SAR vs QSAR or is QSAR different from







- Slides: 21

SAR vs QSAR or “is QSAR different from SAR” Joanna Jaworska Procer & Gamble, Brussels, Belgium and Nina Jeliazkova IPP, Bulgarian Academy of Sciences, Sofia, Bulgaria

SAR vs. QSAR how could we say there is no difference ? n SAR is supposed to be not quantitative concept n SAR is based on the notion of “similarity” : n n n “Similar compounds have similar activity” “Dissimilar compounds have dissimilar activity” QSAR aims to derive a quantitative model of the activity

SAR vs. QSAR Roadmap n n What “similarity” means? A philosophers’ view and implications to the toxicology; Are the basic tenets of SAR true ? What do similarity measures measure ? How does the similarity measure relate to QSAR modeling ?

Similarity : philosophers’ view exploiting the similarity concept is a sign of immature science (Quine) n “it is ill defined to say “A is similar to B” and it is only meaningful to say “A is similar to B with respect to C” n implications for toxicology : A chemical “A” cannot be similar to a chemical “B” in absolute terms but only with respect to some measurable key feature

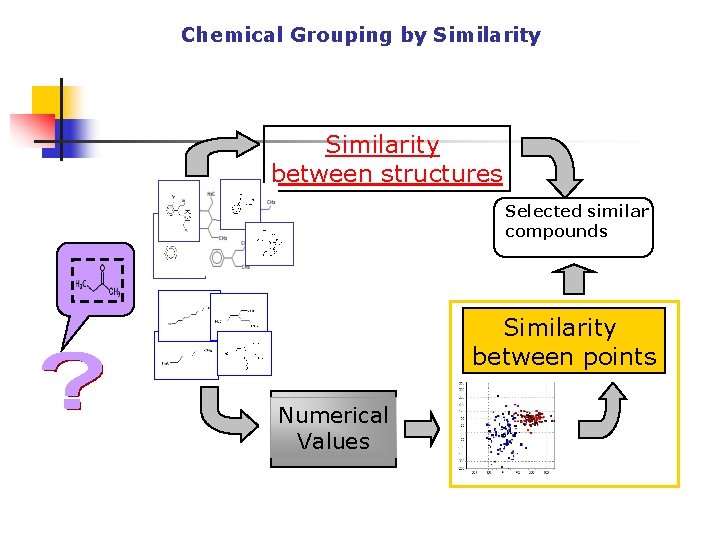
Chemical Grouping by Similarity between structures Selected similar compounds Similarity between points Numerical Values

Structural similarity n Does not imply always similarity in activity n n Martin et al. 2002 J. Med. Chem 45, 4350 -58 Does not always imply similarity in descriptors n Kubinyi, H. , Chemical Similarity and Biological activity (with permission of the author)

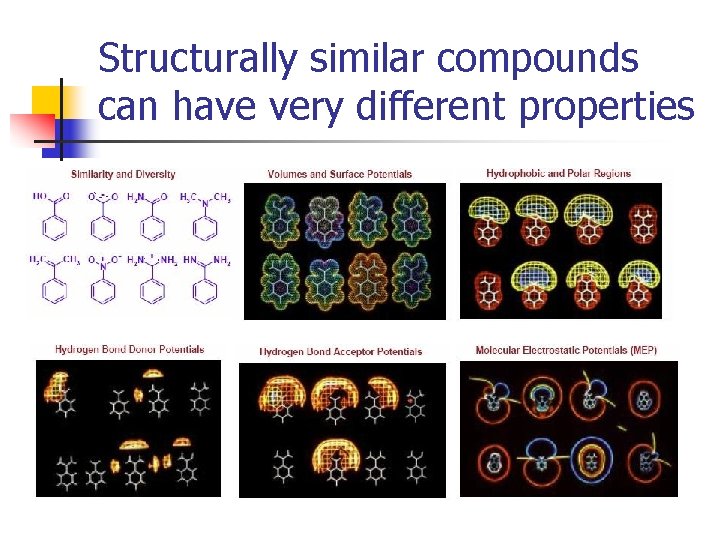
Structurally similar compounds can have very different properties

Example: Y. Martin et al ( 2002) Do structurally similar molecules have similar biological activity ? n n n Set of 1645 chemicals with IC 50 s for monoamine oxidase inhibition Daylight fingertips 1024 bits long ( 0 -7 bonds) Using Tanimoto coeff with a cut off value 0 f 0. 85 only 30 % of actives were detected Cutoff values % of actives detected % False positives J. Med. Chem. 2002, 45, 4350 -4358

How else to measure chemical similarity ? • Describe chemical compounds with a set of numerical values ( fingerprints, diverse descriptors, field values, etc. ) • Set up some measure between values (Euclidean distance, Tanimoto distance, Carbo similarity index, etc. ) What do we actually measure ? And how it is related to the activity ?

What do we measure ? The distance between numerical representations of chemical compounds A few warnings: • The numerical representation is not unique • The numerical representation includes only part of all the information about the compound • A distance measure reflects “closeness” only if the data holds specific assumptions (next slide - example)

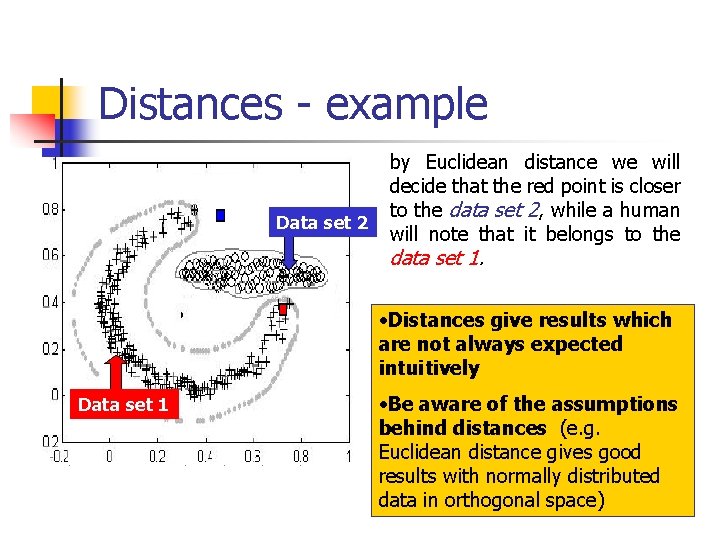
Distances - example Data set 2 by Euclidean distance we will decide that the red point is closer to the data set 2, while a human will note that it belongs to the data set 1. • Distances give results which are not always expected intuitively Data set 1 • Be aware of the assumptions behind distances (e. g. Euclidean distance gives good results with normally distributed data in orthogonal space)

How do we represent a chemical compound ? Fingerprints, Descriptors (more than 3000 available), electron density, various fields, etc. All representations lose information. We should ensure this information is not important. How?

Finding important information n A problem not unique to (Q)SAR Lot of methods available Most popular (e. g. PCA ) not the best Possible solution : look for the most discriminative information (example: descriptors which provide best discrimination between active and inactive compounds)

SAR vs. QSAR how could we say there is no difference ? Two common things to this point: n Both methods use numerical representation of chemical compounds; n Both methods need to decide which representation to use; One more difference : “SAR is a qualitative not a quantitative relationship” Is this true indeed?

Similarity and Activity n Proximity with respect to descriptors does not necessary mean proximity with respect to the activity (example) n n n This is only true if a linear relationship holds between descriptors and activity (examples) The linear relationship is only a special case, given the complexity of biochemical interactions. Its use should be justified in every specific case Structural similarity should be used with care (examples)

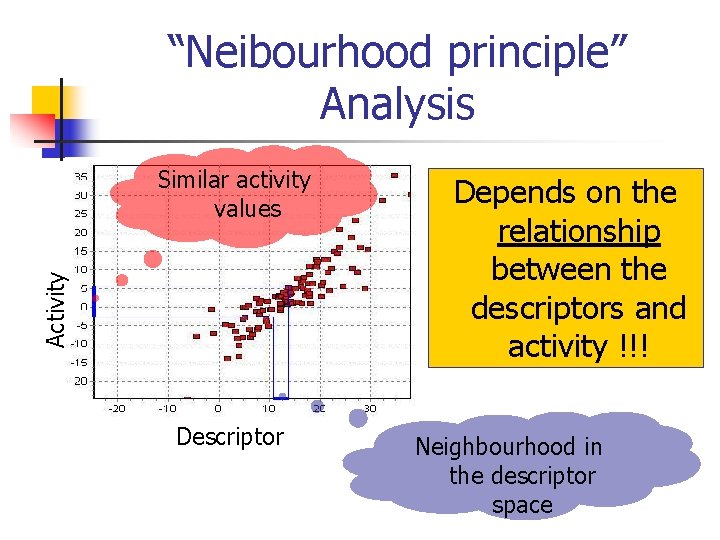
“Neighbourhood principle” n n Molecules in the same local region (“neighbourhood”) of a descriptor space tend to have similar values of a desired property Contradictory evidence exists : both supporting and rejecting

“Neibourhood principle” Analysis Activity Similar activity values Descriptor Depends on the relationship between the descriptors and activity !!! Neighbourhood in the descriptor space

“Neighbourhood principle” Lessons n In order to apply the “neighbourhood principle” the TYPE of the relationship between descriptor and activity should be known; n n The “neighbourhood principle” is genuine only if the relationship is LINEAR; The linear relationship is only a simple special case, given the complexity of biochemical interactions. Its use SHOULD BE JUSTIFIED in every specific case.

SAR vs QSAR n n SAR is based on the “similarity” principle; The principle is assumed, but in the reality it is not always true; n n Similarity of structures Similarity of descriptors The authenticity depends on the type of the relationship between descriptors (numerical representation of chemicals) and activity; The type of the relationship should be known (or derived)

SAR vs. QSAR how could we say there is a difference ? Three common things to this point: n Both methods use numerical representation of chemical compounds; n Both methods need to decide which representation to use; n Both methods need to derive the relationship between numerical representation (descriptors, etc. ) and activity.

Thank you! When you can measure what you are speaking about, and express it in numbers, you know something about it; but when you cannot measure it, when you cannot express it in numbers, your knowledge is of a meager and unsatisfactory kind: it may be the beginning of knowledge, but you have scarcely advanced to the stage of science. William Thomson, Lord Kelvin