Identifying human disease genes Overview Position independent methods









- Slides: 58

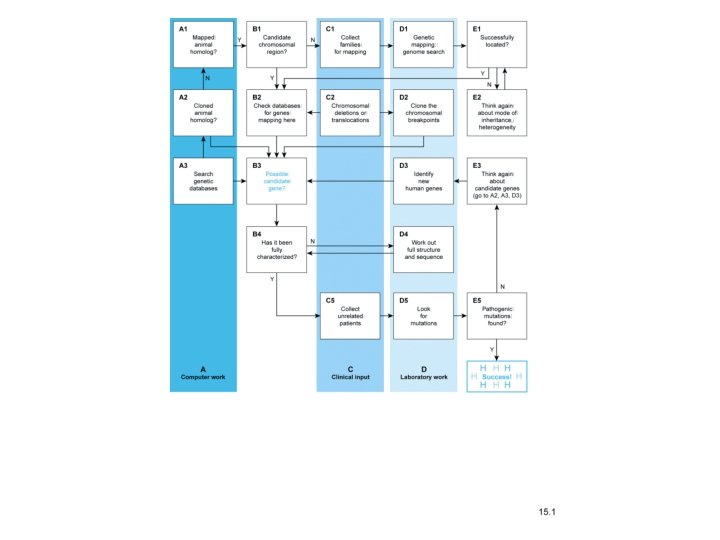
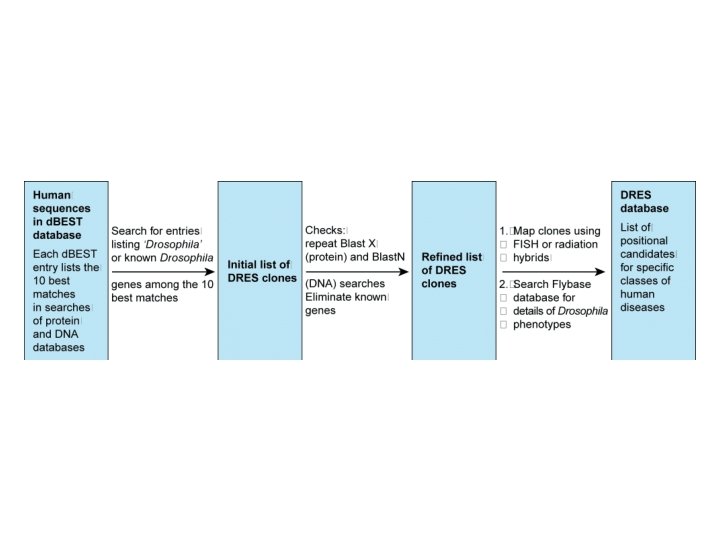
Identifying human disease genes • • • Overview Position independent methods Positional cloning Synteny Drosophila mutants that are positional candidates for human disease genes

Identifying human disease genes • Overview of the process


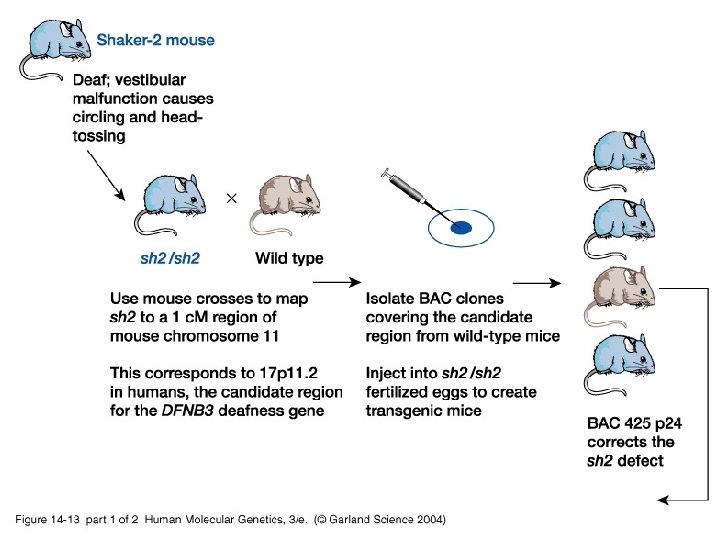
Identifying human disease genes • Position independent methods – Complementation

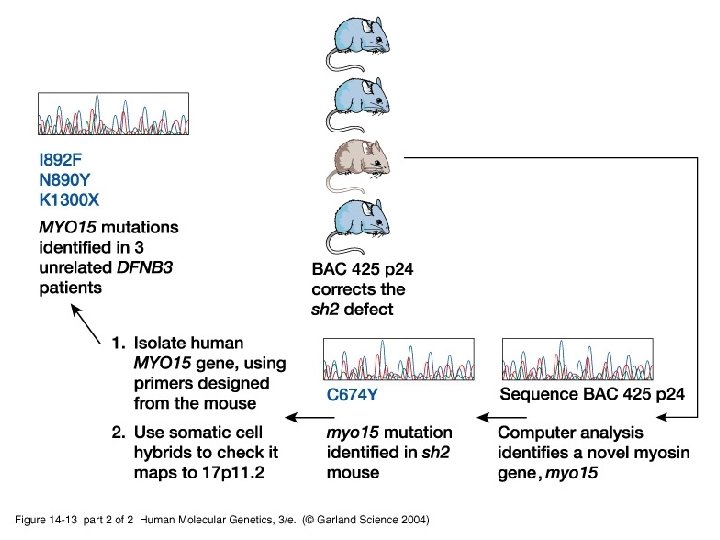
14_13. jpg

14_13_2. jpg

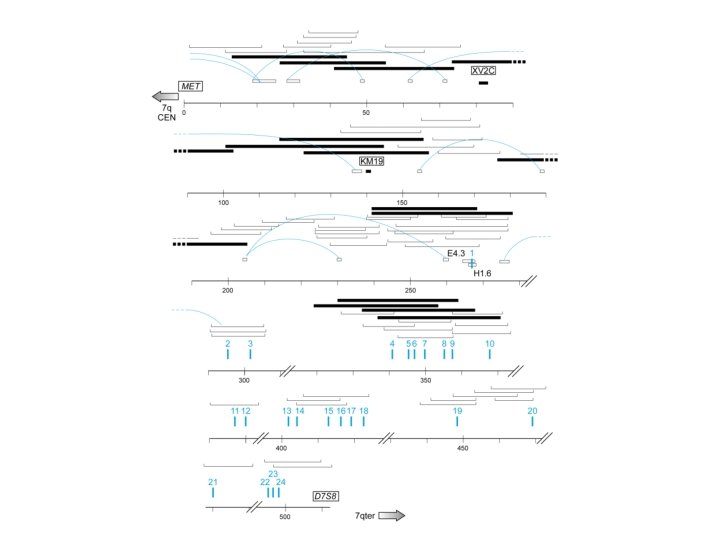
Identifying human disease genes • Position dependent methods – CF gene chromosome jumping and walking


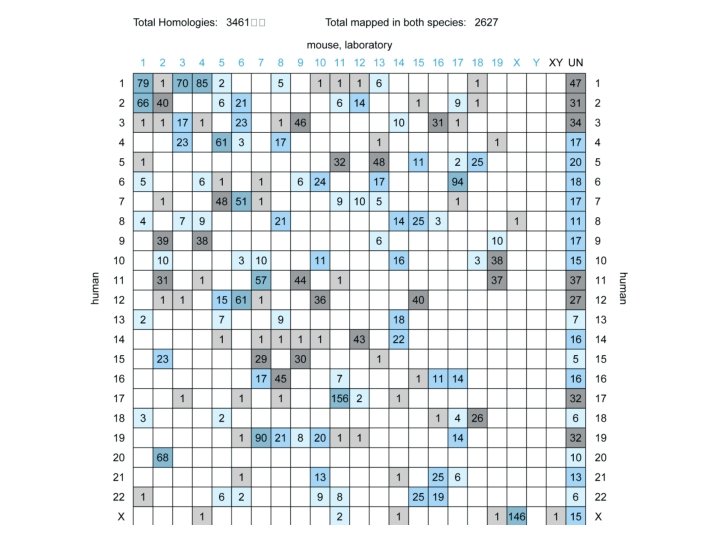
Identifying human disease genes • Mouse and human synteny


Identifying human disease genes • Drosophila and human disorders


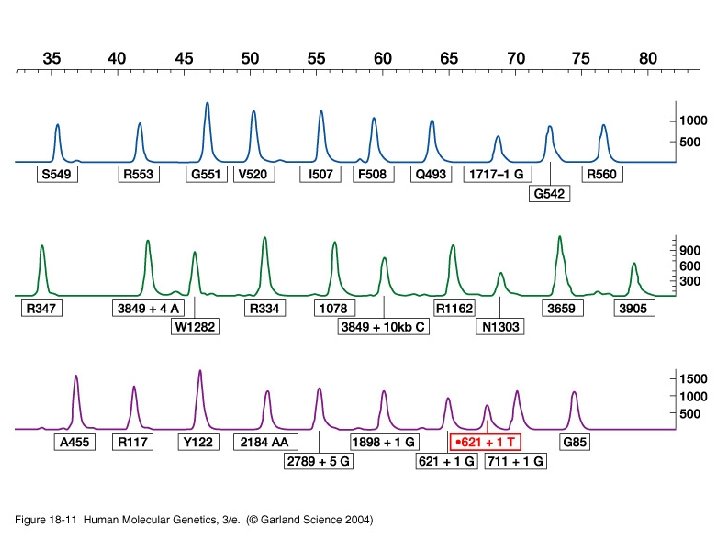
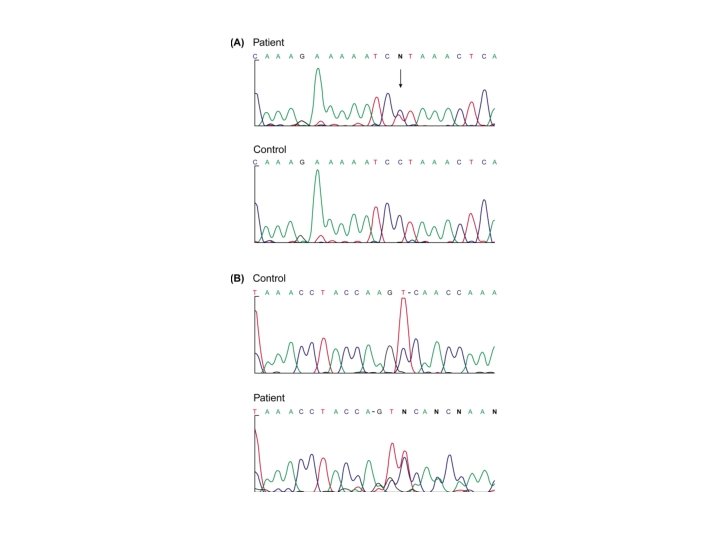
Genetic testing • Overview • Examples CF gene


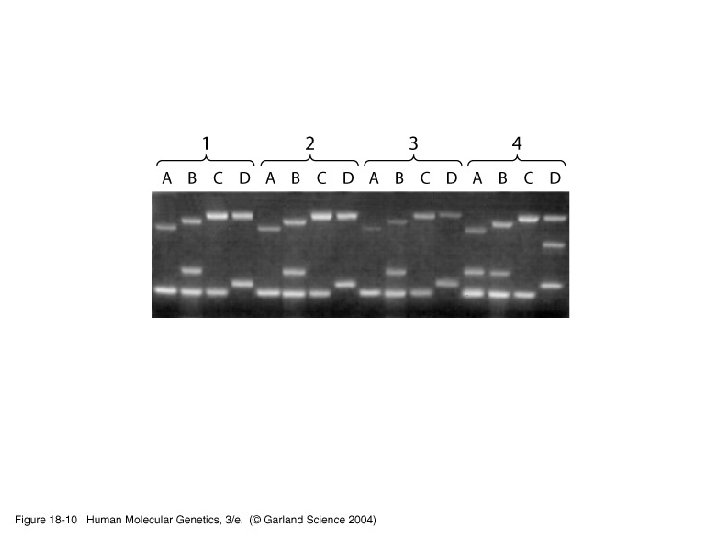
Genetic testing • CF testing – ARMs – OLA – Sequencing – Heteroduplex screening – DGGE – Mismatch cleavage







Genetic testing • Oligonucleotide arrays

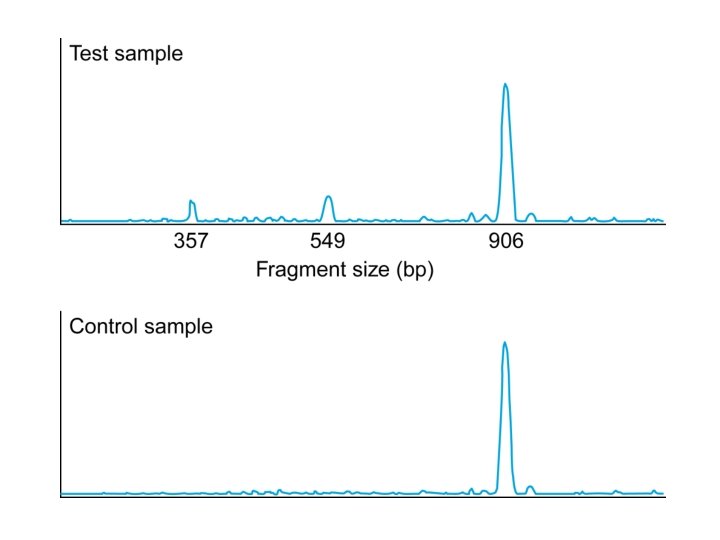
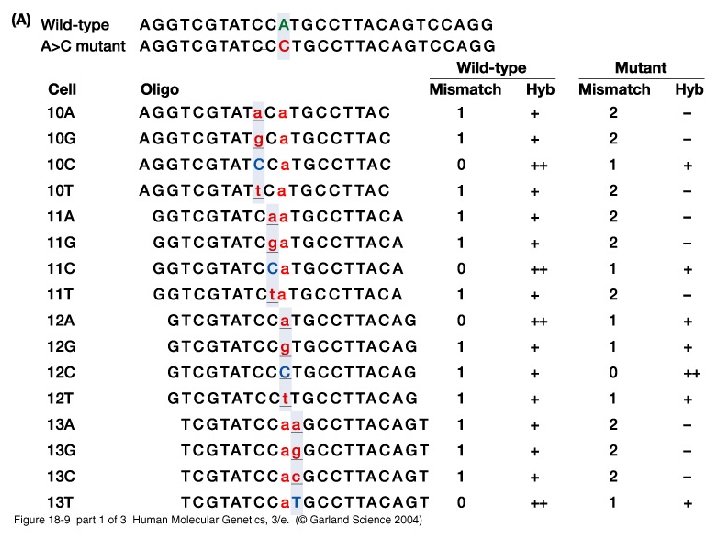
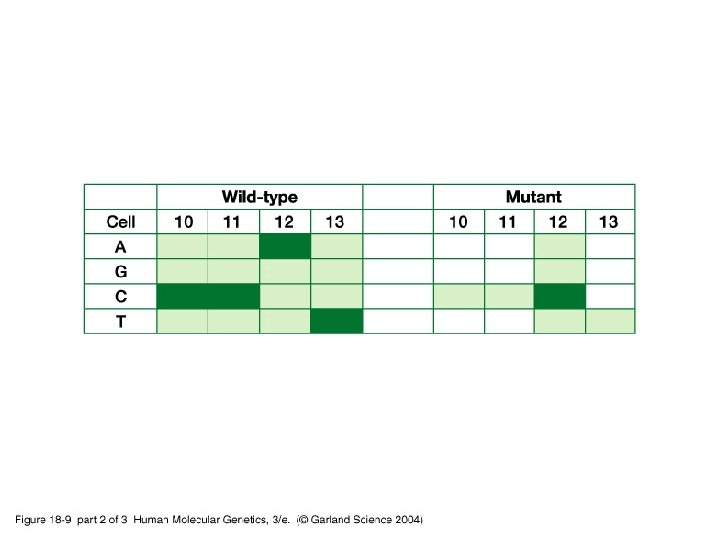
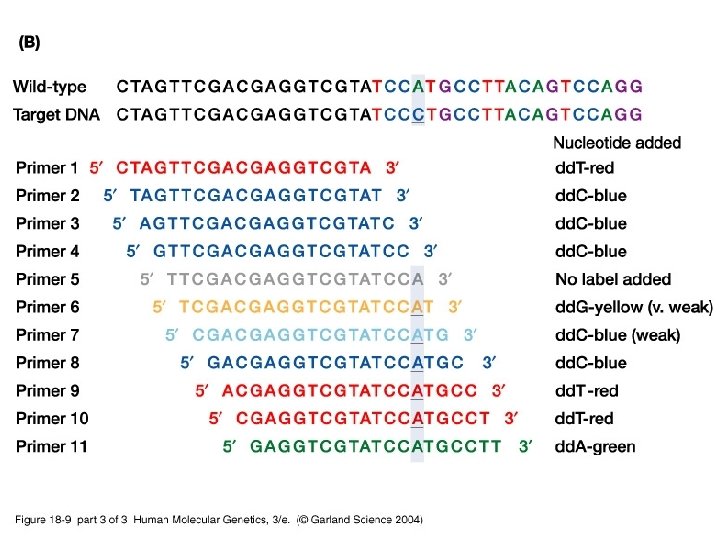
18_09. jpg

18_09_2. jpg

18_09_3. jpg

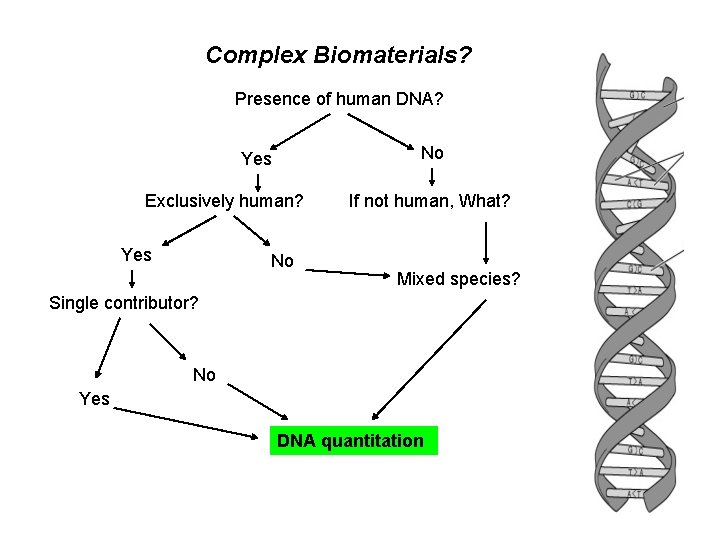
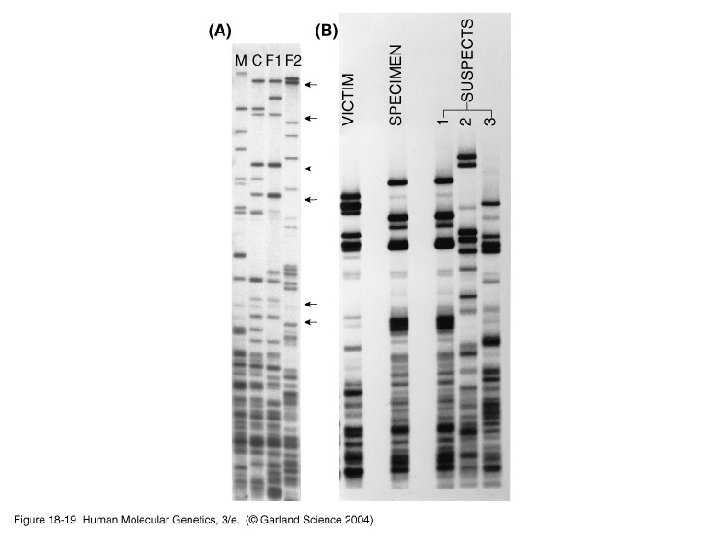
Genetic testing • Forensics – Species identification – Paternity testing – DNA quantitation – Human identification

Complex Biomaterials? Presence of human DNA? No Yes Exclusively human? Yes No If not human, What? Mixed species? Single contributor? No Yes DNA quantitation

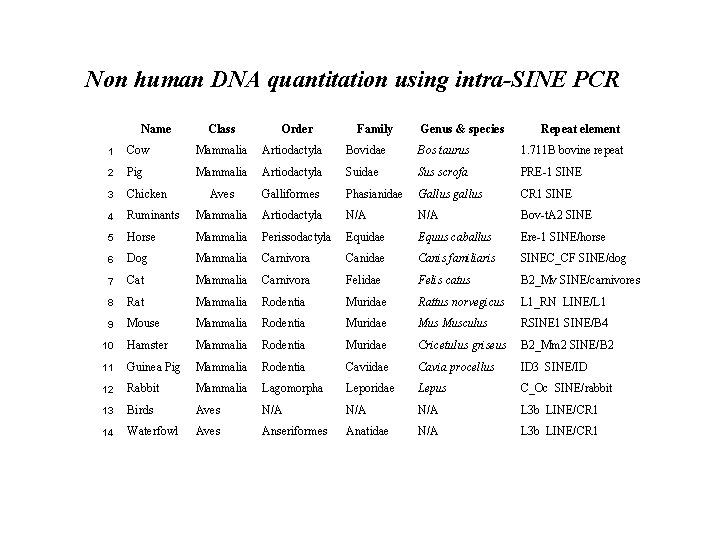
Non human DNA quantitation using intra-SINE PCR Name Class Order Family Genus & species Repeat element 1 Cow Mammalia Artiodactyla Bovidae Bos taurus 1. 711 B bovine repeat 2 Pig Mammalia Artiodactyla Suidae Sus scrofa PRE-1 SINE 3 Chicken Aves Galliformes Phasianidae Gallus gallus CR 1 SINE 4 Ruminants Mammalia Artiodactyla N/A Bov-t. A 2 SINE 5 Horse Mammalia Perissodactyla Equidae Equus caballus Ere-1 SINE/horse 6 Dog Mammalia Carnivora Canidae Canis familiaris SINEC_CF SINE/dog 7 Cat Mammalia Carnivora Felidae Felis catus B 2_Mv SINE/carnivores 8 Rat Mammalia Rodentia Muridae Rattus norvegicus L 1_RN LINE/L 1 9 Mouse Mammalia Rodentia Muridae Musculus RSINE 1 SINE/B 4 10 Hamster Mammalia Rodentia Muridae Cricetulus griseus B 2_Mm 2 SINE/B 2 11 Guinea Pig Mammalia Rodentia Caviidae Cavia procellus ID 3 SINE/ID 12 Rabbit Mammalia Lagomorpha Leporidae Lepus C_Oc SINE/rabbit 13 Birds Aves N/A N/A L 3 b LINE/CR 1 14 Waterfowl Aves Anseriformes Anatidae N/A L 3 b LINE/CR 1

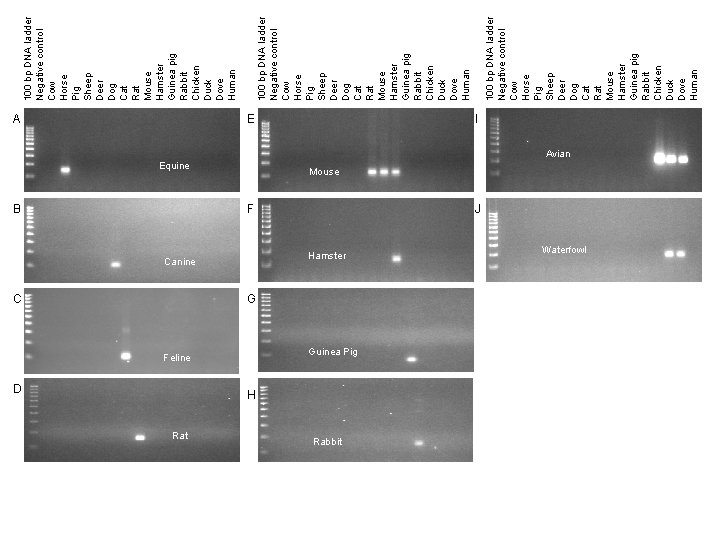
A E Equine B Canine C Feline D Rat I Avian Mouse F J Hamster G Guinea Pig H Rabbit Waterfowl 100 bp DNA ladder Negative control Cow Horse Pig Sheep Deer Dog Cat Rat Mouse Hamster Guinea pig Rabbit Chicken Duck Dove Human


Our Assay Design Objectives 1. Human Specific a. Comparison of genomic sequences b. Testing complex biomaterials 2. Target Specific a. Comparison of target sequences b. Testing for non-specific amplification 3. Multiplex Compatible a. ABI Prism 7000 default PCR conditions b. Experimentally optimized PCR reagents FAM VIC NED

Nuclear DNA

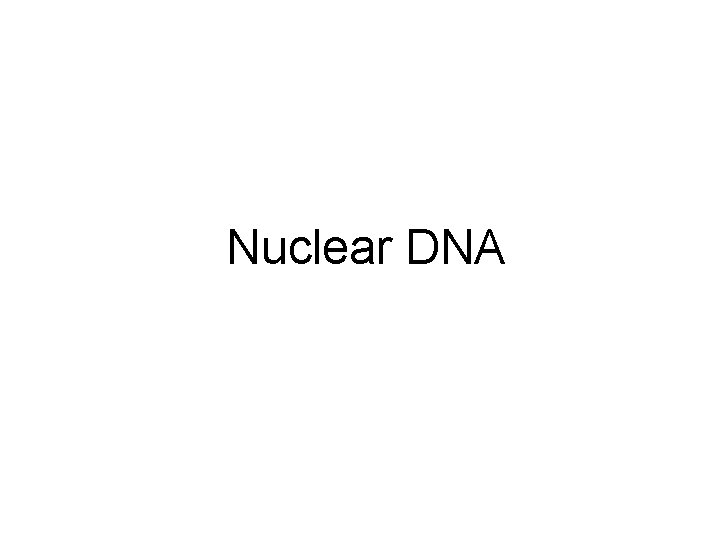
Schematic of inter-Alu and intra-Alu PCR Inter-Alu PCR 5’ Alu 3’ 3’ Alu 5’ tail to tail 5’ Alu 3’ head to head 5’ Alu 3’ tail to head Intra-Alu PCR 5’ Alu Element 3’

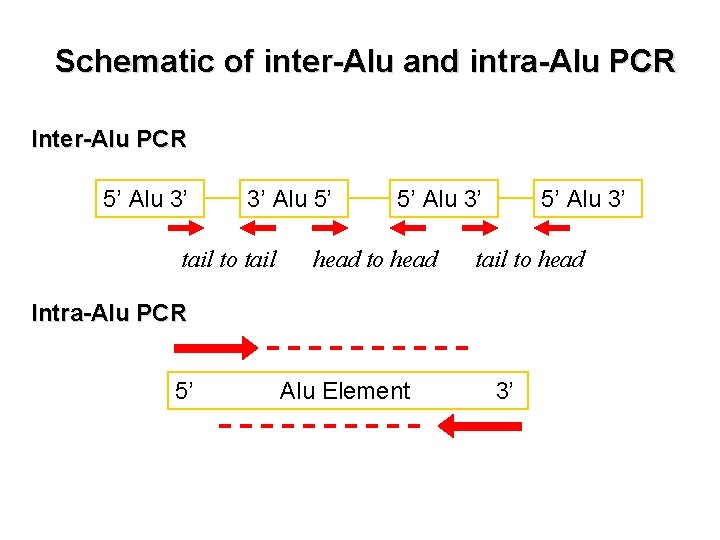
Nuclear DNA target design Intra-Alu Yb 8 Alu. Y GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGA Alu. Yb 8. . . 50 50 Alu. Y GGCGGATCACGAGGTCAGGAGATCGAGACCATCCTGGCTAACACGG Alu. Yb 8. . . T. . . . A. . 100 Alu. Y TGAAACCCCGTCTCTACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGC Alu. Yb 8. . . C. . . 150 Alu. Y GGGCGCCTGTAGTCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATGGCGT Alu. Yb 8. . . 200 Alu. Y GAACCCGGGAGGCGGAGCTTGCAGTGAGCCGAGATCGCGCCACTGCACTC Alu. Yb 8. . A. . . T. . . G. . 250 Alu. Y CA-------GCCTGGGCGACAGAGCGAGACTCCGTCTCAAAAAA Alu. Yb 8. GCAGTCCG. . . . . 287 294 PCR primers Taq. Man-MGB probe

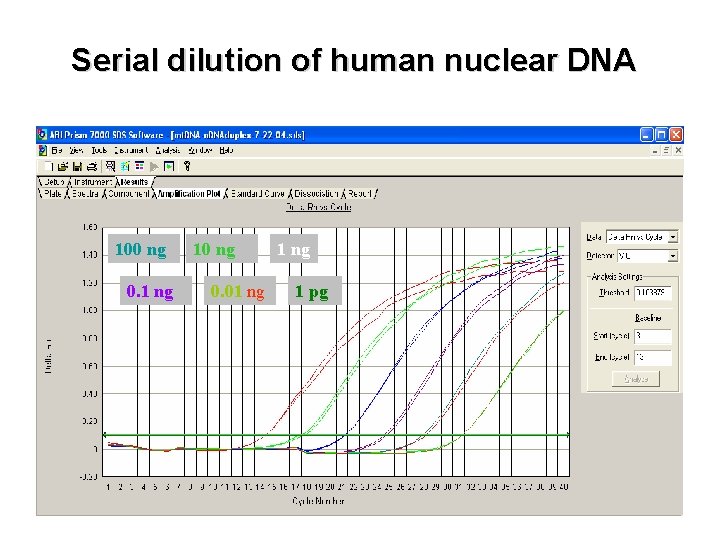
Serial dilution of human nuclear DNA 100 ng 0. 1 ng 10 ng 0. 01 ng 1 pg

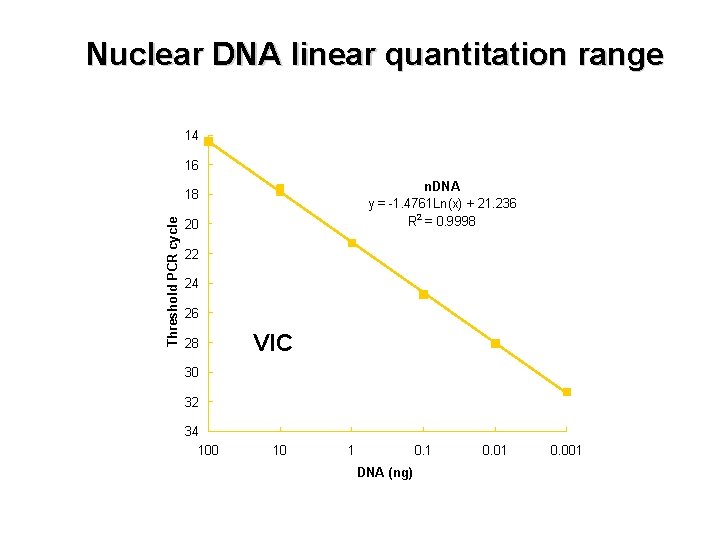
Nuclear DNA linear quantitation range 14 16 n. DNA y = -1. 4761 Ln(x) + 21. 236 R 2 = 0. 9998 Threshold PCR cycle 18 20 22 24 26 28 VIC 30 32 34 100 10 1 0. 1 DNA (ng) 0. 01 0. 001

Mitochondrial DNA

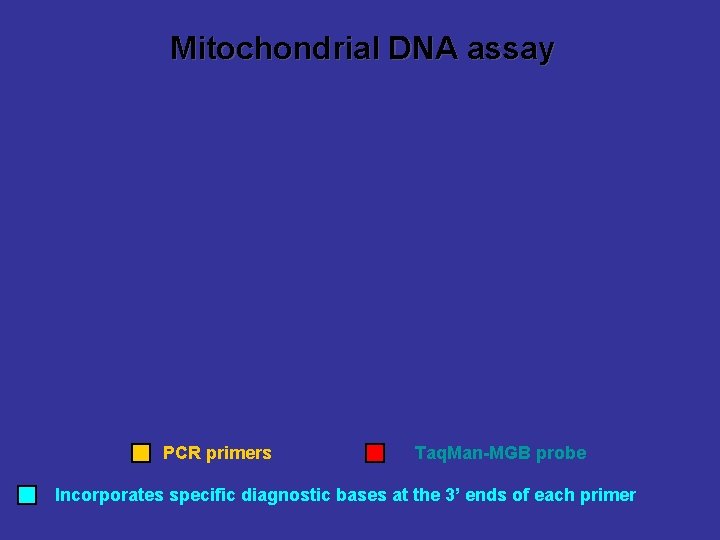
Mitochondrial DNA assay PCR primers Taq. Man-MGB probe Incorporates specific diagnostic bases at the 3’ ends of each primer

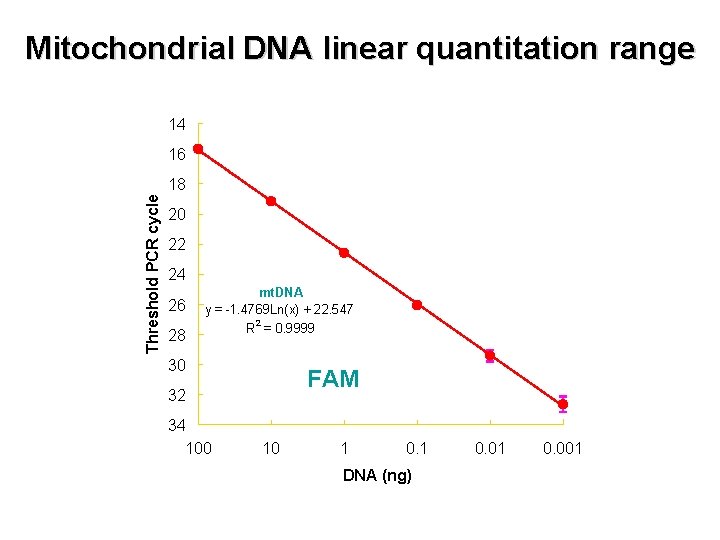
Mitochondrial DNA linear quantitation range 14 16 Threshold PCR cycle 18 20 22 24 26 28 mt. DNA y = -1. 4769 Ln(x) + 22. 547 R 2 = 0. 9999 30 FAM 32 34 100 10 1 0. 1 DNA (ng) 0. 01 0. 001

Y chromosome DNA

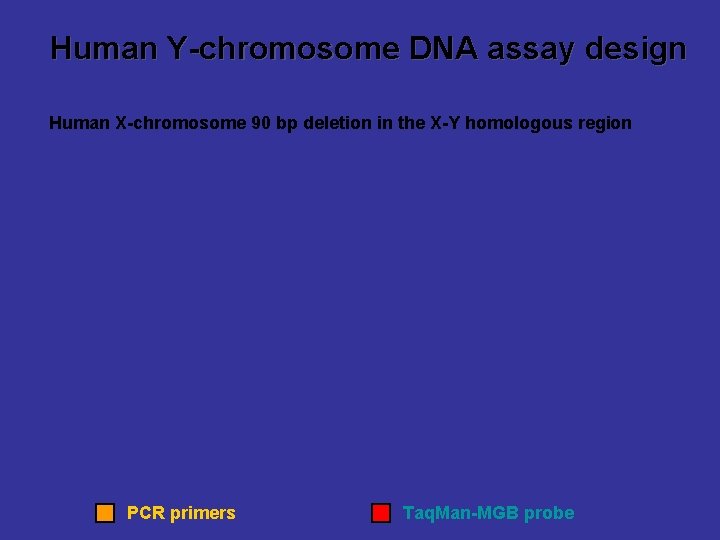
Human Y-chromosome DNA assay design Human X-chromosome 90 bp deletion in the X-Y homologous region PCR primers Taq. Man-MGB probe

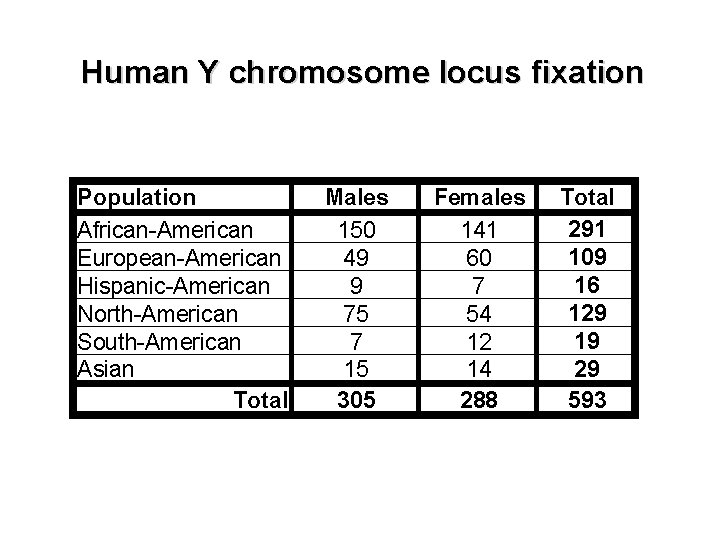
Human Y chromosome locus fixation Population African-American European-American Hispanic-American North-American South-American Asian Total Males 150 49 9 75 7 15 305 Females 141 60 7 54 12 14 288 Total 291 109 16 129 19 29 593

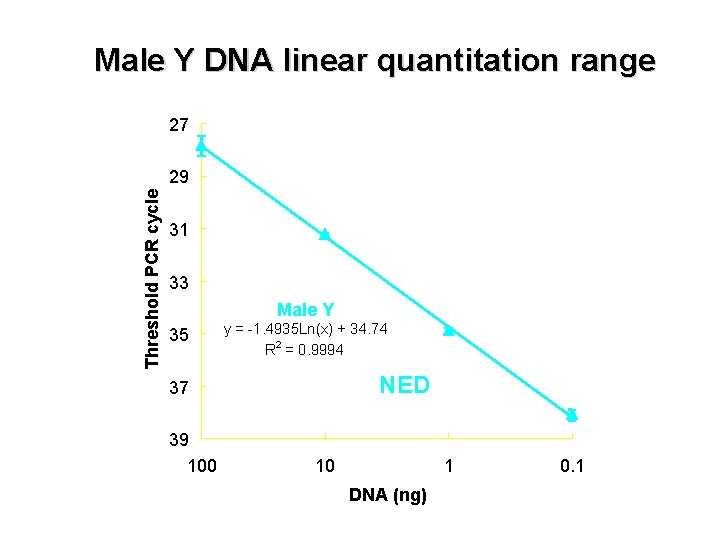
Male Y DNA linear quantitation range 27 Threshold PCR cycle 29 31 33 Male Y 35 y = -1. 4935 Ln(x) + 34. 74 2 R = 0. 9994 NED 37 39 100 10 1 DNA (ng) 0. 1

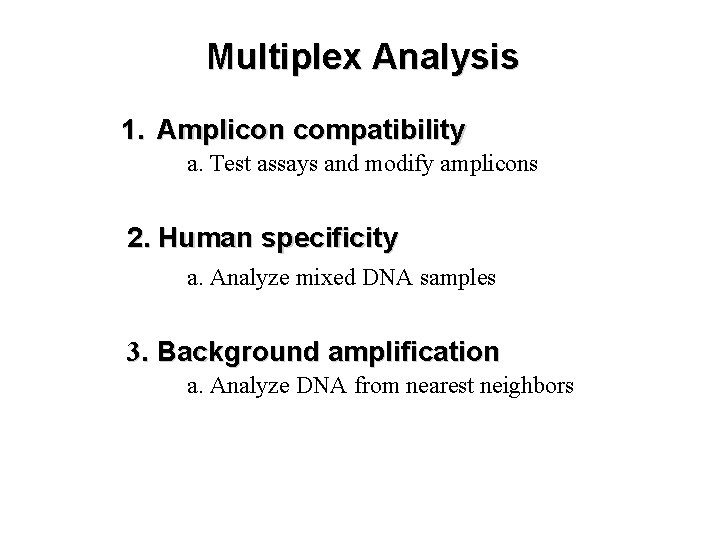
Multiplex Analysis

Multiplex Analysis 1. Amplicon compatibility a. Test assays and modify amplicons 2. Human specificity a. Analyze mixed DNA samples 3. Background amplification a. Analyze DNA from nearest neighbors

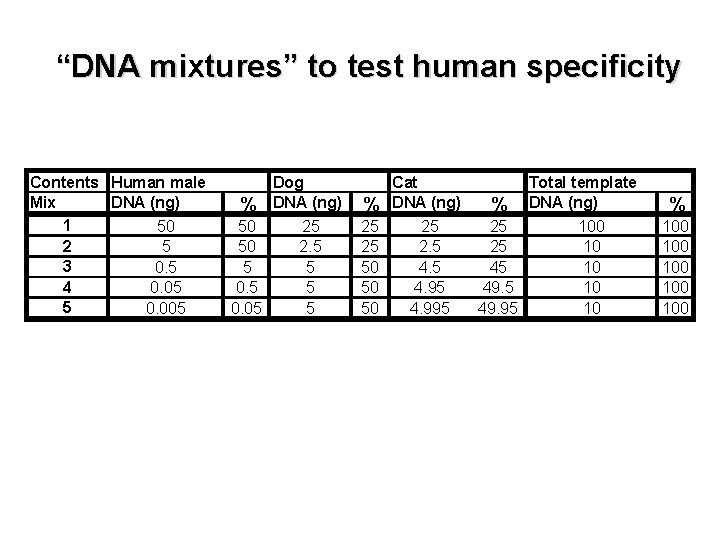
“DNA mixtures” to test human specificity Contents Human male Mix DNA (ng) 1 50 2 5 3 0. 5 4 0. 05 5 0. 005 Dog % DNA (ng) 50 25 50 2. 5 5 5 0. 05 5 % 25 25 50 50 50 Cat DNA (ng) 25 2. 5 4. 95 4. 995 Total template % DNA (ng) 25 100 25 10 49. 95 10 % 100 100 100

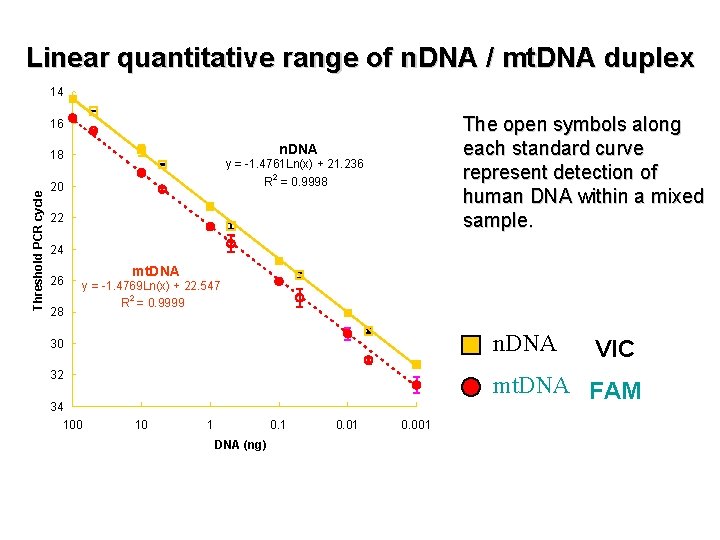
Linear quantitative range of n. DNA / mt. DNA duplex 14 The open symbols along each standard curve represent detection of human DNA within a mixed sample. 16 n. DNA Threshold PCR cycle 18 y = -1. 4761 Ln(x) + 21. 236 2 R = 0. 9998 20 22 24 26 28 mt. DNA y = -1. 4769 Ln(x) + 22. 547 2 R = 0. 9999 n. DNA 30 32 34 100 VIC mt. DNA FAM 10 1 0. 1 DNA (ng) 0. 01 0. 001

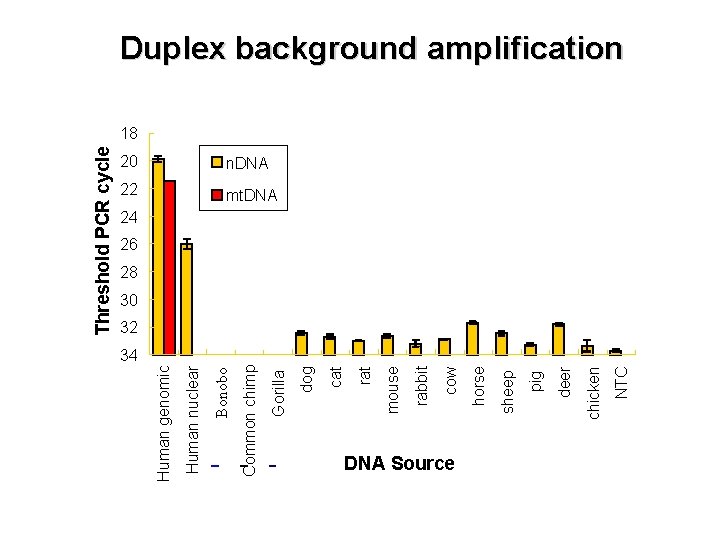
DNA Source NTC chicken deer pig sheep horse cow rabbit mt. DNA mouse 22 rat n. DNA cat 20 dog Gorilla Common chimp Bonobo Human nuclear Human genomic Threshold PCR cycle Duplex background amplification 18 24 26 28 30 32 34

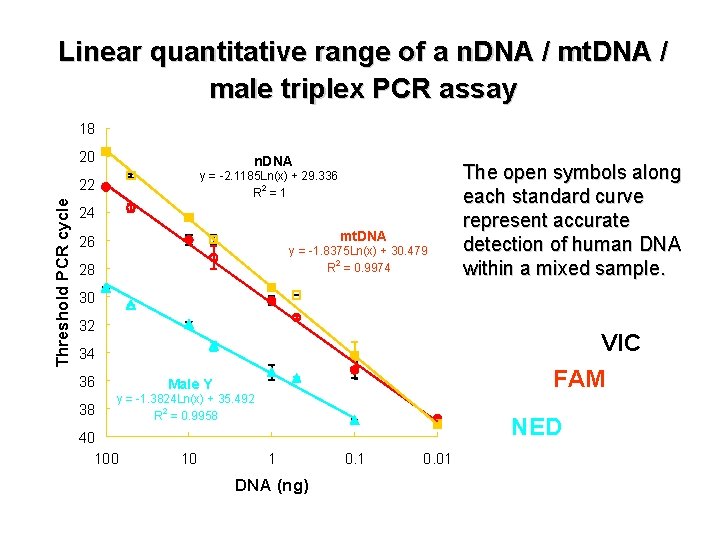
Linear quantitative range of a n. DNA / mt. DNA / male triplex PCR assay 18 20 n. DNA y = -2. 1185 Ln(x) + 29. 336 2 R =1 Threshold PCR cycle 22 24 mt. DNA 26 y = -1. 8375 Ln(x) + 30. 479 2 R = 0. 9974 28 The open symbols along each standard curve represent accurate detection of human DNA within a mixed sample. 30 32 VIC FAM 34 36 38 Male Y y = -1. 3824 Ln(x) + 35. 492 2 R = 0. 9958 40 10 NED 1 DNA (ng) 0. 1 0. 01

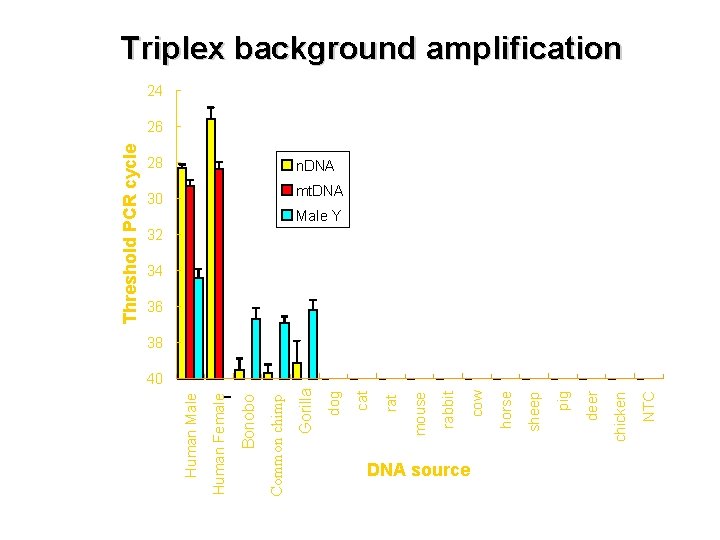
DNA source NTC chicken deer pig sheep horse cow rabbit mouse rat cat 30 dog 28 Gorilla Common chimp Bonobo Human Female Human Male Threshold PCR cycle Triplex background amplification 24 26 n. DNA mt. DNA Male Y 32 34 36 38 40

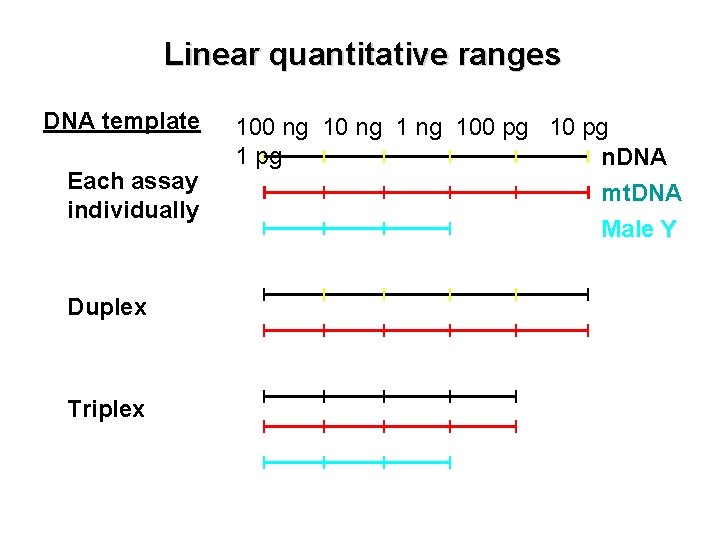
Linear quantitative ranges DNA template Each assay individually Duplex Triplex 100 ng 100 pg 1 pg n. DNA mt. DNA Male Y

Duplex PCR for Human Sex Typing X PCR primers Y PCR primers Taq. Man-MGB probes

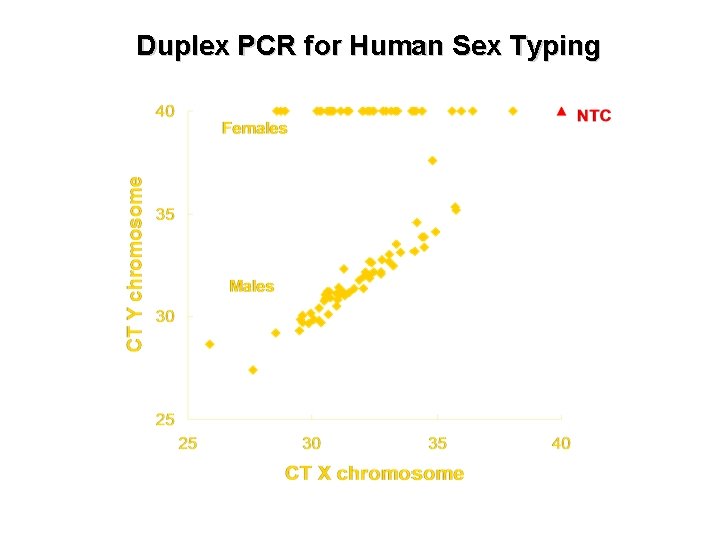
Duplex PCR for Human Sex Typing

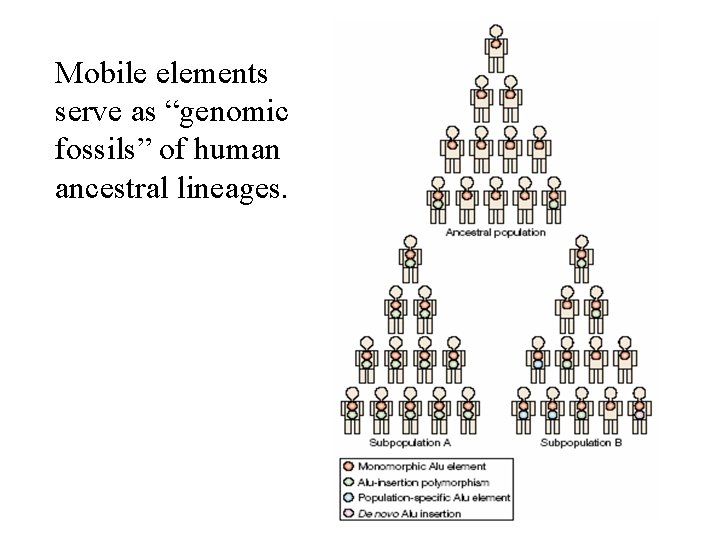
Mobile elements serve as “genomic fossils” of human ancestral lineages.

Human Population Biology and Investigative Forensics 1. Since the dispersal from Africa, mobile elements have continued to expand in the human genome. 2. Many elements are polymorphic and occur at high/low frequencies in human populations. 3. These elements can be exploited to examine human population history. 4. Display-based PCR methods can be used to “extract” recent, population-indicative elements.

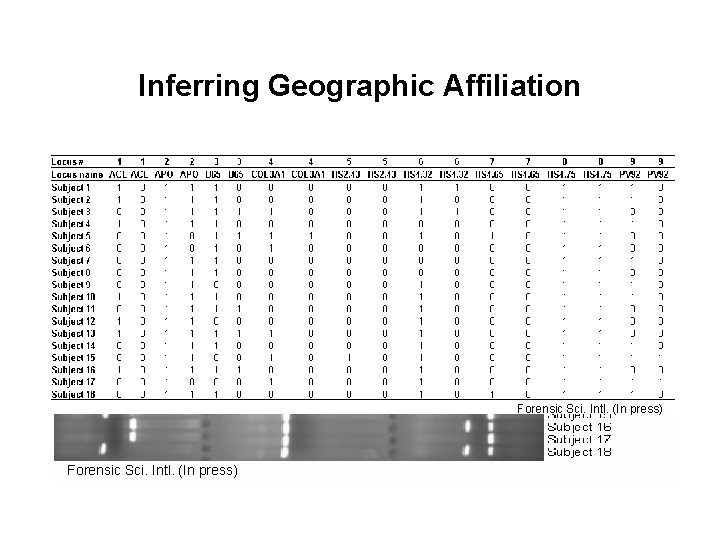
Inferring Geographic Affiliation 1) Series of genetic markers (100 Alu loci) 2) Database of human variation (currently 715 individuals of known ancestry) 3) Genotype unknown sample Forensic Sci. Intl. (In press) 4) Analytical approach (Structure analysis) Forensic Sci. Intl. (In press)

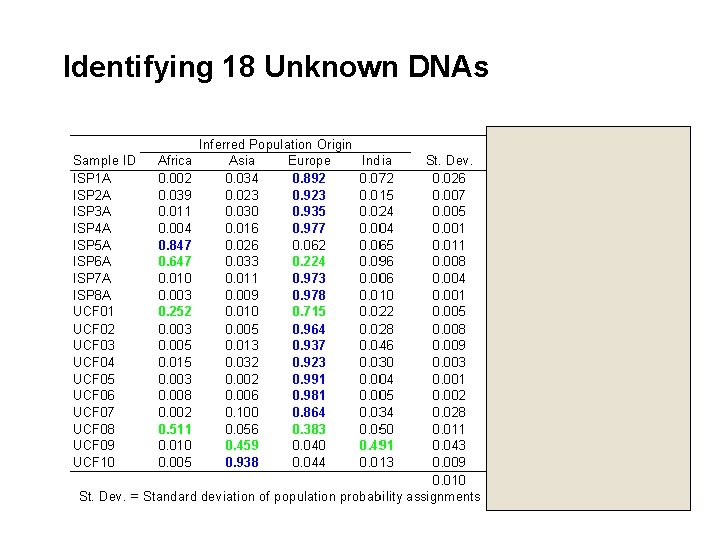
Identifying 18 Unknown DNAs Forensic Sci. Intl. (In press)
