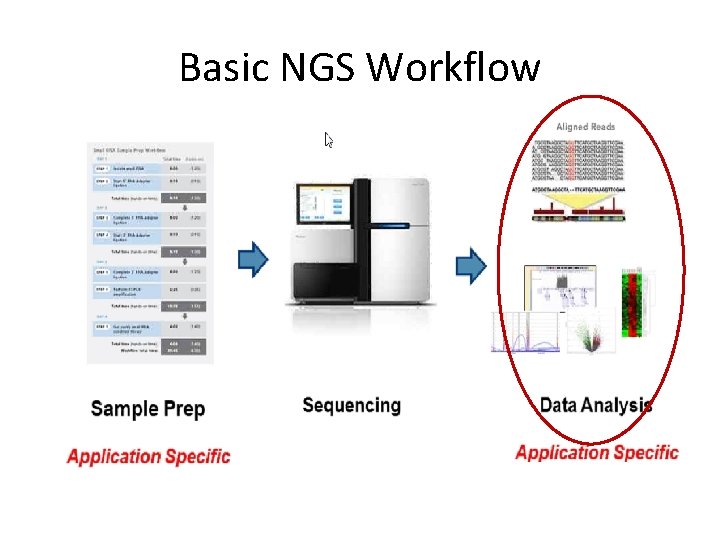
Basic NGS Workflow

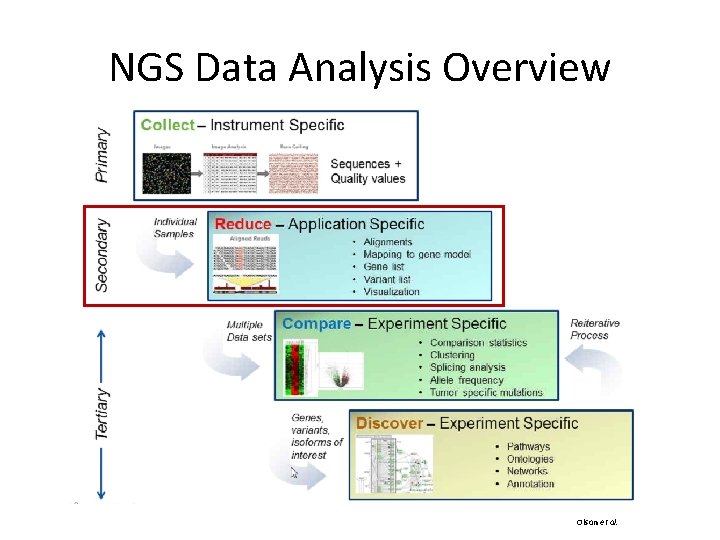
NGS Data Analysis Overview Olson et al.

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

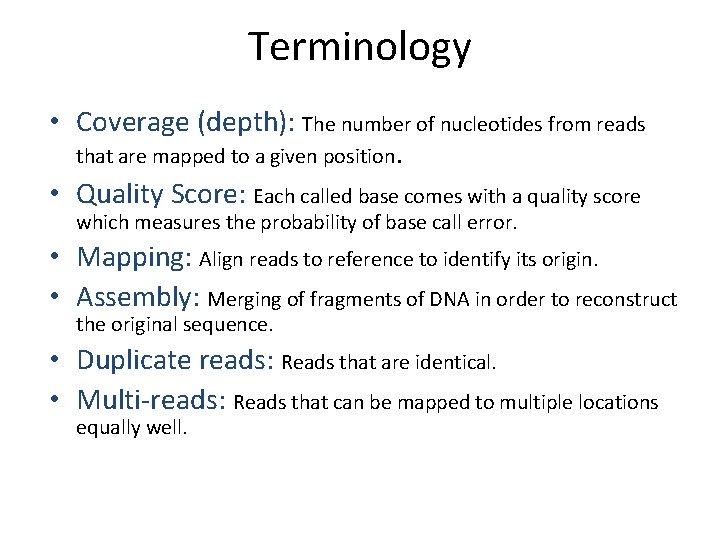
Terminology • Coverage (depth): The number of nucleotides from reads that are mapped to a given position. • Quality Score: Each called base comes with a quality score which measures the probability of base call error. • Mapping: Align reads to reference to identify its origin. • Assembly: Merging of fragments of DNA in order to reconstruct the original sequence. • Duplicate reads: Reads that are identical. • Multi‐reads: Reads that can be mapped to multiple locations equally well.

What does the data look like? Common NGS Data Formats

FASTA Format (Reference Seq)

FASTQ Format (reads)

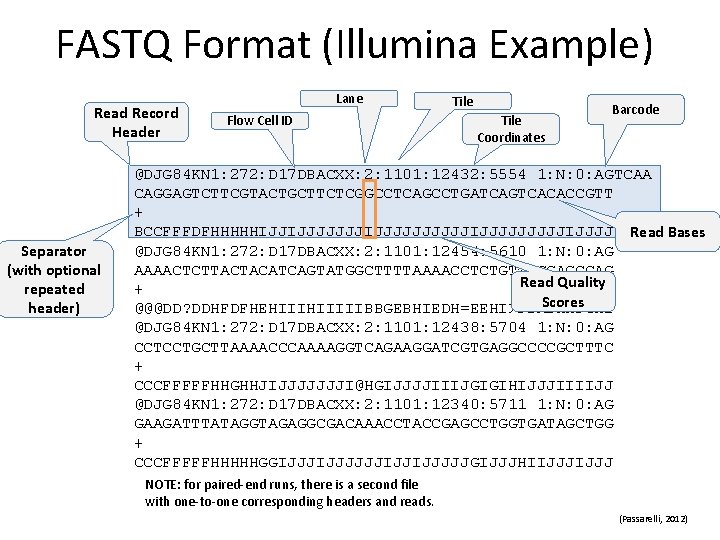
FASTQ Format (Illumina Example) Read Record Header Separator (with optional repeated header) Lane Flow Cell ID Tile Coordinates Barcode @DJG 84 KN 1: 272: D 17 DBACXX: 2: 1101: 12432: 5554 1: N: 0: AGTCAA CAGGAGTCTTCGTACTGCTTCTCGGCCTCAGCCTGATCAGTCACACCGTT + BCCFFFDFHHHHHIJJIJJJJJJJJJJIJJJJ Read Bases @DJG 84 KN 1: 272: D 17 DBACXX: 2: 1101: 12454: 5610 1: N: 0: AG AAAACTCTTACTACATCAGTATGGCTTTTAAAACCTCTGTTTGGAGCCAG Read Quality + Scores @@@DD? DDHFDFHEHIIIIIBBGEBHIEDH=EEHI>FDABHHFGH 2 @DJG 84 KN 1: 272: D 17 DBACXX: 2: 1101: 12438: 5704 1: N: 0: AG CCTCCTGCTTAAAACCCAAAAGGTCAGAAGGATCGTGAGGCCCCGCTTTC + CCCFFFFFHHGHHJIJJJJJJJI@HGIJJJJIIIJGIGIHIJJJIIIIJJ @DJG 84 KN 1: 272: D 17 DBACXX: 2: 1101: 12340: 5711 1: N: 0: AG GAAGATTTATAGGTAGAGGCGACAAACCTACCGAGCCTGGTGATAGCTGG + CCCFFFFFHHHHHGGIJJJJJJIJJIJJJJJGIJJJHIIJJJ NOTE: for paired‐end runs, there is a second file with one‐to‐one corresponding headers and reads. (Passarelli, 2012)

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow, – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

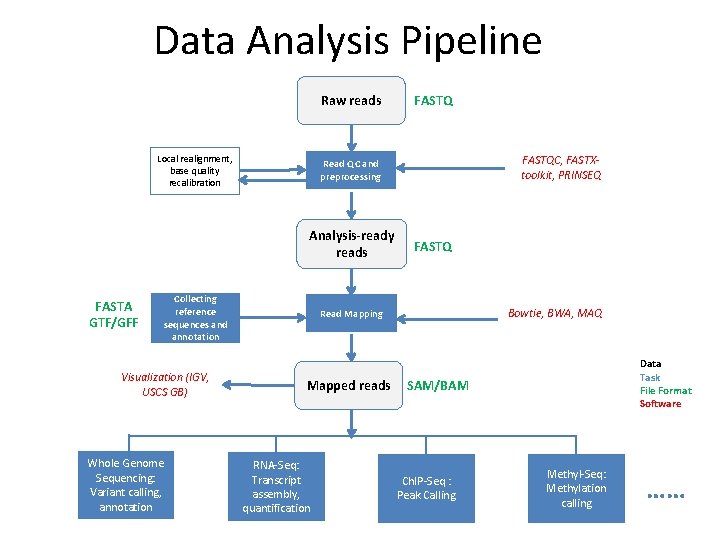
Data Analysis Pipeline Raw reads Local realignment, base quality recalibration Collecting reference sequences and annotation Visualization (IGV, USCS GB) Whole Genome Sequencing: Variant calling, annotation FASTQC, FASTXtoolkit, PRINSEQ Read QC and preprocessing Analysis-ready reads FASTA GTF/GFF FASTQ Bowtie, BWA, MAQ Read Mapping Mapped reads RNA‐Seq: Transcript assembly, quantification Data Task File Format Software SAM/BAM Ch. IP‐Seq : Peak Calling Methyl‐Seq: Methylation calling ……

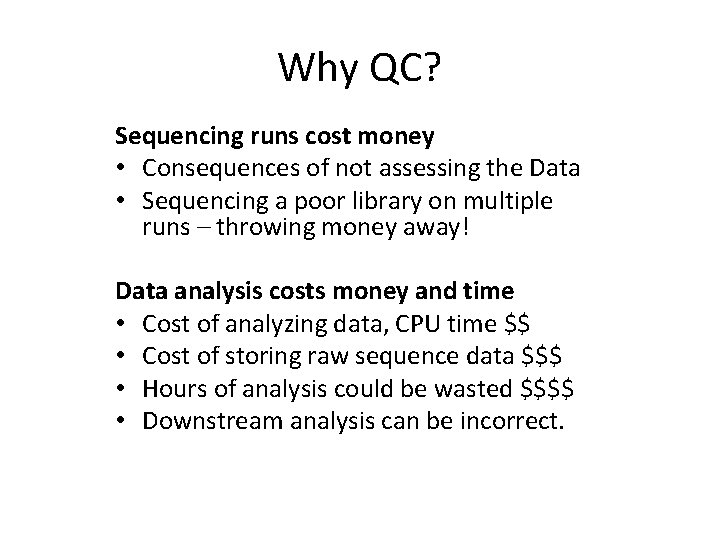
Why QC? Sequencing runs cost money • Consequences of not assessing the Data • Sequencing a poor library on multiple runs – throwing money away! Data analysis costs money and time • Cost of analyzing data, CPU time $$ • Cost of storing raw sequence data $$$ • Hours of analysis could be wasted $$$$ • Downstream analysis can be incorrect.

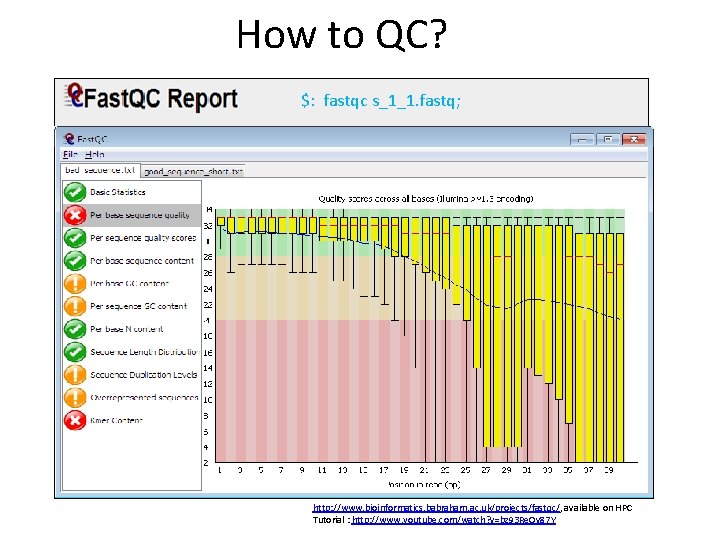
How to QC? $: fastqc s_1_1. fastq; http: //www. bioinformatics. babraham. ac. uk/projects/fastqc/, available on HPC Tutorial : http: //www. youtube. com/watch? v=bz 93 Re. Ov 87 Y

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow, – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

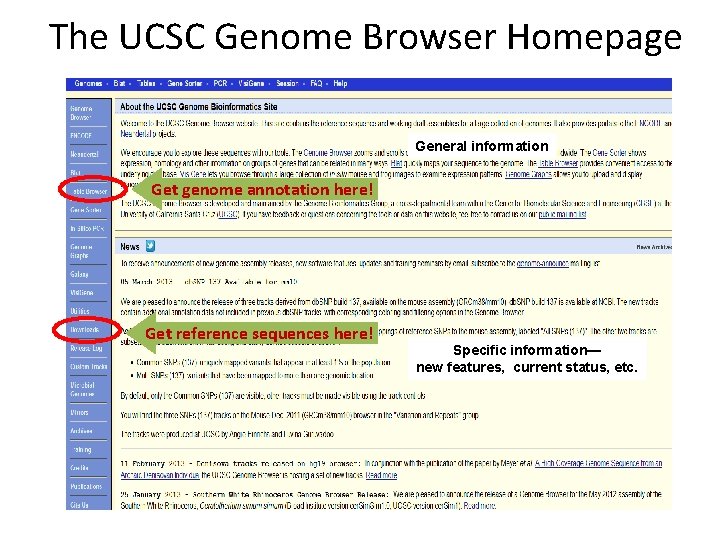
The UCSC Genome Browser Homepage General information Get genome annotation here! Get reference sequences here! Specific information— new features, current status, etc.

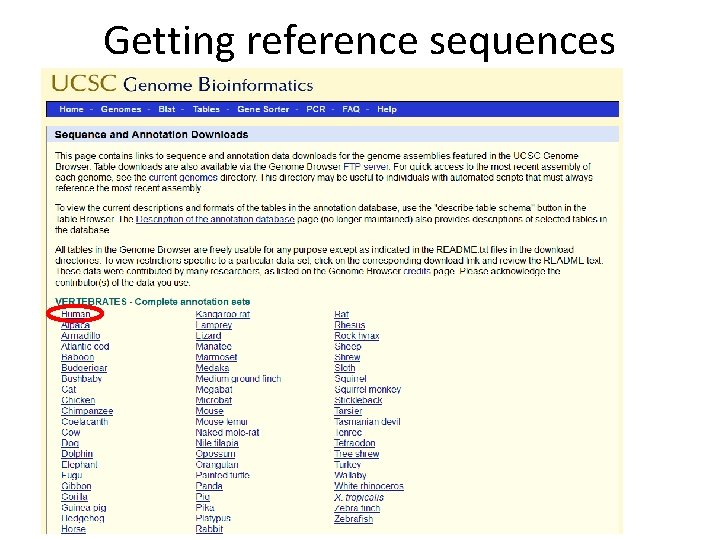
Getting reference sequences

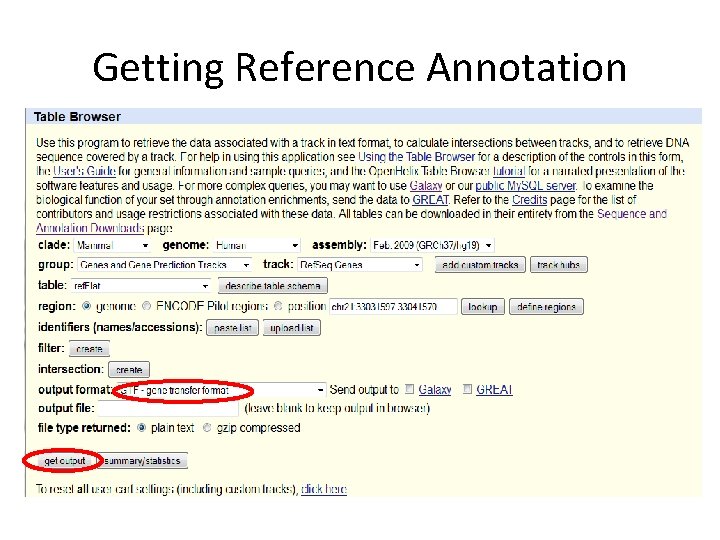
Getting Reference Annotation

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow, – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

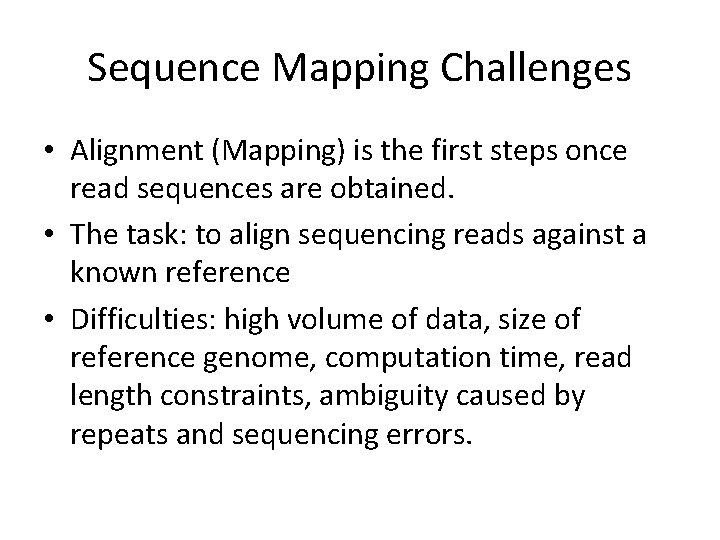
Sequence Mapping Challenges • Alignment (Mapping) is the first steps once read sequences are obtained. • The task: to align sequencing reads against a known reference • Difficulties: high volume of data, size of reference genome, computation time, read length constraints, ambiguity caused by repeats and sequencing errors.

Short Read Alignment Olson et al.

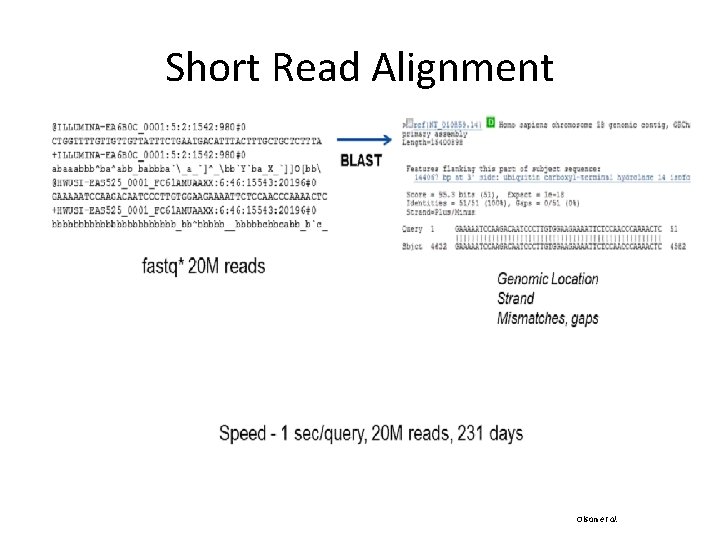
Short Read Alignment Software

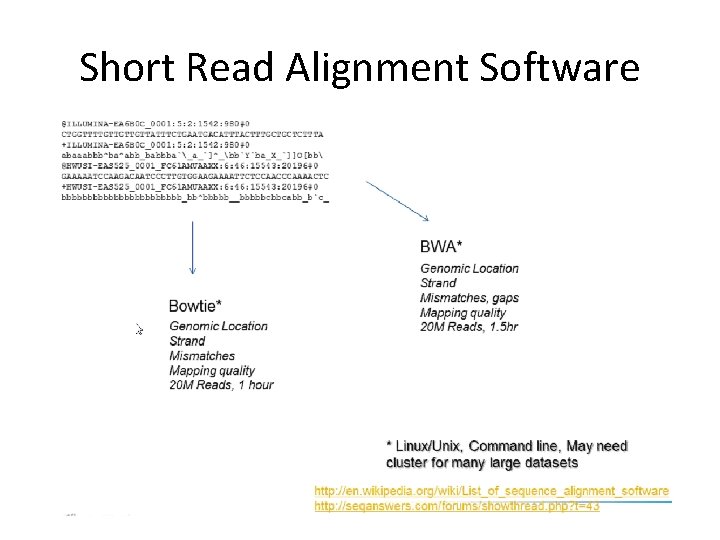
Short Reads Mapping Software

How to choose an aligner? • There are many aligners and they vary a lot in performance (accuracy, memory usage, speed, etc). • Factors to consider : application, platform, read length, downstream analysis, etc. • Constant trade off between speed and sensitivity (e. g. MAQ vs. Bowtie) • Guaranteed high accuracy will take longer.

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow, – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

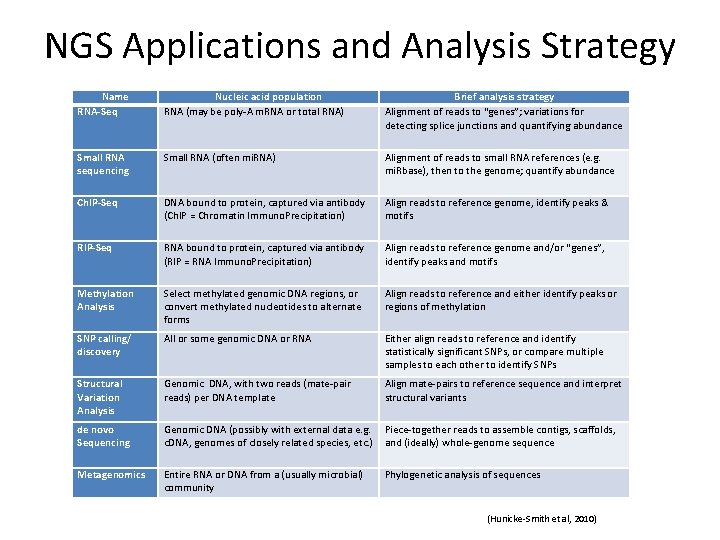
NGS Applications and Analysis Strategy Name RNA-Seq Nucleic acid population RNA (may be poly‐A m. RNA or total RNA) Brief analysis strategy Alignment of reads to “genes”; variations for detecting splice junctions and quantifying abundance Small RNA sequencing Small RNA (often mi. RNA) Alignment of reads to small RNA references (e. g. mi. Rbase), then to the genome; quantify abundance Ch. IP-Seq DNA bound to protein, captured via antibody (Ch. IP = Chromatin Immuno. Precipitation) Align reads to reference genome, identify peaks & motifs RIP-Seq RNA bound to protein, captured via antibody (RIP = RNA Immuno. Precipitation) Align reads to reference genome and/or “genes”, identify peaks and motifs Methylation Analysis Select methylated genomic DNA regions, or convert methylated nucleotides to alternate forms Align reads to reference and either identify peaks or regions of methylation SNP calling/ discovery All or some genomic DNA or RNA Either align reads to reference and identify statistically significant SNPs, or compare multiple samples to each other to identify SNPs Structural Variation Analysis Genomic DNA, with two reads (mate‐pair reads) per DNA template Align mate‐pairs to reference sequence and interpret structural variants de novo Sequencing Genomic DNA (possibly with external data e. g. c. DNA, genomes of closely related species, etc. ) Piece‐together reads to assemble contigs, scaffolds, and (ideally) whole‐genome sequence Metagenomics Entire RNA or DNA from a (usually microbial) community Phylogenetic analysis of sequences (Hunicke‐Smith et al, 2010)

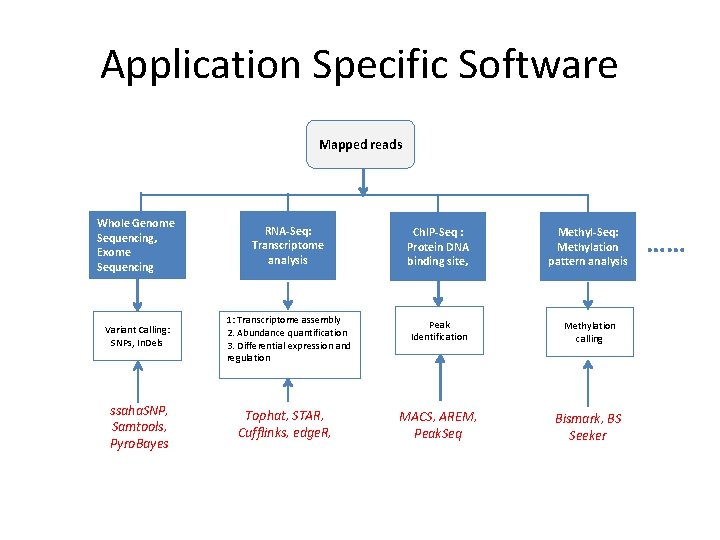
Application Specific Software Mapped reads Whole Genome Sequencing, Exome Sequencing RNA-Seq: Transcriptome analysis Ch. IP-Seq : Protein DNA binding site, Methyl-Seq: Methylation pattern analysis Variant Calling: SNPs, In. Dels 1: Transcriptome assembly 2. Abundance quantification 3. Differential expression and regulation Peak Identification Methylation calling MACS, AREM, Peak. Seq Bismark, BS Seeker ssaha. SNP, Samtools, Pyro. Bayes Tophat, STAR, Cufflinks, edge. R, ……

Outline • Goals • Bioinformatics Challenges in NGS data analysis – Basics: terminology, data file formats, general workflow, – Analysis Pipeline • • Sequence QC and preprocessing Obtaining and preparing reference Sequence mapping Downstream analysis workflow and software • RNA‐Seq analysis with Tuxedo protocol • Summary and future plan

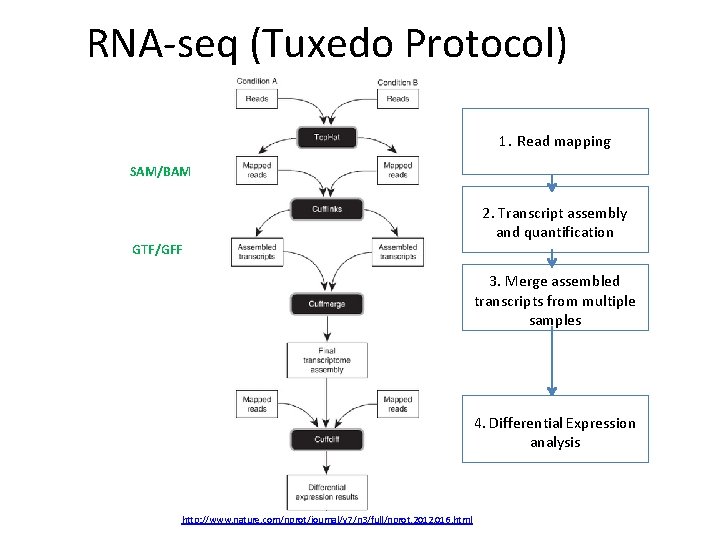
RNA‐seq (Tuxedo Protocol) 1. Read mapping SAM/BAM GTF/GFF 2. Transcript assembly and quantification 3. Merge assembled transcripts from multiple samples 4. Differential Expression analysis http: //www. nature. com/nprot/journal/v 7/n 3/full/nprot. 2012. 016. html

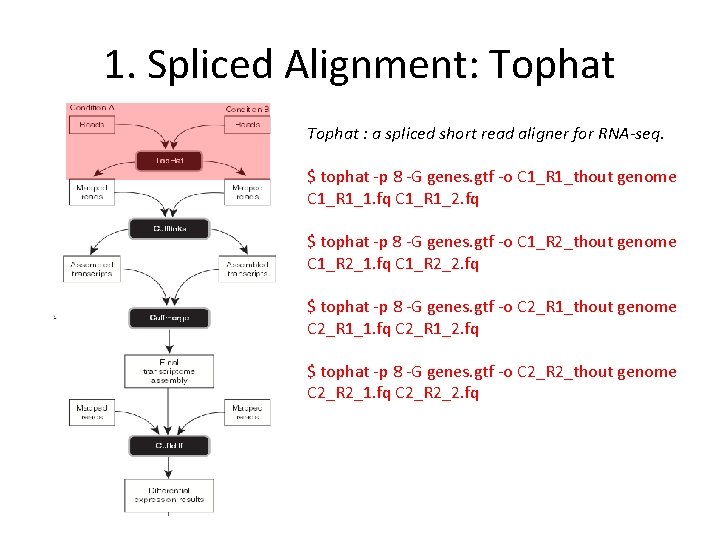
1. Spliced Alignment: Tophat : a spliced short read aligner for RNA-seq. $ tophat ‐p 8 ‐G genes. gtf ‐o C 1_R 1_thout genome C 1_R 1_1. fq C 1_R 1_2. fq $ tophat ‐p 8 ‐G genes. gtf ‐o C 1_R 2_thout genome C 1_R 2_1. fq C 1_R 2_2. fq $ tophat ‐p 8 ‐G genes. gtf ‐o C 2_R 1_thout genome C 2_R 1_1. fq C 2_R 1_2. fq $ tophat ‐p 8 ‐G genes. gtf ‐o C 2_R 2_thout genome C 2_R 2_1. fq C 2_R 2_2. fq

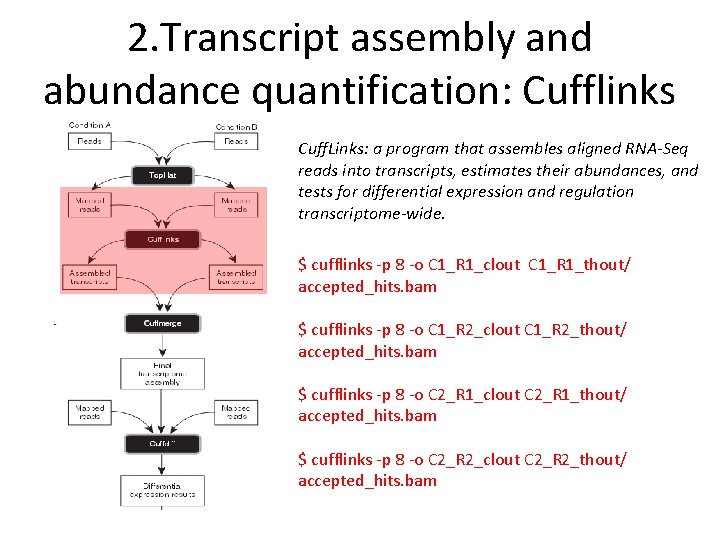
2. Transcript assembly and abundance quantification: Cufflinks Cuff. Links: a program that assembles aligned RNA-Seq reads into transcripts, estimates their abundances, and tests for differential expression and regulation transcriptome-wide. $ cufflinks ‐p 8 ‐o C 1_R 1_clout C 1_R 1_thout/ accepted_hits. bam $ cufflinks ‐p 8 ‐o C 1_R 2_clout C 1_R 2_thout/ accepted_hits. bam $ cufflinks ‐p 8 ‐o C 2_R 1_clout C 2_R 1_thout/ accepted_hits. bam $ cufflinks ‐p 8 ‐o C 2_R 2_clout C 2_R 2_thout/ accepted_hits. bam

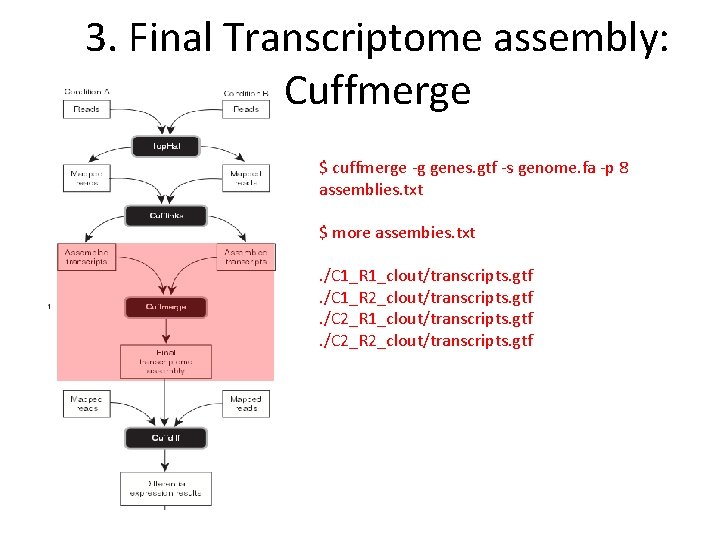
3. Final Transcriptome assembly: Cuffmerge $ cuffmerge ‐g genes. gtf ‐s genome. fa ‐p 8 assemblies. txt $ more assembies. txt. /C 1_R 1_clout/transcripts. gtf. /C 1_R 2_clout/transcripts. gtf. /C 2_R 1_clout/transcripts. gtf. /C 2_R 2_clout/transcripts. gtf

4. Differential Expression: Cuffdiff Cuff. Diff: a program that compares transcript abundance between samples. $ cuffdiff ‐o diff_out ‐b genome. fa ‐p 8 –L C 1, C 2 ‐u merged_asm/merged. gtf. /C 1_R 1_thout/accepted_hits. bam, . /C 1_R 2 _thout/accepted_hits. bam, . /C 2_R 1_thout/accepted_hits. bam, . /C 2_R 2 _thout/accepted_hits. bam

Integrative Genomics Viewer (IGV) http: //www. broadinstitute. org/igv

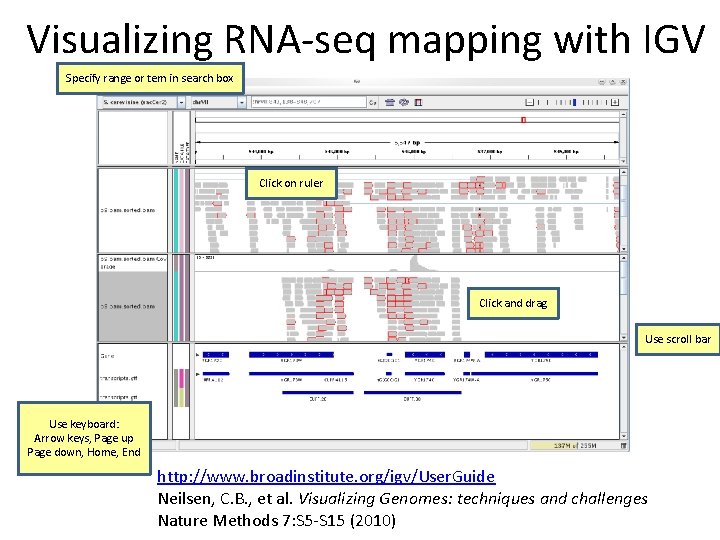
Visualizing RNA‐seq mapping with IGV Specify range or tem in search box Click on ruler Click and drag Use scroll bar Use keyboard: Arrow keys, Page up Page down, Home, End http: //www. broadinstitute. org/igv/User. Guide Neilsen, C. B. , et al. Visualizing Genomes: techniques and challenges Nature Methods 7: S 5‐S 15 (2010)

Summary • NGS technologies are transforming molecular biology. • Bioinformatics analysis is a crucial part in NGS applications – Data formats, terminology, general workflow – Analysis pipeline – Software for various NGS applications • RNA‐seq with Tuxedo suite Thank you!