in silico 3 D QSAR Quantitative StructureActivity Relationship

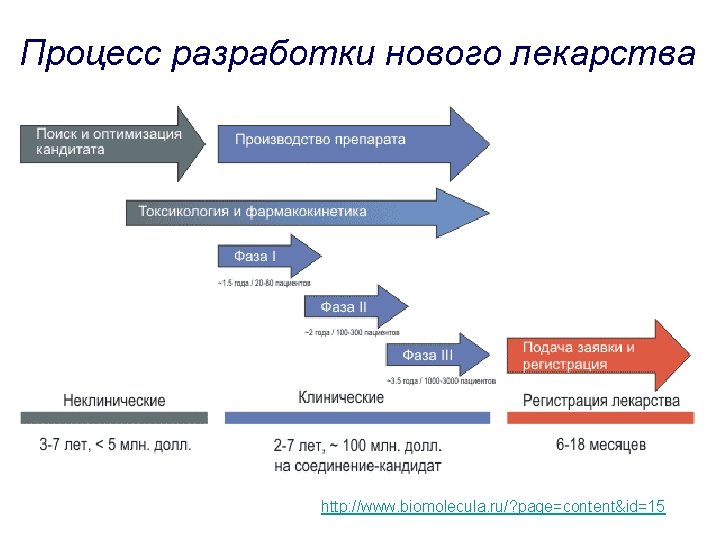






















- Slides: 25















Конструирование ФАВ in silico Если 3 D-структура мишени неизвестна: Данные: структура гомологов мишени Метод: моделирование структуры мишени по гомологии Данные: структура уже известных лигандов Метод: QSAR (Quantitative Structure-Activity Relationship) «Структура определяет свойства» : QSAR represents an attempt to correlate structural or property descriptors of compounds with activities. These physicochemical descriptors, which include parameters to account for


Конструирование ФАВ in silico Если 3 D-структура мишени известна: Данные: структура мишени и лиганда Метод: молекулярный докинг, молекулярная динамика Трудности докинга: “Traditional” atom-atom scoring functions (knowledge-based or empirical): not accurate enough More adequate physical models (e. g. , Quantum Mechanical Polarizable Force Field ): too complicated for direct computation Трудности ММ: очень медленно

Конструирование ФАВ in silico Если 3 D-структура мишени известна: § Search for binding site in known protein 3 D structure § De novo ligand construction from a chemical space § Ligand self-docking: positioning of a known ligand in a binding site of a protein § Virtual screening of a set of known ligands against a target protein binding site § Ligand optimization: search for replacement for a certain chemical group of

Ligand virtual screening: performance evaluation on the HSV -1 thymidine kinase Target: 990 drug-like molecules + 10 real inhibitors screened vs. HSV-1 thymidine kinase (PDB ID 1 kim) Performance evaluation: A number of real inhibitors in a certain top fraction of the energy scores SURFLEX software: 8 true hits in top-50 [Bissantz et al. , J Med Chem 2000; 43: 47594767]

Ligand optimization: problem definition Given (1) known ligand, (2) its position in the binding site (known from X-ray or docking), (3) ligand “side group” (fragment) to be replaced, Find a small number (~10 -100) of replacing side groups aimed at the improvement of (a) binding affinity, or (b) synthesizeability, (c) solubility, (d) side effects, etc. , depending on the type of the optimized feature

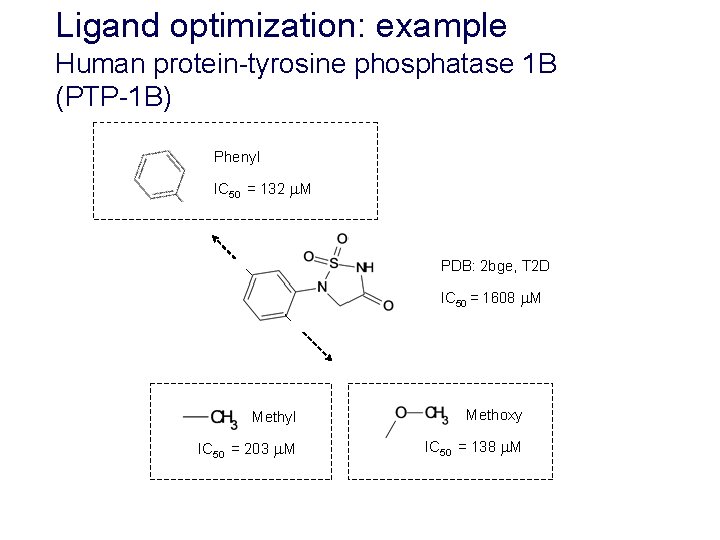
Ligand optimization: example Human protein-tyrosine phosphatase 1 B (PTP-1 B) Phenyl IC 50 = 132 M PDB: 2 bge, T 2 D IC 50 = 1608 M Methyl Methoxy IC 50 = 203 M IC 50 = 138 M



