Introduction to Affymetrix SNP technology Jonathan Pevsner Ph

![Across the genome, there are four possible SNP calls: [1] homozygous (AA) [2] homozygous Across the genome, there are four possible SNP calls: [1] homozygous (AA) [2] homozygous](https://slidetodoc.com/presentation_image_h2/14a290ea1aa71666b9dbcd3d571df049/image-6.jpg)

- Slides: 32

Introduction to Affymetrix SNP technology Jonathan Pevsner, Ph. D. SNP group at the Genome Cafe July 27, 2005

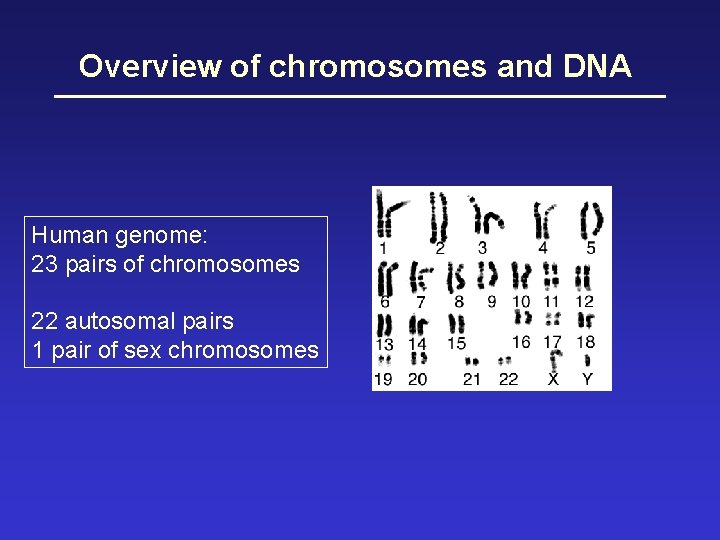
Overview of chromosomes and DNA Human genome: 23 pairs of chromosomes 22 autosomal pairs 1 pair of sex chromosomes

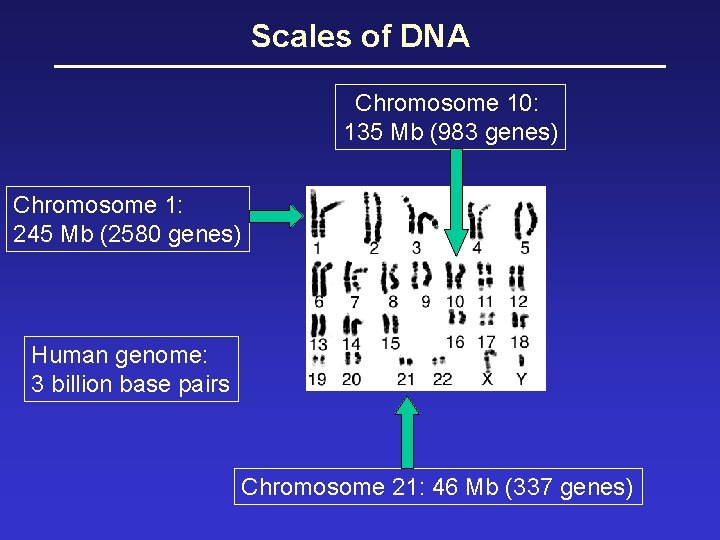
Scales of DNA Chromosome 10: 135 Mb (983 genes) Chromosome 1: 245 Mb (2580 genes) Human genome: 3 billion base pairs Chromosome 21: 46 Mb (337 genes)

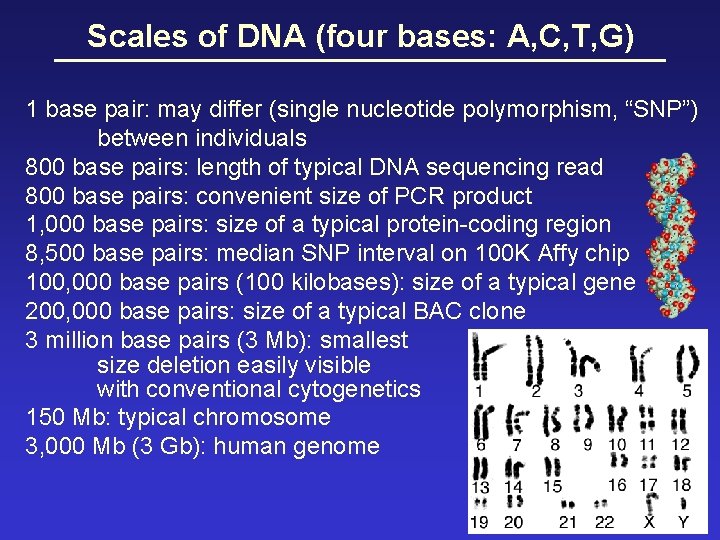
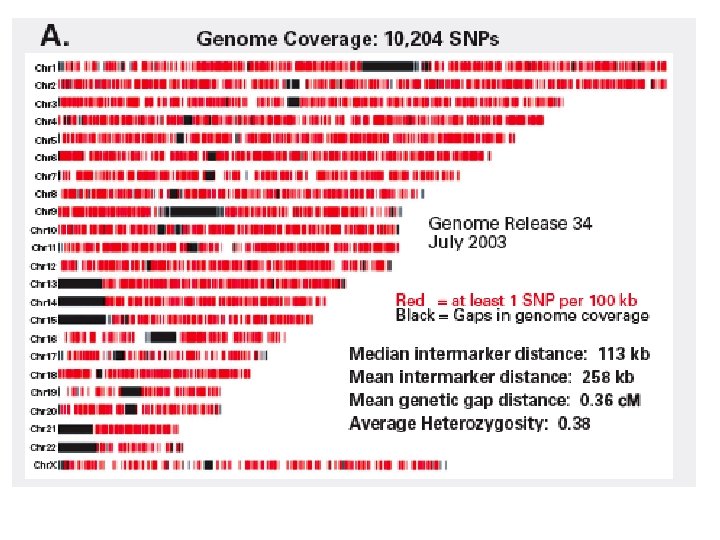
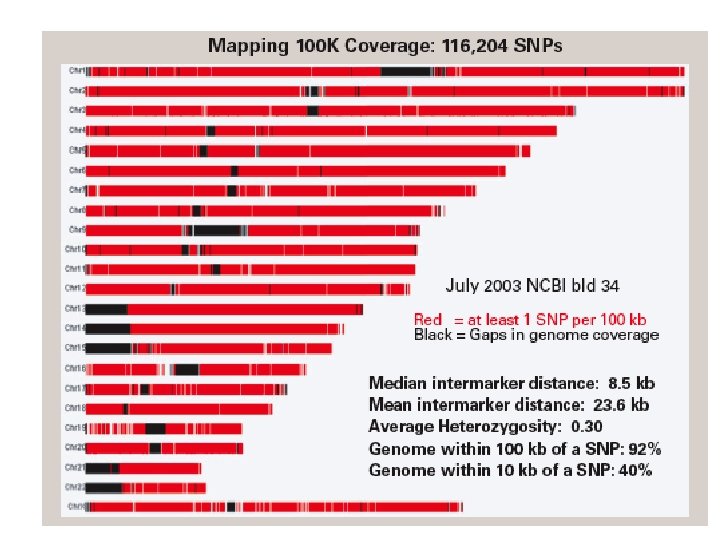
Scales of DNA (four bases: A, C, T, G) 1 base pair: may differ (single nucleotide polymorphism, “SNP”) between individuals 800 base pairs: length of typical DNA sequencing read 800 base pairs: convenient size of PCR product 1, 000 base pairs: size of a typical protein-coding region 8, 500 base pairs: median SNP interval on 100 K Affy chip 100, 000 base pairs (100 kilobases): size of a typical gene 200, 000 base pairs: size of a typical BAC clone 3 million base pairs (3 Mb): smallest size deletion easily visible with conventional cytogenetics 150 Mb: typical chromosome 3, 000 Mb (3 Gb): human genome

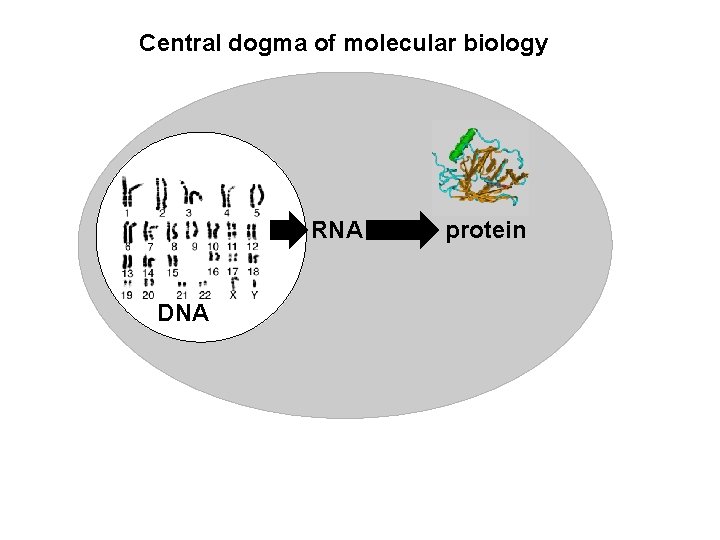
Central dogma of molecular biology RNA DNA protein
![Across the genome there are four possible SNP calls 1 homozygous AA 2 homozygous Across the genome, there are four possible SNP calls: [1] homozygous (AA) [2] homozygous](https://slidetodoc.com/presentation_image_h2/14a290ea1aa71666b9dbcd3d571df049/image-6.jpg)
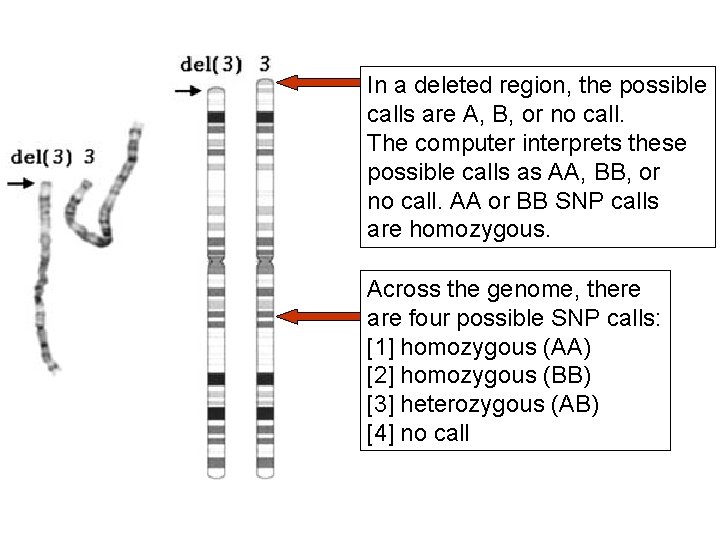
Across the genome, there are four possible SNP calls: [1] homozygous (AA) [2] homozygous (BB) [3] heterozygous (AB) [4] no call

In a deleted region, the possible calls are A, B, or no call. The computer interprets these possible calls as AA, BB, or no call. AA or BB SNP calls are homozygous. Across the genome, there are four possible SNP calls: [1] homozygous (AA) [2] homozygous (BB) [3] heterozygous (AB) [4] no call


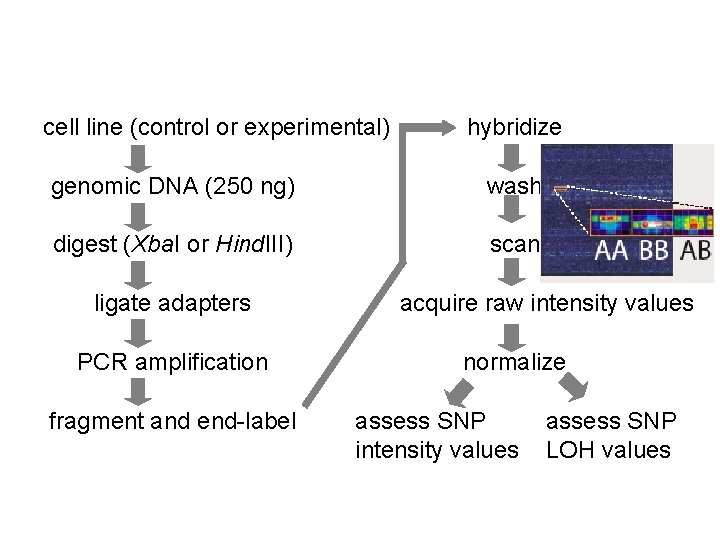
cell line (control or experimental) hybridize genomic DNA (250 ng) wash digest (Xba. I or Hind. III) scan ligate adapters PCR amplification fragment and end-label acquire raw intensity values normalize assess SNP intensity values assess SNP LOH values

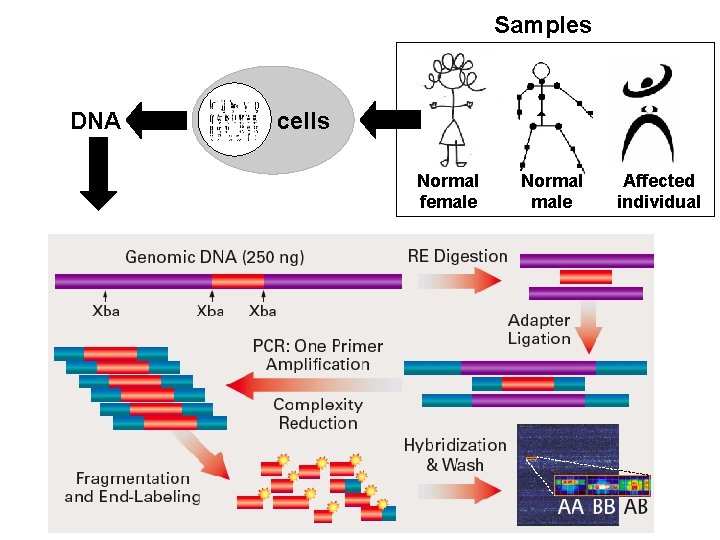
Samples DNA cells Normal female Normal male Affected individual



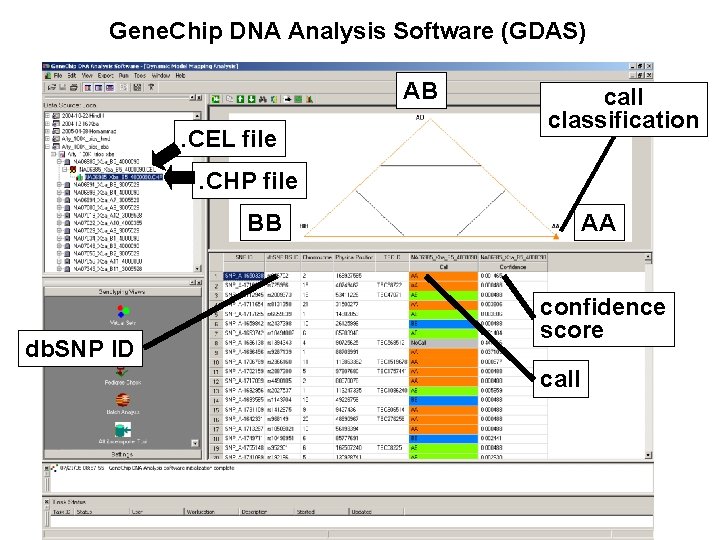
Gene. Chip DNA Analysis Software (GDAS): . CEL file



Gene. Chip DNA Analysis Software (GDAS) AB. CEL file call classification . CHP file BB db. SNP ID AA confidence score call

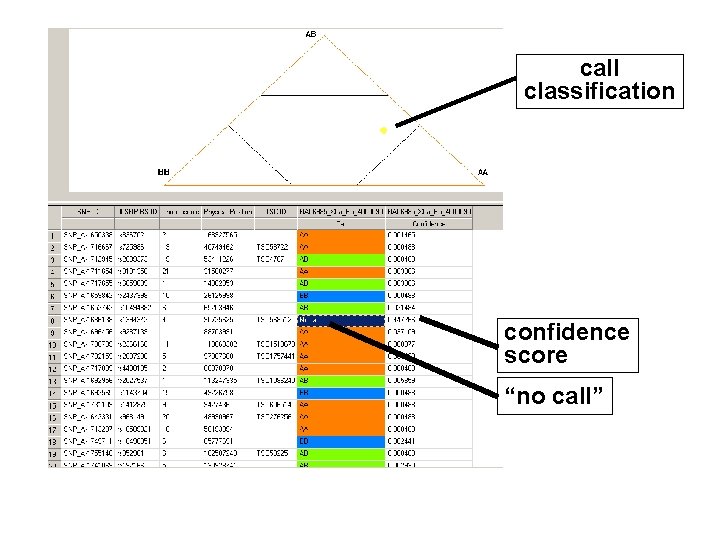
call classification confidence score “no call”


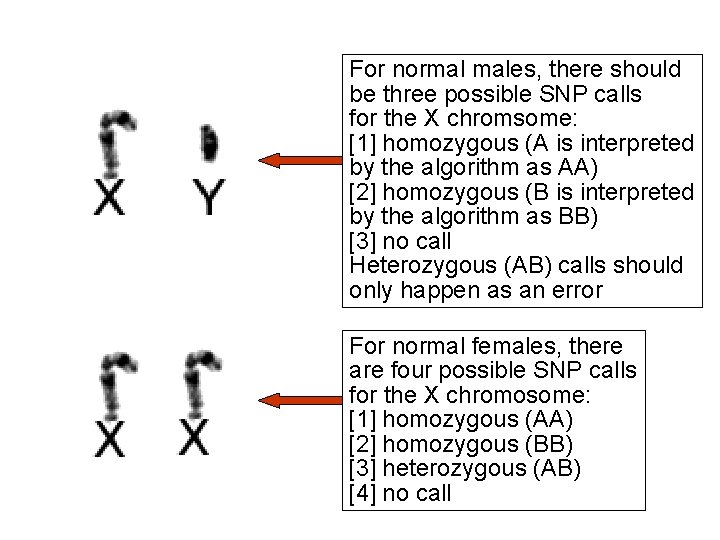
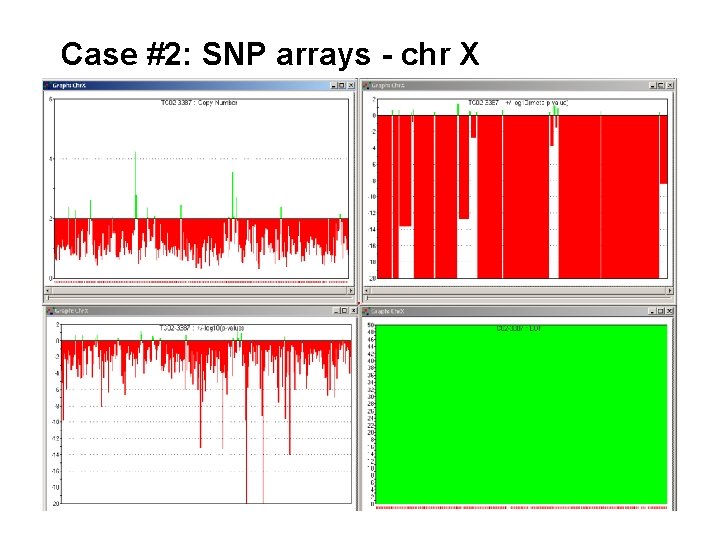
For normal males, there should be three possible SNP calls for the X chromsome: [1] homozygous (A is interpreted by the algorithm as AA) [2] homozygous (B is interpreted by the algorithm as BB) [3] no call Heterozygous (AB) calls should only happen as an error For normal females, there are four possible SNP calls for the X chromosome: [1] homozygous (AA) [2] homozygous (BB) [3] heterozygous (AB) [4] no call

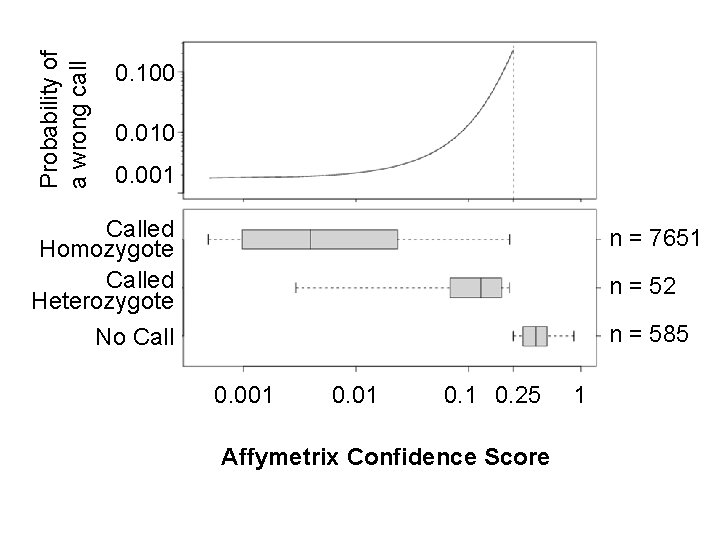
Probability of a wrong call 0. 100 0. 010 0. 001 Called Homozygote Called Heterozygote n = 7651 n = 52 n = 585 No Call 0. 001 0. 25 Affymetrix Confidence Score 1


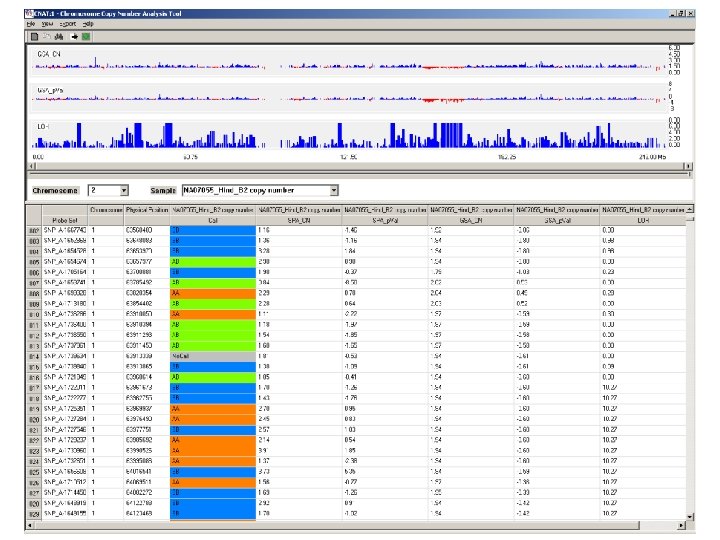
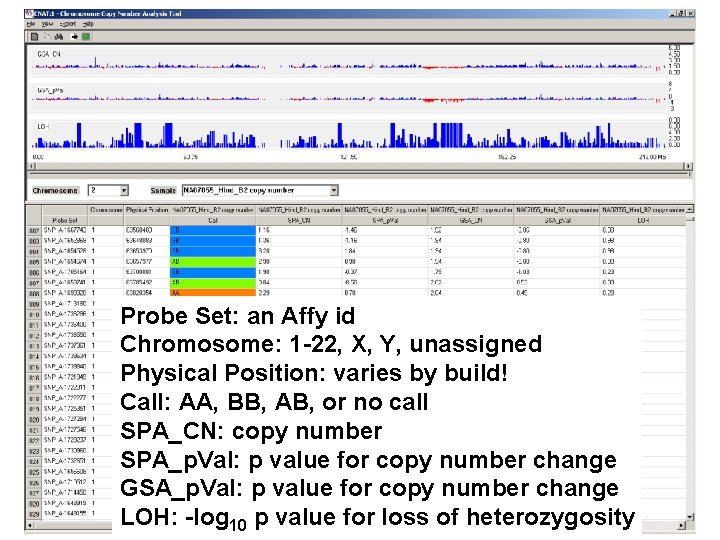
Probe Set: an Affy id Chromosome: 1 -22, X, Y, unassigned Physical Position: varies by build! Call: AA, BB, AB, or no call SPA_CN: copy number SPA_p. Val: p value for copy number change GSA_p. Val: p value for copy number change LOH: -log 10 p value for loss of heterozygosity


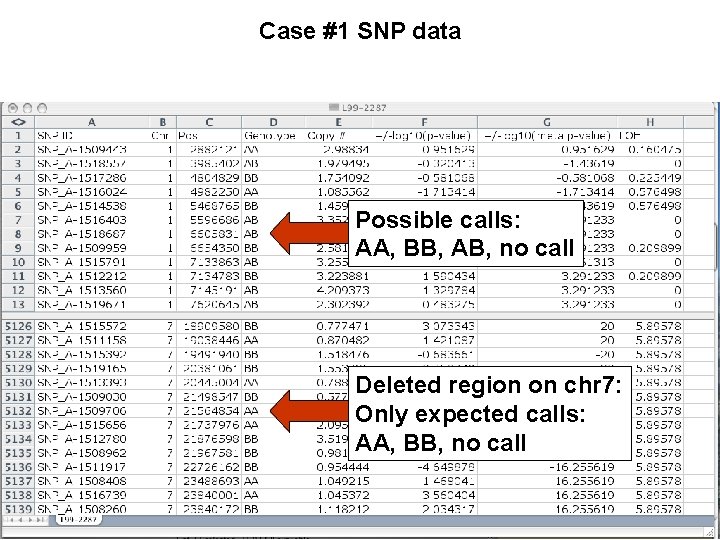
Case #1 SNP data Possible calls: AA, BB, AB, no call Deleted region on chr 7: Only expected calls: AA, BB, no call

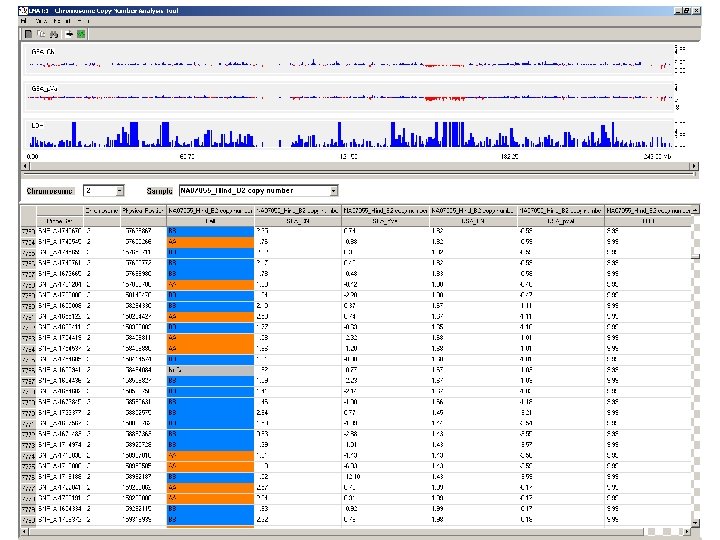
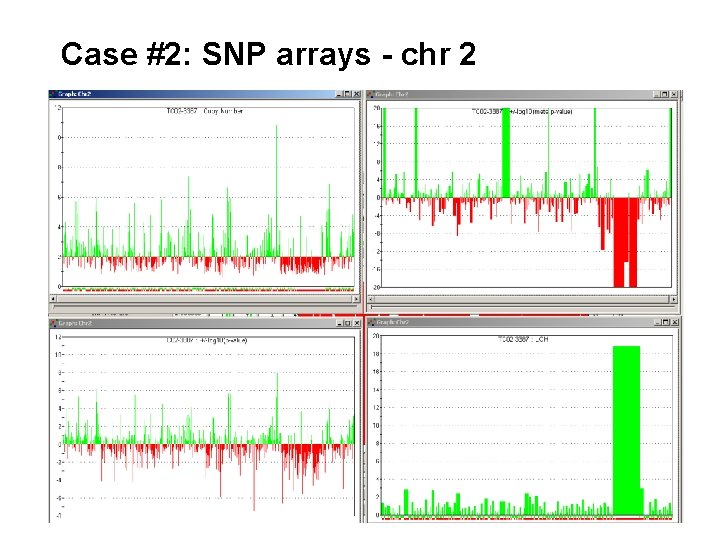
Case #2: SNP arrays - chr 2

Case #2: SNP arrays - chr X

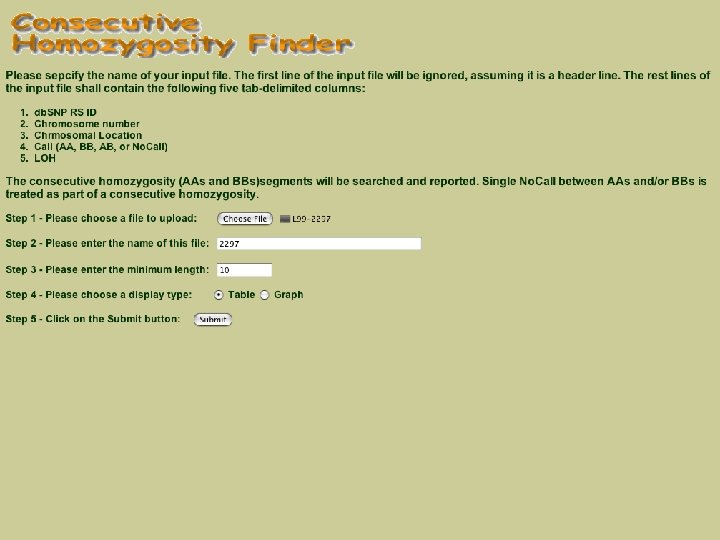
Web site

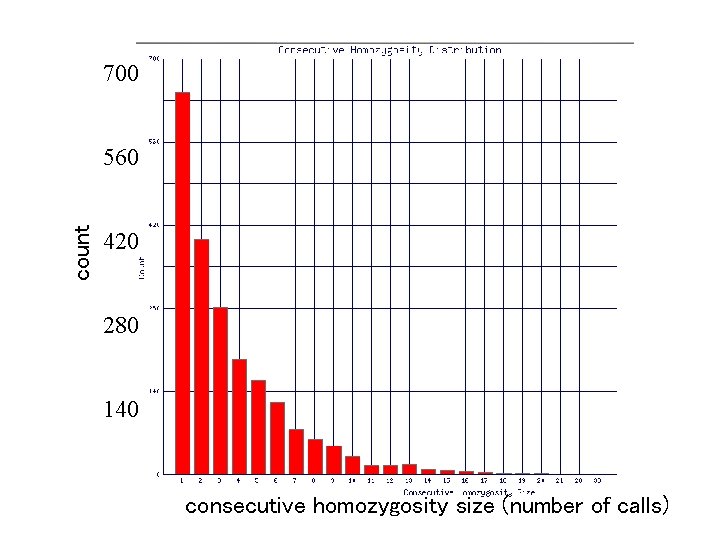
700 count 560 420 280 140 consecutive homozygosity size (number of calls)

Web site

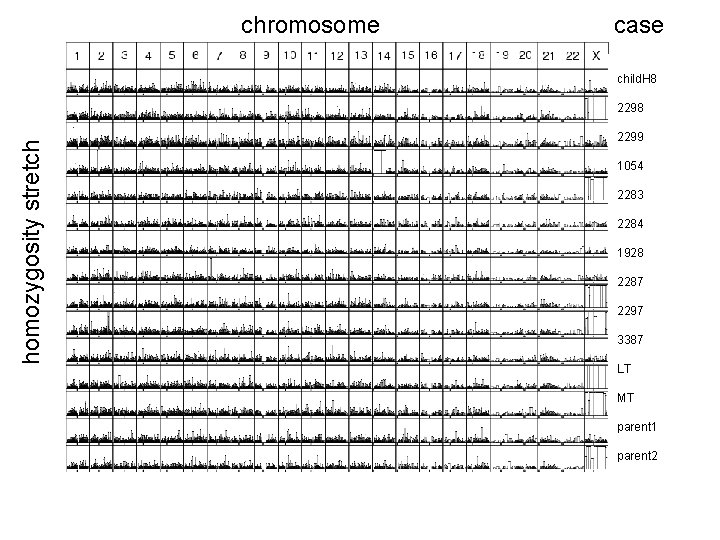
chromosome case child. H 8 homozygosity stretch 2298 2299 1054 2283 2284 1928 2287 2297 3387 LT MT parent 1 parent 2

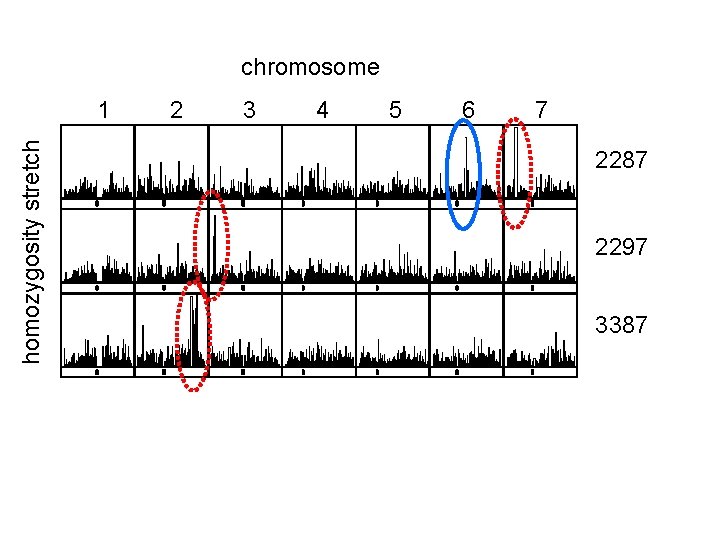
chromosome homozygosity stretch 1 2 3 4 5 6 7 2287 2297 3387

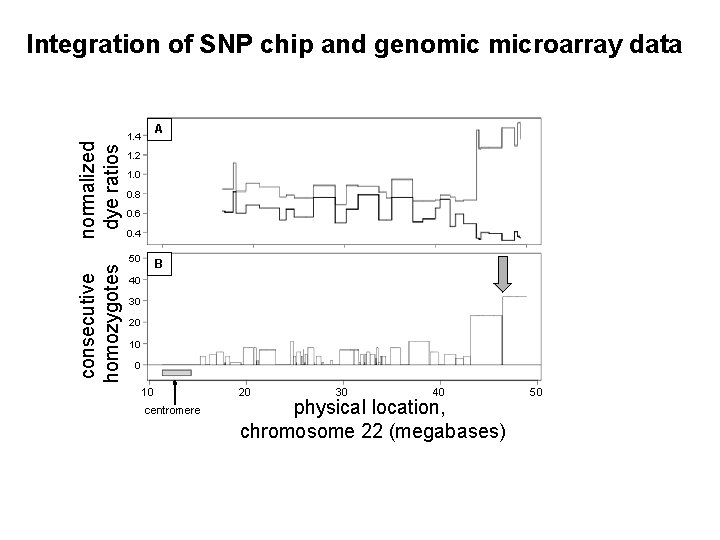
consecutive homozygotes normalized dye ratios Integration of SNP chip and genomic microarray data A 1. 4 1. 2 1. 0 0. 8 0. 6 0. 4 50 B 40 30 20 10 centromere 20 30 40 physical location, chromosome 22 (megabases) 50