Quantitative StructureActivity Relationship QSAR Chapter 8 Lecture 16

- Slides: 14

Quantitative Structure-Activity Relationship (QSAR) Chapter 8 Lecture 16 Dr Brajesh Kumar 1

Introduction Quantitative structure-activity relationships, collectively referred to as QSARs, are theoretical models that can be used to predict the physicochemical and biological properties of molecules. A structure-activity relationship (SAR) is a (qualitative) association between a chemical substructure and the potential of a chemical containing the substructure to exhibit a certain biological effect - similar structures –similar effects - more potency or improved side effects A quantitative structure-activity relationship (QSAR) is a mathematical model that relates a quantitative measure of chemical structure (e. g. a physicochemical property) to a physical property or to a biological effect (e. g. a toxicological endpoint) -similar structures –similar effects but uses parameters to describe the potency -parameters – anything (related to drug action) that can be represented by a numerical values 2

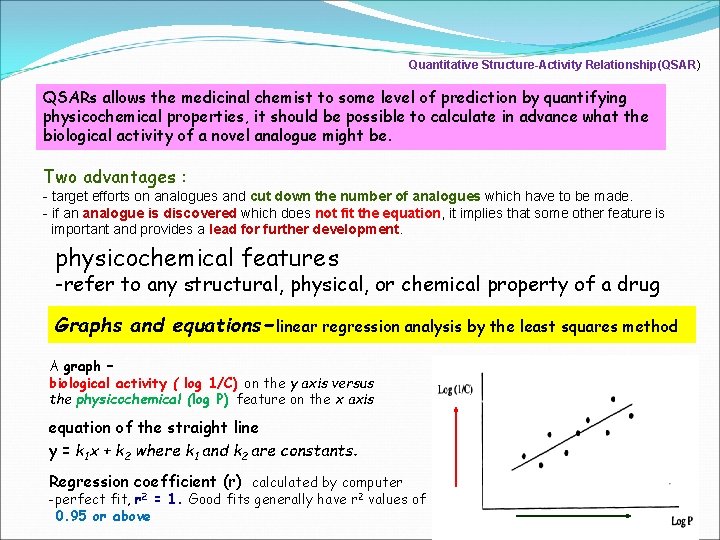
Quantitative Structure-Activity Relationship(QSAR) QSARs allows the medicinal chemist to some level of prediction by quantifying physicochemical properties, it should be possible to calculate in advance what the biological activity of a novel analogue might be. Two advantages : - target efforts on analogues and cut down the number of analogues which have to be made. - if an analogue is discovered which does not fit the equation, it implies that some other feature is important and provides a lead for further development. physicochemical features -refer to any structural, physical, or chemical property of a drug Graphs and equations-linear regression analysis by the least squares method A graph – biological activity ( log 1/C) on the y axis versus the physicochemical (log P) feature on the x axis equation of the straight line y = k 1 x + k 2 where k 1 and k 2 are constants. Regression coefficient (r) calculated by computer -perfect fit, r 2 = 1. Good fits generally have r 2 values of 0. 95 or above 3

Quantitative Structure-Activity Relationship(QSAR) QSAR – mathematical relationship (equations) -biological effect vs. physicochemical parameters Three most studied physicochemical properties - Hydrophobicity/lipophilicity - Electronic effects/electron distribution - Steric Factors - shape - size - Other physicochemical parameters Biological activity = F {parameters (s)} 4

Quantitative Structure-Activity Relationship(QSAR) Hydrophobicity /lipophilicity Hydrophobicity/ lipophilicity -Crucial to cross cell membranes and may in receptor interactions. -Changing substituent's -significant effects on hydrophobic character and its biological activity. Hydrophobic/Lipophilic parameters Hydrophobic character of a drug – measured experimentally – relative distribution -in an octanol/water mixture Relative distribution known as the partition coefficient (P) Partition coefficient log (1/C) = k 1 log P +k 2 equation shows -binding of drugs to serum albumin increases as log P increases. hydrophobic drugs bind more strongly to serum albumin than hydrophilic drug s 5

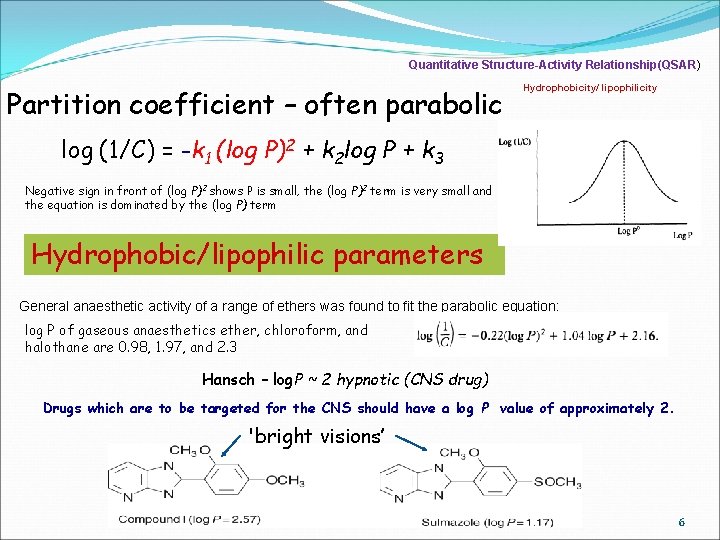
Quantitative Structure-Activity Relationship(QSAR) Partition coefficient – often parabolic Hydrophobicity/ lipophilicity log (1/C) = -k 1 (log P)2 + k 2 log P + k 3 Negative sign in front of (log P)2 shows P is small, the (log P)2 term is very small and the equation is dominated by the (log P) term Hydrophobic/lipophilic parameters General anaesthetic activity of a range of ethers was found to fit the parabolic equation: log P of gaseous anaesthetics ether, chloroform, and halothane are 0. 98, 1. 97, and 2. 3 Hansch – log. P ~ 2 hypnotic (CNS drug) Drugs which are to be targeted for the CNS should have a log P value of approximately 2. 'bright visions’ 6

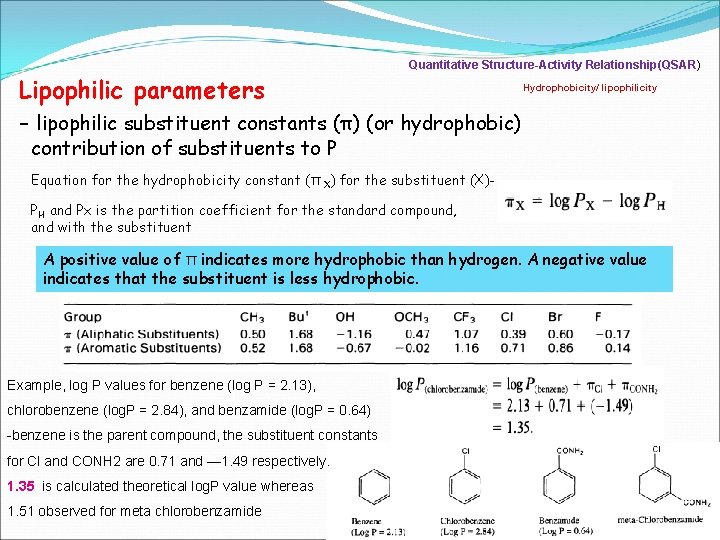
Quantitative Structure-Activity Relationship(QSAR) Hydrophobicity/ lipophilicity Lipophilic parameters - lipophilic substituent constants (π) (or hydrophobic) contribution of substituents to P Equation for the hydrophobicity constant (π X) for the substituent (X)PH and Px is the partition coefficient for the standard compound, and with the substituent A positive value of π indicates more hydrophobic than hydrogen. A negative value indicates that the substituent is less hydrophobic. Example, log P values for benzene (log P = 2. 13), chlorobenzene (log. P = 2. 84), and benzamide (log. P = 0. 64) -benzene is the parent compound, the substituent constants for Cl and CONH 2 are 0. 71 and — 1. 49 respectively. 1. 35 is calculated theoretical log. P value whereas 1. 51 observed for meta chlorobenzamide 7

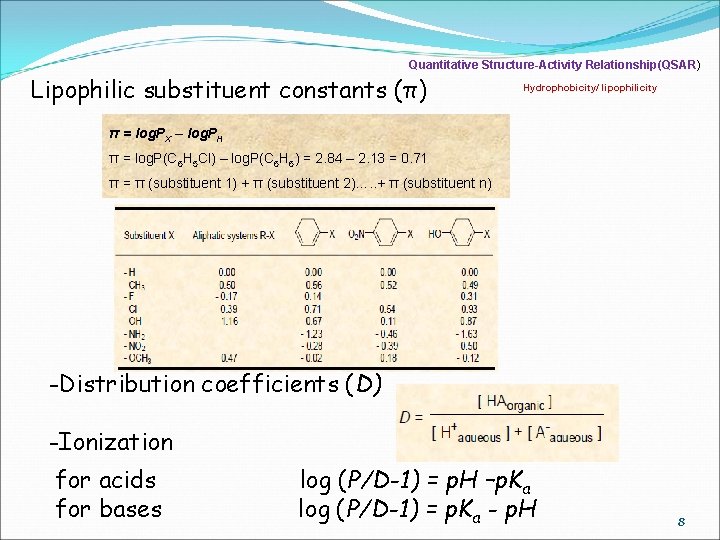
Quantitative Structure-Activity Relationship(QSAR) Lipophilic substituent constants (π) Hydrophobicity/ lipophilicity π = log. PX – log. PH π = log. P(C 6 H 5 Cl) – log. P(C 6 H 6) = 2. 84 – 2. 13 = 0. 71 π = π (substituent 1) + π (substituent 2)…. . + π (substituent n) -Distribution coefficients (D) -Ionization for acids for bases log (P/D-1) = p. H –p. Ka log (P/D-1) = p. Ka - p. H 8

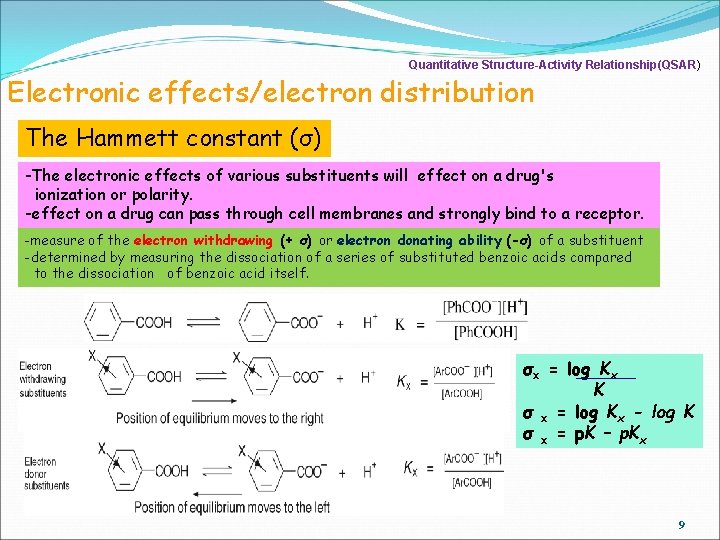
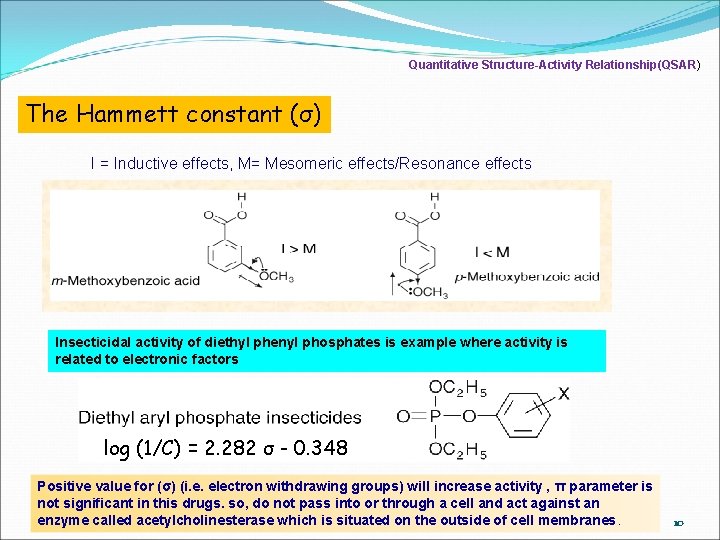
Quantitative Structure-Activity Relationship(QSAR) Electronic effects/electron distribution The Hammett constant (σ) -The electronic effects of various substituents will effect on a drug's ionization or polarity. -effect on a drug can pass through cell membranes and strongly bind to a receptor. -measure of the electron withdrawing (+ σ) or electron donating ability (-σ) of a substituent -determined by measuring the dissociation of a series of substituted benzoic acids compared to the dissociation of benzoic acid itself. σx = log Kx K σ x = log Kx - log K σ x = p. K – p. Kx 9

Quantitative Structure-Activity Relationship(QSAR) The Hammett constant (σ) I = Inductive effects, M= Mesomeric effects/Resonance effects Insecticidal activity of diethyl phenyl phosphates is example where activity is related to electronic factors log (1/C) = 2. 282 σ - 0. 348 Positive value for (σ) (i. e. electron withdrawing groups) will increase activity , π parameter is not significant in this drugs. so, do not pass into or through a cell and act against an enzyme called acetylcholinesterase which is situated on the outside of cell membranes. 10

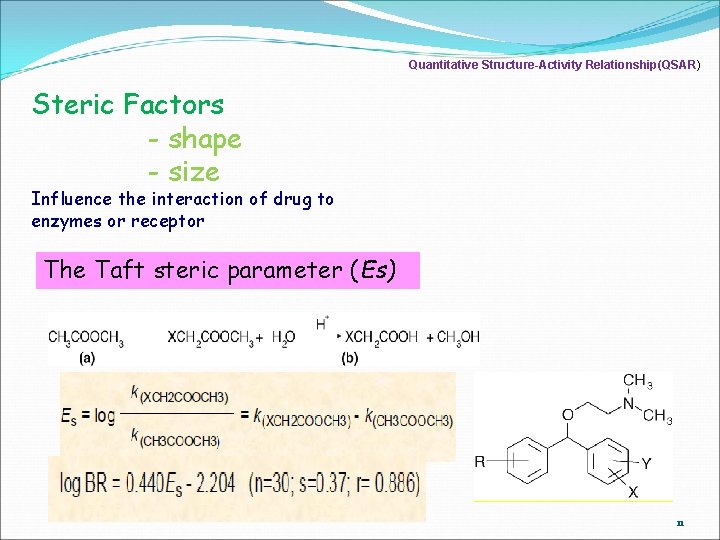
Quantitative Structure-Activity Relationship(QSAR) Steric Factors - shape - size Influence the interaction of drug to enzymes or receptor The Taft steric parameter (Es) 11

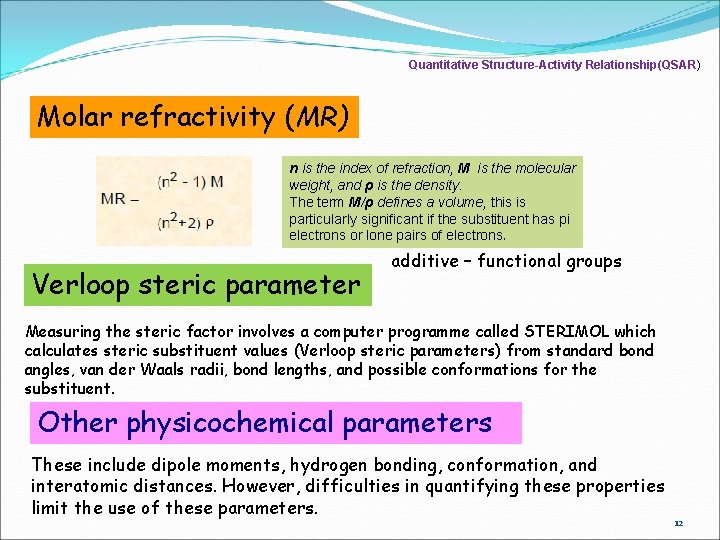
Quantitative Structure-Activity Relationship(QSAR) Molar refractivity (MR) n is the index of refraction, M is the molecular weight, and ρ is the density. The term M/ρ defines a volume, this is particularly significant if the substituent has pi electrons or lone pairs of electrons. Verloop steric parameter additive – functional groups Measuring the steric factor involves a computer programme called STERIMOL which calculates steric substituent values (Verloop steric parameters) from standard bond angles, van der Waals radii, bond lengths, and possible conformations for the substituent. Other physicochemical parameters These include dipole moments, hydrogen bonding, conformation, and interatomic distances. However, difficulties in quantifying these properties limit the use of these parameters. 12

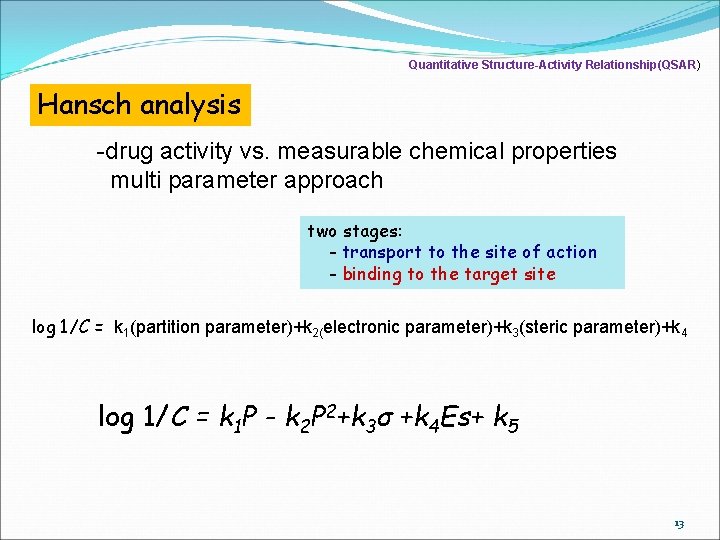
Quantitative Structure-Activity Relationship(QSAR) Hansch analysis -drug activity vs. measurable chemical properties multi parameter approach two stages: - transport to the site of action - binding to the target site log 1/C = k 1(partition parameter)+k 2(electronic parameter)+k 3(steric parameter)+k 4 log 1/C = k 1 P - k 2 P 2+k 3σ +k 4 Es+ k 5 13

Quantitative Structure-Activity Relationship(QSAR) Accuracy : - Greater number of analogs – n=5 x ; (x= number of parameters) - biological data - the choice of parameters Use: - Asses the factors controlling the activity - predict optimum activity (ideal parameter values) Sources of parameters - CRC, CAS, Merck Index, etc. 14