I corsi vengono integrati e conterranno grosso modo






















- Slides: 66

I corsi vengono integrati e conterranno grosso modo due moduli: SB: systems biology ML: machine learning Alternativa 1 Alternativa 2 Mattina (Fariselli) Pomeriggio (Martelli) Lun 1 Ott SVM (ML) Grafi (SB) Mar 2 Ott System Biology (SB) Probabilita (ML) Mer 3 Ott System Biology (SB) HMM (ML) Ven 5 Ott Lun 8 Ott System Biology (SB) HMM (ML) Lun 8 Ott Mar 9 Ott Linux e python Mar 9 Ott Mer 10 Ott Esercitazione su SVM Mer 10 Ott Gio 11 Ott Esercitazione su HMM

Systems Biology. What Is It? v A branch of science that seeks to integrate different levels of information to understand how biological systems function. v L. Hood: “Systems biology defines and analyses the interrelationships of all of the elements in a functioning system in order to understand how the system works. ” v It is not (only) the number and properties of system elements but their relations!!

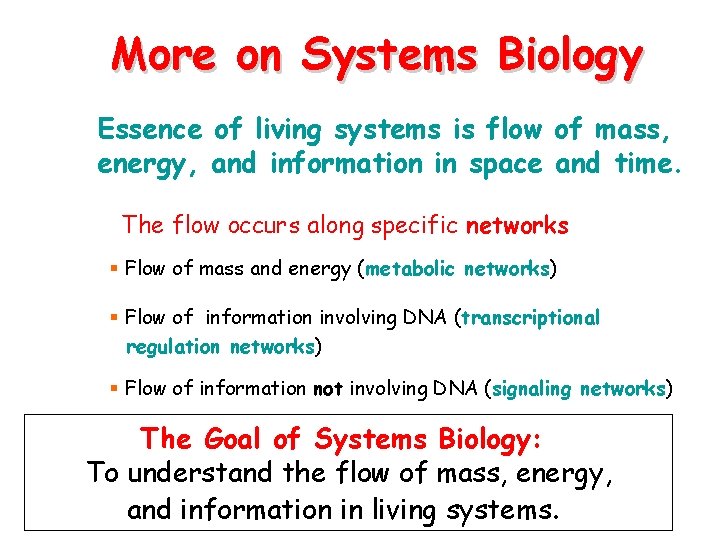
More on Systems Biology Essence of living systems is flow of mass, energy, and information in space and time. The flow occurs along specific networks § Flow of mass and energy (metabolic networks) § Flow of information involving DNA (transcriptional regulation networks) § Flow of information not involving DNA (signaling networks) The Goal of Systems Biology: To understand the flow of mass, energy, and information in living systems.

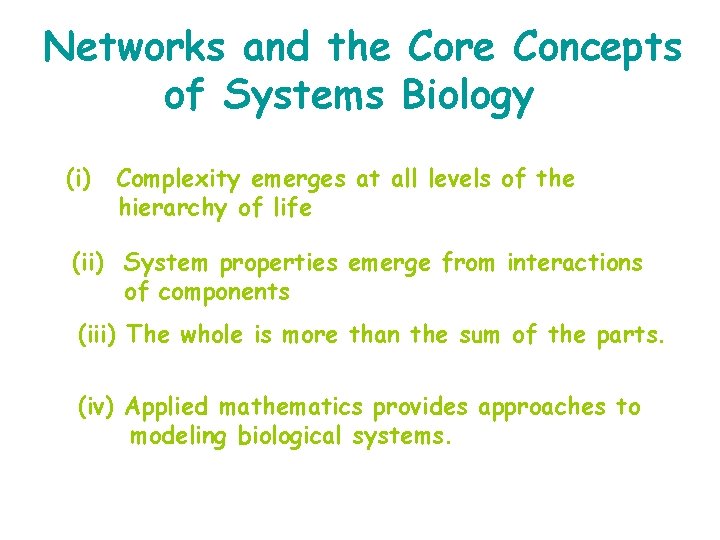
Networks and the Core Concepts of Systems Biology (i) Complexity emerges at all levels of the hierarchy of life (ii) System properties emerge from interactions of components (iii) The whole is more than the sum of the parts. (iv) Applied mathematics provides approaches to modeling biological systems.

How to Describe a System As a Whole? Networks - The Language of Complex Systems

Air Transportation Network

The World Wide Web

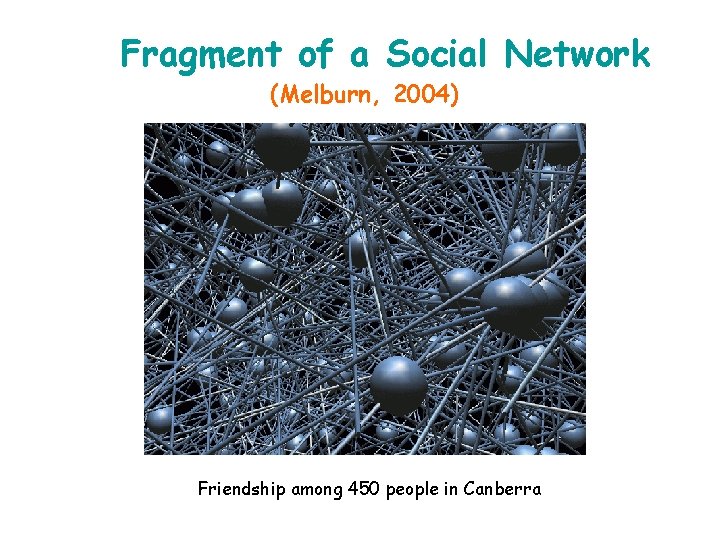
Fragment of a Social Network (Melburn, 2004) Friendship among 450 people in Canberra

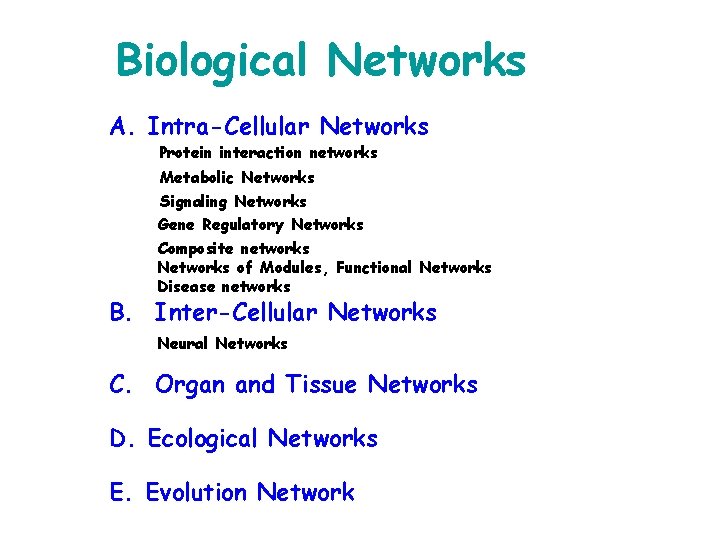
Biological Networks A. Intra-Cellular Networks Protein interaction networks Metabolic Networks Signaling Networks Gene Regulatory Networks Composite networks Networks of Modules, Functional Networks Disease networks B. Inter-Cellular Networks Neural Networks C. Organ and Tissue Networks D. Ecological Networks E. Evolution Network

The Protein Interaction Network of Yeast two hybrid Uetz et al, Nature 2000

Metabolic Networks Source: Ex. PASy


Gene Regulation Networks

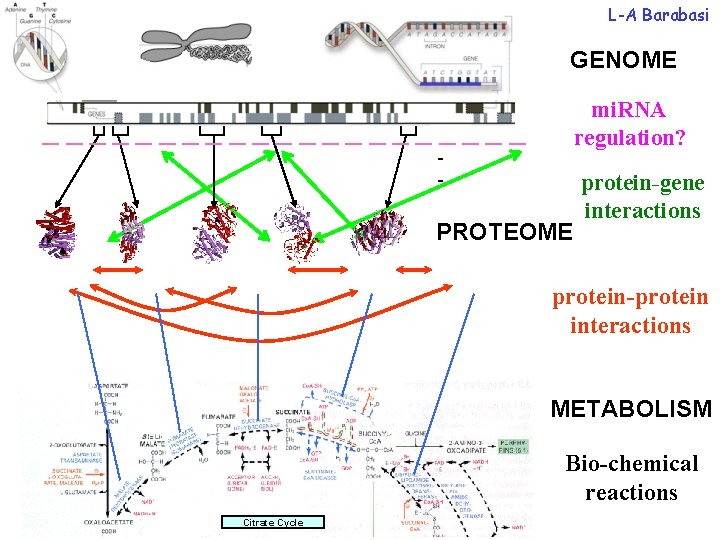
L-A Barabasi GENOME mi. RNA regulation? ___________ - PROTEOME protein-gene interactions protein-protein interactions METABOLISM Bio-chemical reactions Citrate Cycle

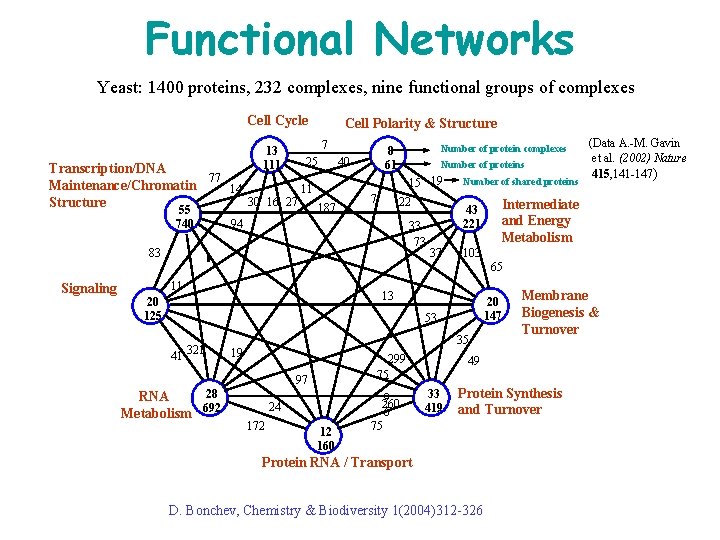
Functional Networks Yeast: 1400 proteins, 232 complexes, nine functional groups of complexes Cell Cycle 13 111 Cell Polarity & Structure 7 25 Transcription/DNA 77 Maintenance/Chromatin 14 11 30 16 27 Structure 187 55 740 Number of protein complexes 8 61 40 Number of proteins 15 19 7 22 94 33 73 83 Number of shared proteins 43 221 37 (Data A. -M. Gavin et al. (2002) Nature 415, 141 -147) Intermediate and Energy Metabolism 103 65 Signaling 11 13 20 125 20 147 53 41 321 35 19 97 28 RNA Metabolism 692 24 172 12 160 299 75 5 9 260 6 75 Membrane Biogenesis & Turnover 49 33 419 Protein Synthesis and Turnover Protein RNA / Transport D. Bonchev, Chemistry & Biodiversity 1(2004)312 -326

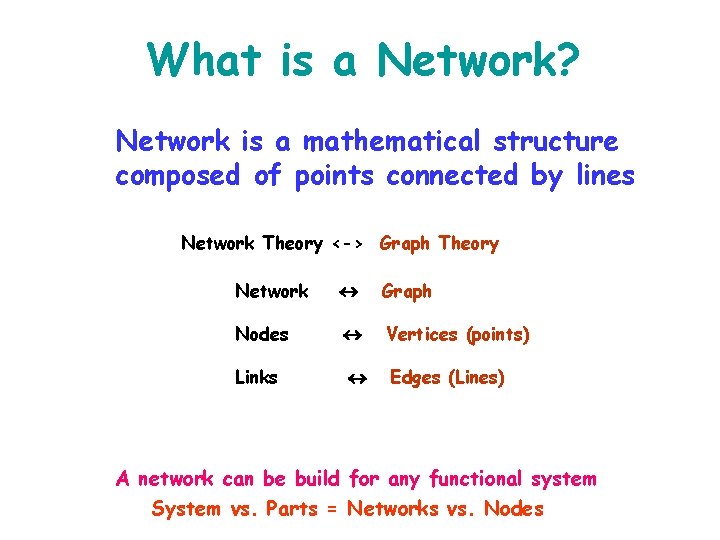
What is a Network? Network is a mathematical structure composed of points connected by lines Network Theory <-> Graph Theory Network Graph Nodes Vertices (points) Links Edges (Lines) A network can be build for any functional system System vs. Parts = Networks vs. Nodes

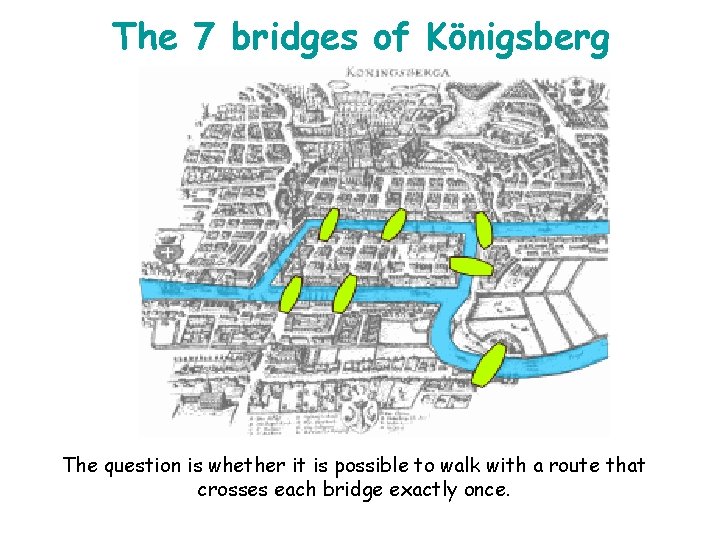
The 7 bridges of Königsberg The question is whether it is possible to walk with a route that crosses each bridge exactly once.

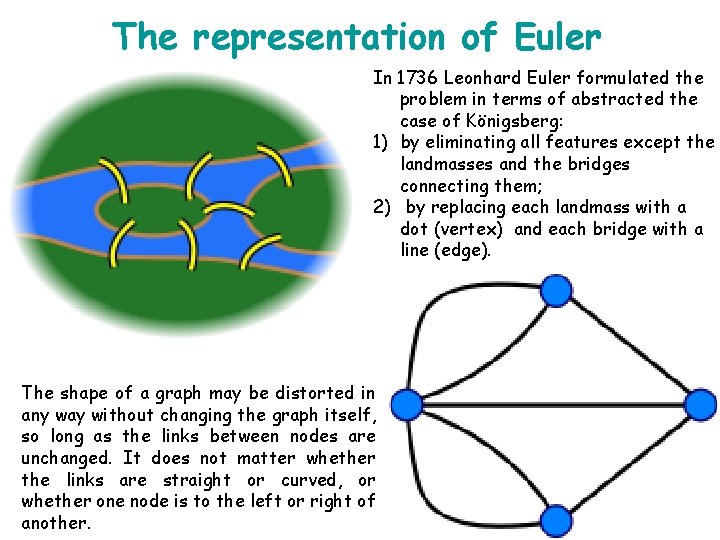
The representation of Euler In 1736 Leonhard Euler formulated the problem in terms of abstracted the case of Königsberg: 1) by eliminating all features except the landmasses and the bridges connecting them; 2) by replacing each landmass with a dot (vertex) and each bridge with a line (edge). The shape of a graph may be distorted in any way without changing the graph itself, so long as the links between nodes are unchanged. It does not matter whether the links are straight or curved, or whether one node is to the left or right of another.

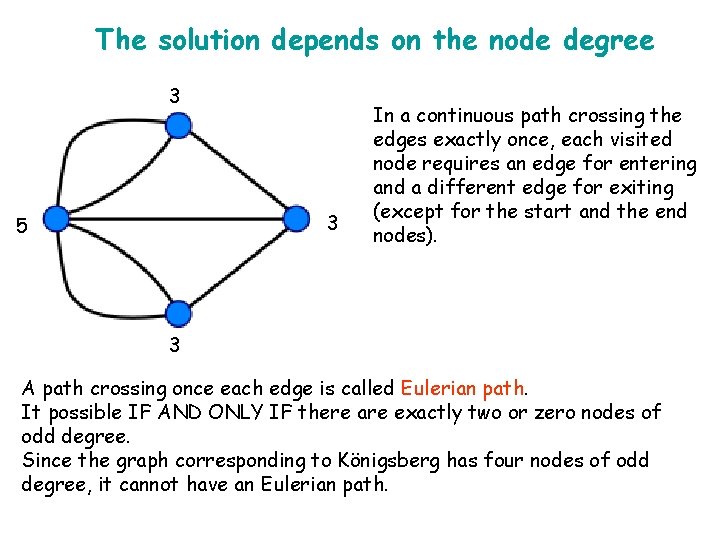
The solution depends on the node degree 3 3 5 In a continuous path crossing the edges exactly once, each visited node requires an edge for entering and a different edge for exiting (except for the start and the end nodes). 3 A path crossing once each edge is called Eulerian path. It possible IF AND ONLY IF there are exactly two or zero nodes of odd degree. Since the graph corresponding to Königsberg has four nodes of odd degree, it cannot have an Eulerian path.

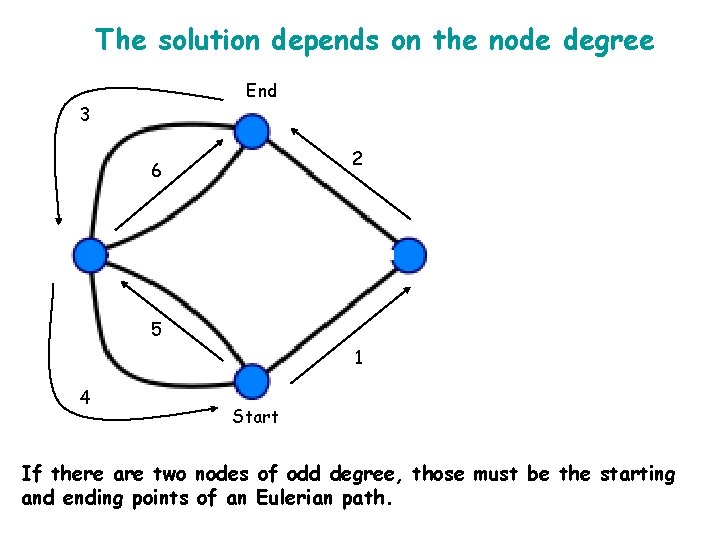
The solution depends on the node degree End 3 2 6 5 1 4 Start If there are two nodes of odd degree, those must be the starting and ending points of an Eulerian path.

Hamiltonian paths Find a path visiting each node exactly one Conditions of existence for Hamiltonian paths are not simple

Hamiltonian paths

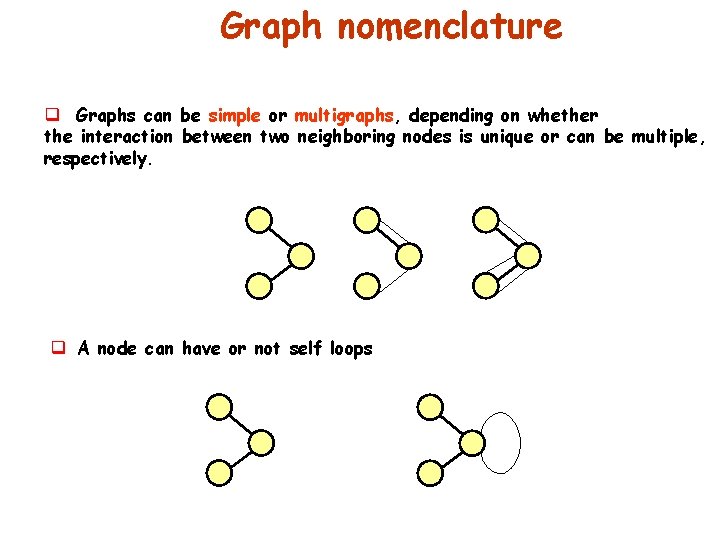
Graph nomenclature q Graphs can be simple or multigraphs, depending on whether the interaction between two neighboring nodes is unique or can be multiple, respectively. q A node can have or not self loops

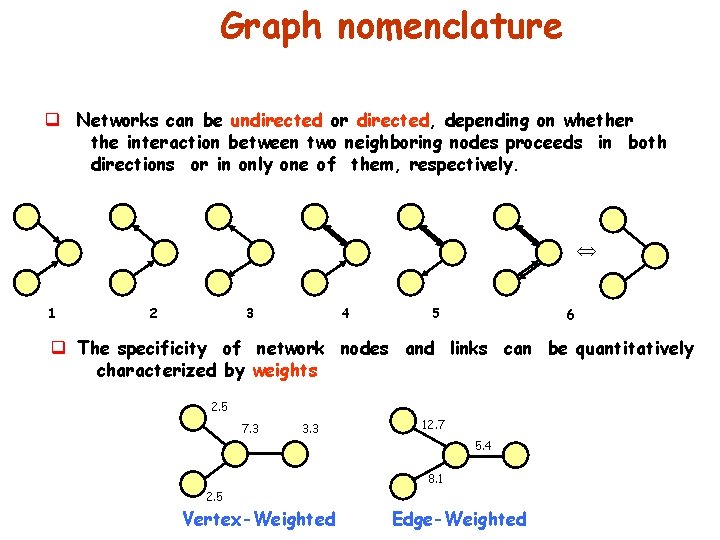
Graph nomenclature q Networks can be undirected or directed, depending on whether the interaction between two neighboring nodes proceeds in both directions or in only one of them, respectively. 1 2 3 4 5 6 q The specificity of network nodes and links can be quantitatively characterized by weights 2. 5 7. 3 3. 3 12. 7 5. 4 8. 1 2. 5 Vertex-Weighted Edge-Weighted

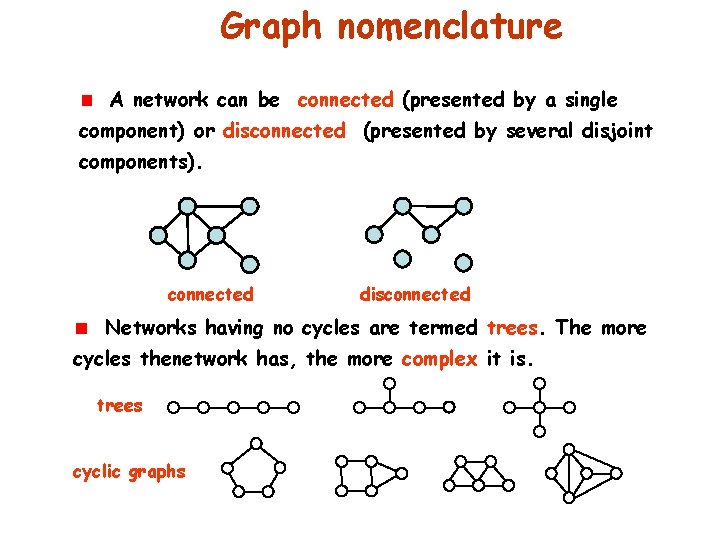
Graph nomenclature A network can be connected (presented by a single component) or disconnected (presented by several disjoint components). connected disconnected Networks having no cycles are termed trees. The more cycles thenetwork has, the more complex it is. trees cyclic graphs

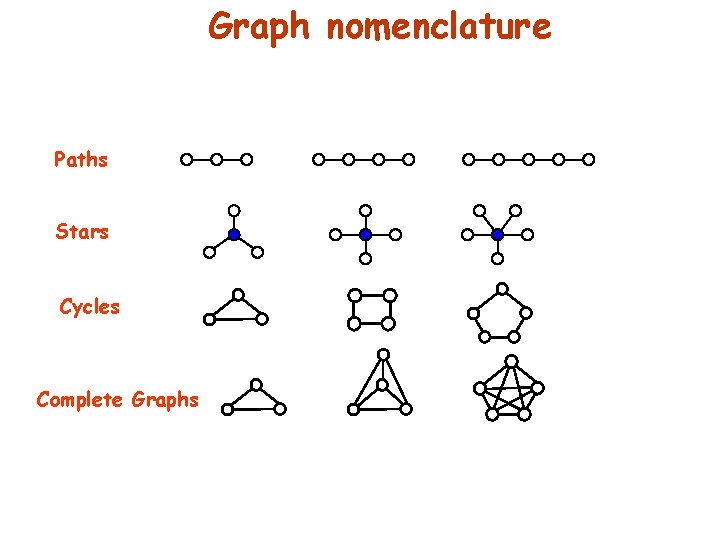
Graph nomenclature Paths Stars Cycles Complete Graphs

Large graphs = Networks

Statistical features of networks • Vertex degree distribution (the degree of a vertex is the number of vertices connected with it via an edge)

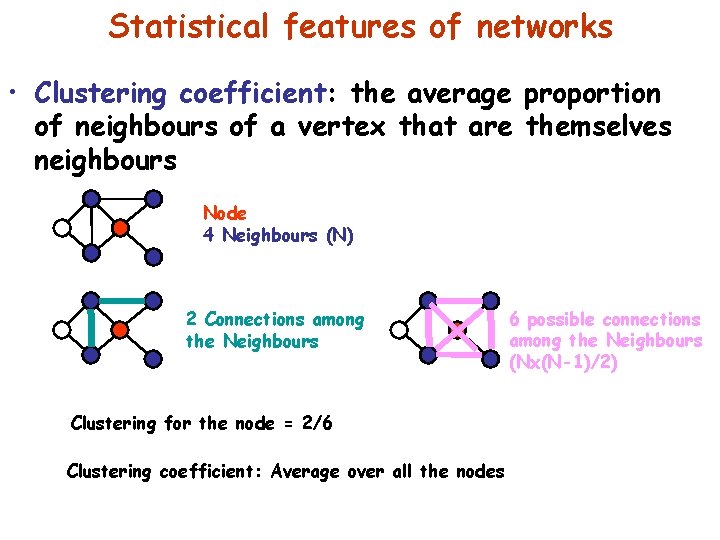
Statistical features of networks • Clustering coefficient: the average proportion of neighbours of a vertex that are themselves neighbours Node 4 Neighbours (N) 2 Connections among the Neighbours Clustering for the node = 2/6 Clustering coefficient: Average over all the nodes 6 possible connections among the Neighbours (Nx(N-1)/2)

Statistical features of networks • Clustering coefficient: the average proportion of neighbours of a vertex that are themselves neighbours C=0 C=0 C=1

Statistical features of networks Given a pair of nodes, compute the shortest path between them • Average shortest distance between two vertices • Diameter: maximal shortest distance How many degrees of separation are they between two random people in the world, when friendship networks are considered?

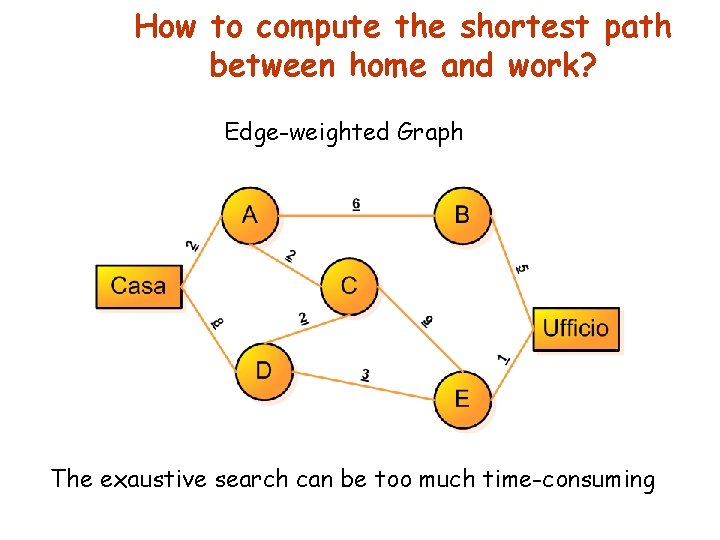
How to compute the shortest path between home and work? Edge-weighted Graph The exaustive search can be too much time-consuming

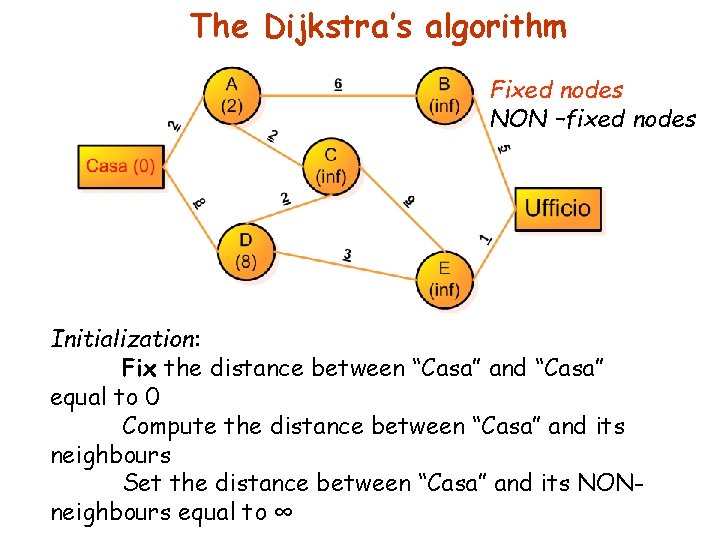
The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Initialization: Fix the distance between “Casa” and “Casa” equal to 0 Compute the distance between “Casa” and its neighbours Set the distance between “Casa” and its NONneighbours equal to ∞

The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Iteration (1): Search the node with the minimum distance among the NON-fixed nodes and Fix its distance, memorizing the incoming direction

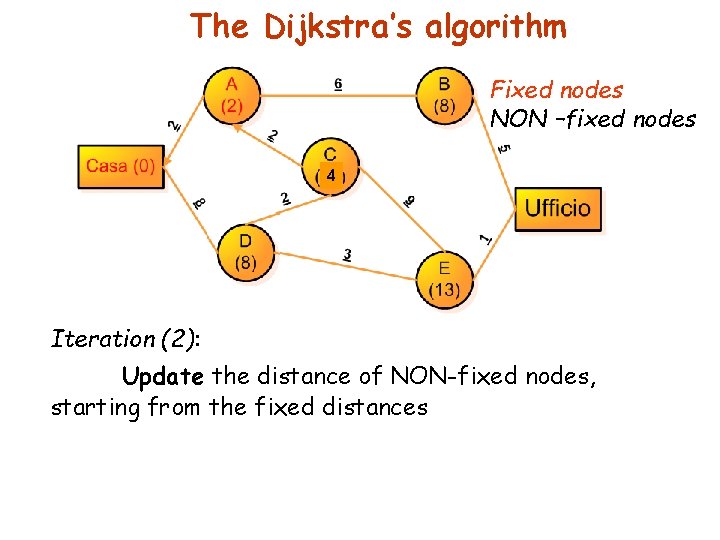
The Dijkstra’s algorithm Fixed nodes NON –fixed nodes 4 Iteration (2): Update the distance of NON-fixed nodes, starting from the fixed distances

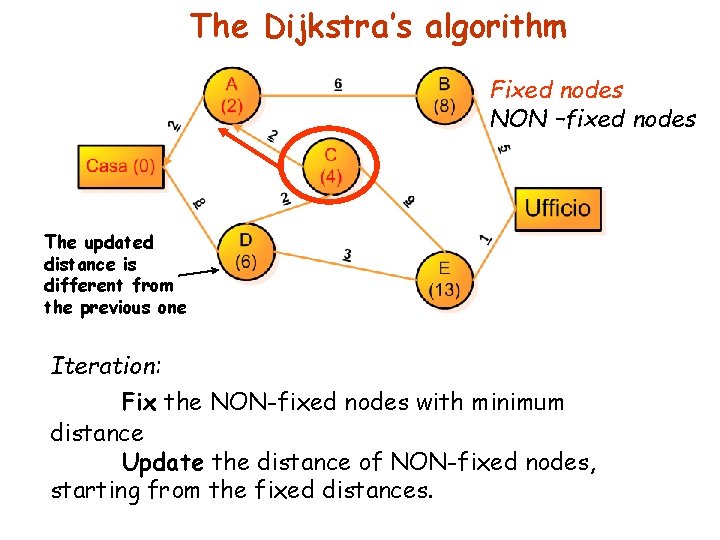
The Dijkstra’s algorithm Fixed nodes NON –fixed nodes The updated distance is different from the previous one Iteration: Fix the NON-fixed nodes with minimum distance Update the distance of NON-fixed nodes, starting from the fixed distances.

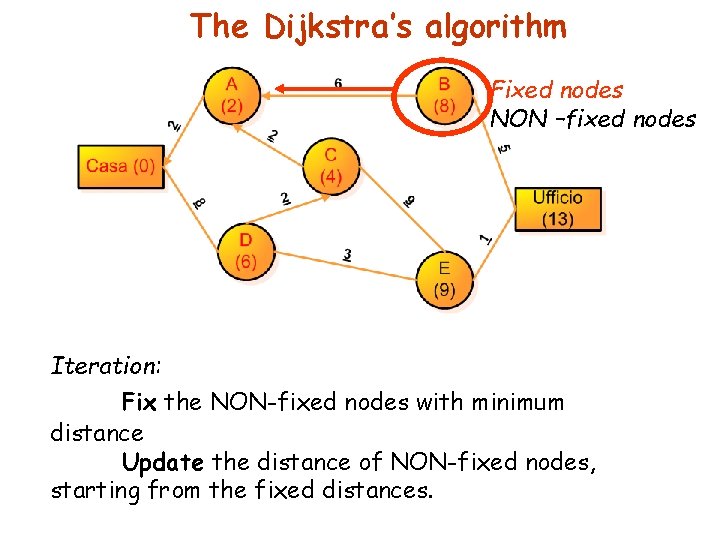
The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Iteration: Fix the NON-fixed nodes with minimum distance Update the distance of NON-fixed nodes, starting from the fixed distances.

The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Iteration: Fix the NON-fixed nodes with minimum distance Update the distance of NON-fixed nodes, starting from the fixed distances.

The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Iteration: Fix the NON-fixed nodes with minimum distance Update the distance of NON-fixed nodes, starting from the fixed distances.

The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Iteration: Fix the NON-fixed nodes with minimum distance Update the distance of NON-fixed nodes, starting from the fixed distances.

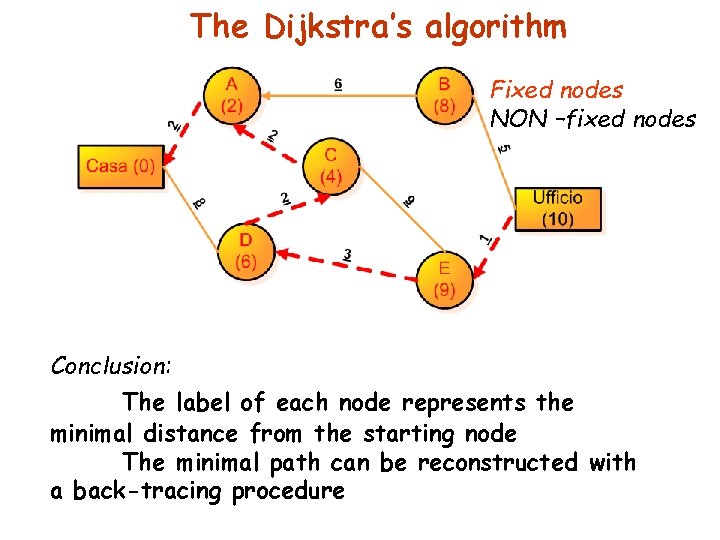
The Dijkstra’s algorithm Fixed nodes NON –fixed nodes Conclusion: The label of each node represents the minimal distance from the starting node The minimal path can be reconstructed with a back-tracing procedure

Statistical features of networks • Vertex degree distribution • Clustering coefficient • Average shortest distance between two vertices • Diameter: maximal shortest distance

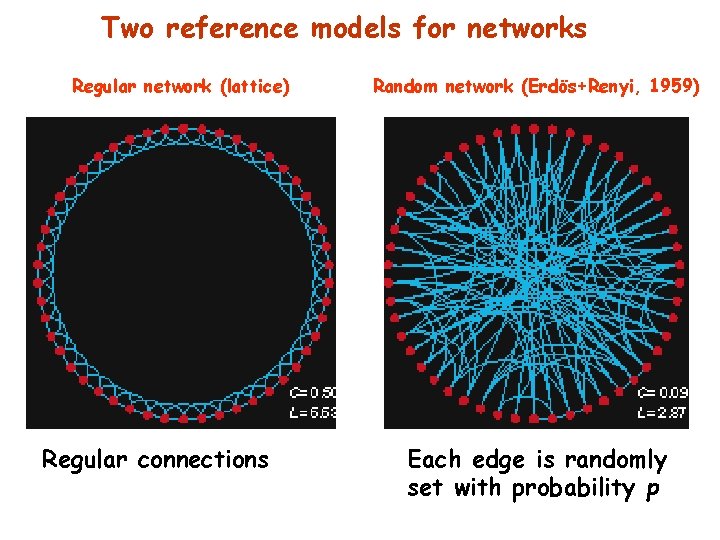
Two reference models for networks Regular network (lattice) Regular connections Random network (Erdös+Renyi, 1959) Each edge is randomly set with probability p

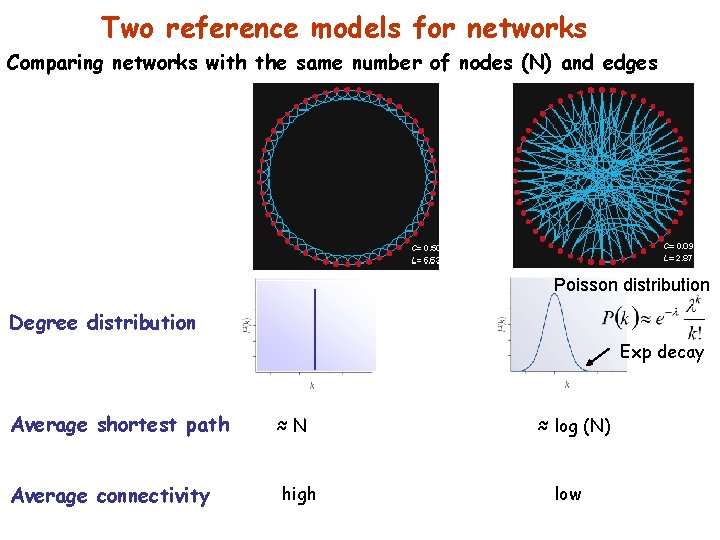
Two reference models for networks Comparing networks with the same number of nodes (N) and edges Poisson distribution Degree distribution Exp decay Average shortest path Average connectivity ≈N high ≈ log (N) low

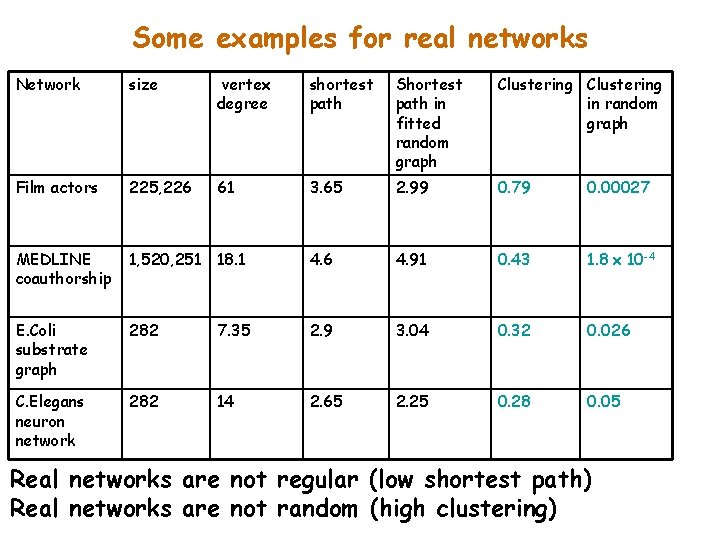
Some examples for real networks Network size vertex degree shortest path Shortest path in fitted random graph Clustering in random graph Film actors 225, 226 61 3. 65 2. 99 0. 79 0. 00027 MEDLINE coauthorship 1, 520, 251 18. 1 4. 6 4. 91 0. 43 1. 8 x 10 -4 E. Coli substrate graph 282 7. 35 2. 9 3. 04 0. 32 0. 026 C. Elegans neuron network 282 14 2. 65 2. 25 0. 28 0. 05 Real networks are not regular (low shortest path) Real networks are not random (high clustering)

Adding randomness in a regular network Random changes in edges OR Addition of random links

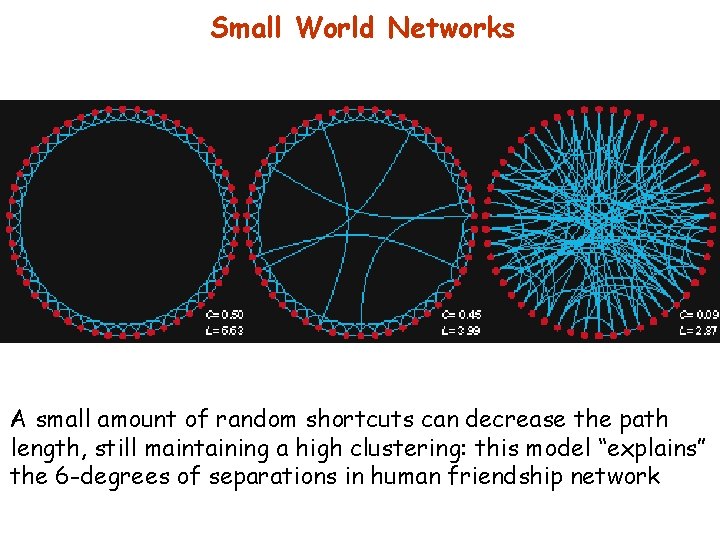
Adding randomness in a regular network (rewiring) Networks with high clustering (like regular ones) and low path length (like random ones) can be obtained: SMALL WORLD NETWORKS (Strogatz and Watts, 1999)

Small World Networks A small amount of random shortcuts can decrease the path length, still maintaining a high clustering: this model “explains” the 6 -degrees of separations in human friendship network

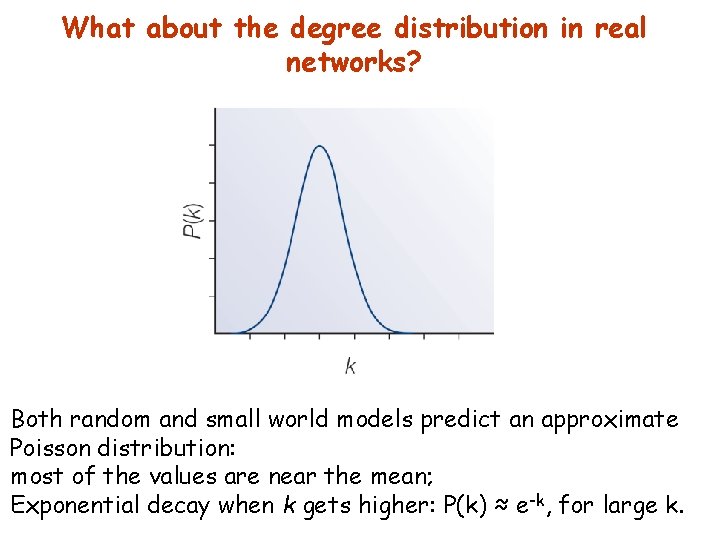
What about the degree distribution in real networks? Both random and small world models predict an approximate Poisson distribution: most of the values are near the mean; Exponential decay when k gets higher: P(k) ≈ e-k, for large k.

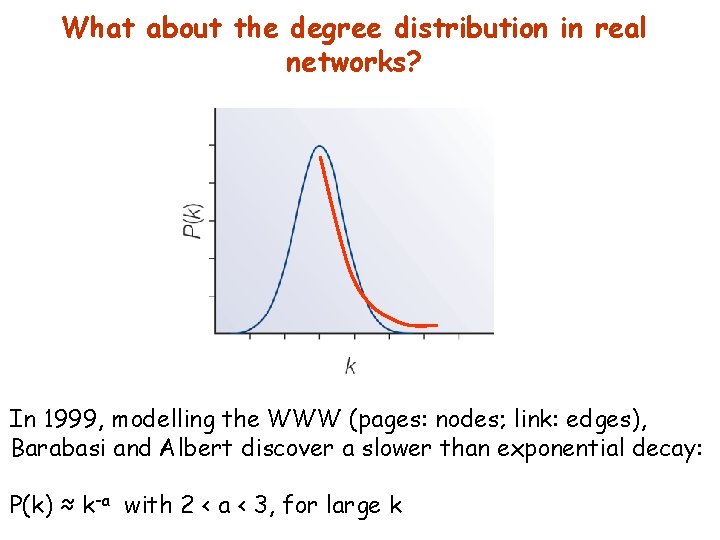
What about the degree distribution in real networks? In 1999, modelling the WWW (pages: nodes; link: edges), Barabasi and Albert discover a slower than exponential decay: P(k) ≈ k-a with 2 < a < 3, for large k

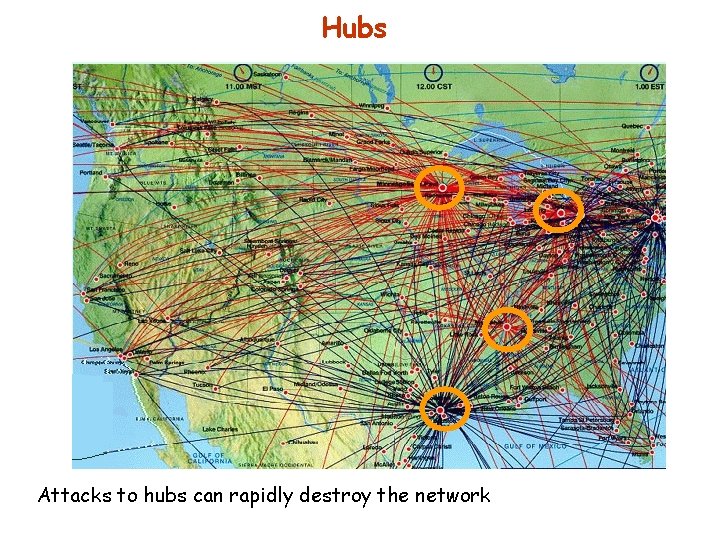
Scale-free networks Networks that are characterized by a power-law degree distribution are highly non-uniform: most of the nodes have only a few links. A few nodes with a very large number of links, which are often called hubs, hold these nodes together. Networks with a power degree distribution are called scale-free hubs It is the same distribution of wealth following Pareto’s 20 -80 law: Few people (20%) possess most of the wealth (80%), most of the people (80%) possess the rest (20%)

Hubs Attacks to hubs can rapidly destroy the network

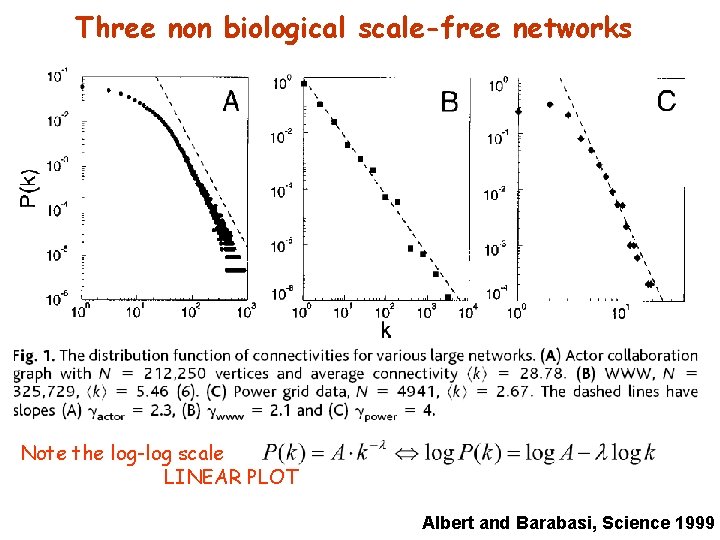
Three non biological scale-free networks Note the log-log scale LINEAR PLOT Albert and Barabasi, Science 1999

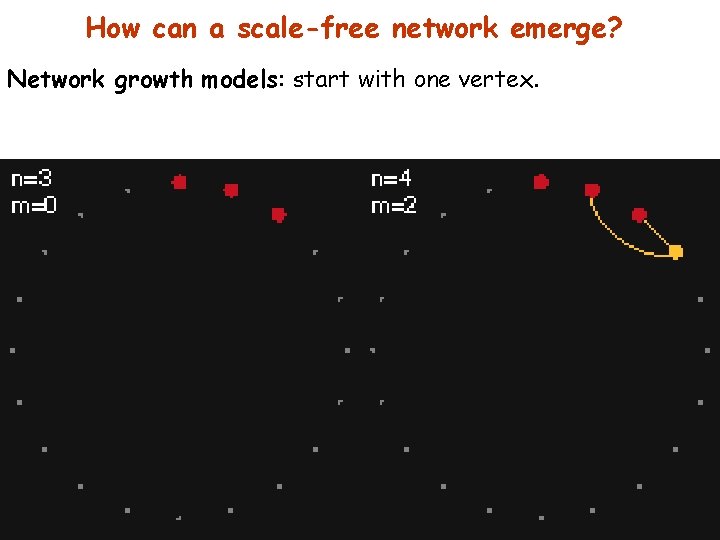
How can a scale-free network emerge? Network growth models: start with one vertex.

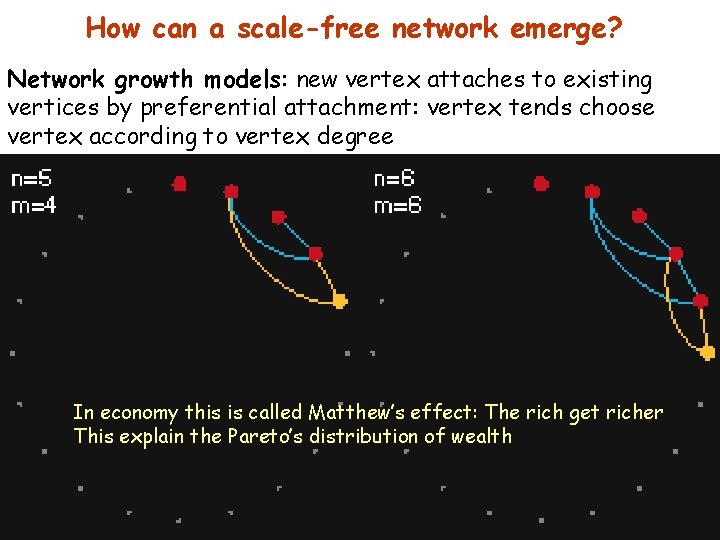
How can a scale-free network emerge? Network growth models: new vertex attaches to existing vertices by preferential attachment: vertex tends choose vertex according to vertex degree In economy this is called Matthew’s effect: The rich get richer This explain the Pareto’s distribution of wealth

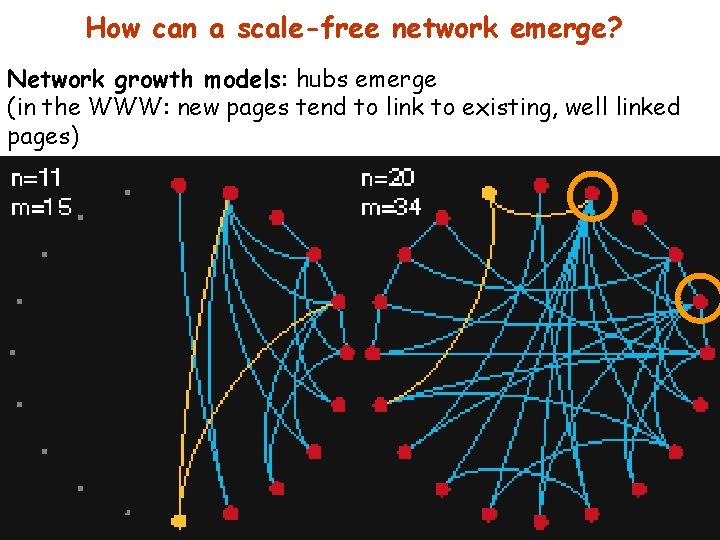
How can a scale-free network emerge? Network growth models: hubs emerge (in the WWW: new pages tend to link to existing, well linked pages)

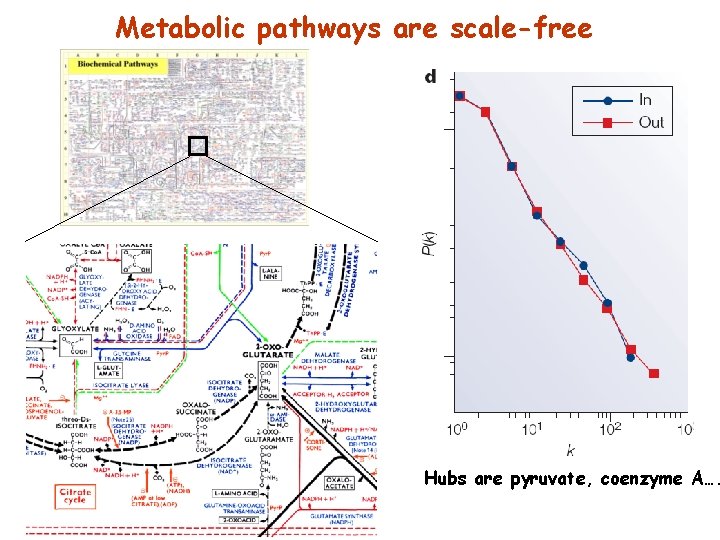
Metabolic pathways are scale-free Hubs are pyruvate, coenzyme A….

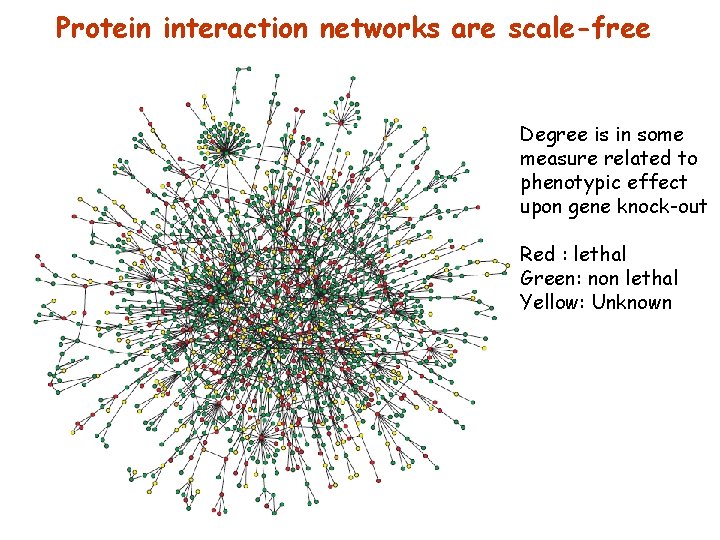
Protein interaction networks are scale-free Degree is in some measure related to phenotypic effect upon gene knock-out Red : lethal Green: non lethal Yellow: Unknown

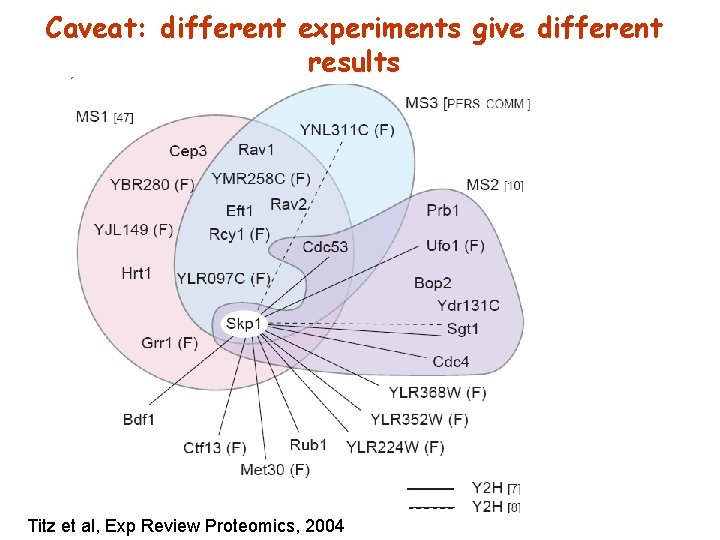
Caveat: different experiments give different results Titz et al, Exp Review Proteomics, 2004

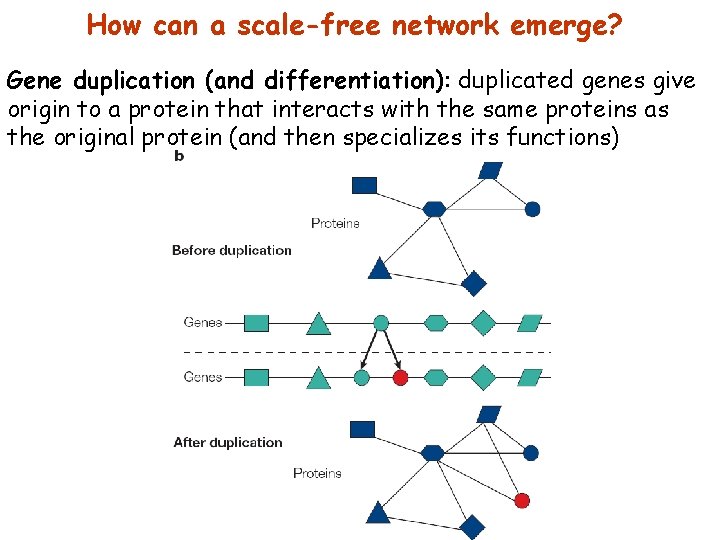
How can a scale-free network emerge? Gene duplication (and differentiation): duplicated genes give origin to a protein that interacts with the same proteins as the original protein (and then specializes its functions)

Caveat on the use of the scale-free theory The same noisy data can be fitted in different ways A sub-net of a non-free-scale network can have a scale-free behaviour Finding a scale-free behaviour do NOT imply the growth with preferential attachment mechanism Keller, Bio. Essays 2006

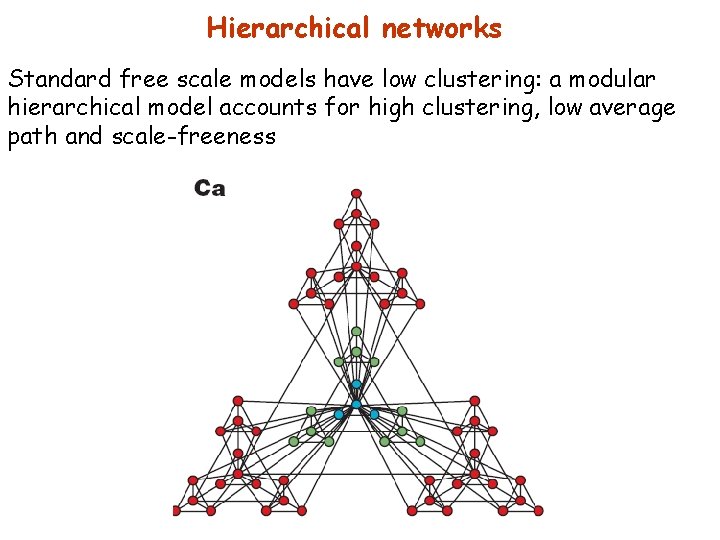
Hierarchical networks Standard free scale models have low clustering: a modular hierarchical model accounts for high clustering, low average path and scale-freeness

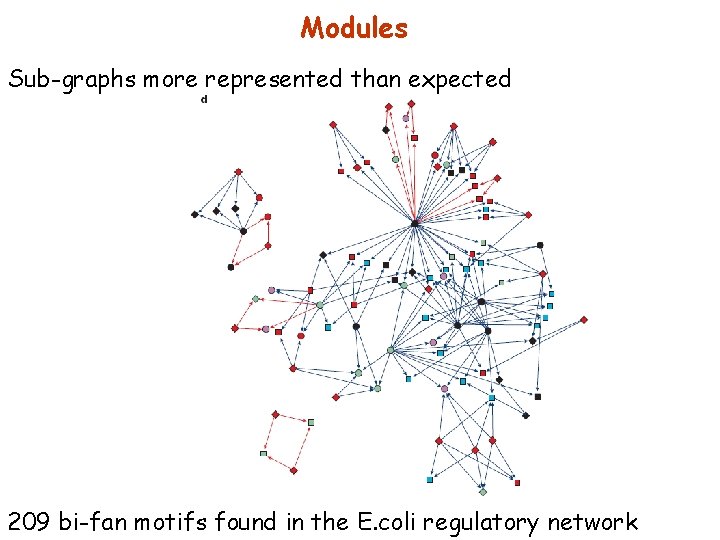
Modules Sub-graphs more represented than expected 209 bi-fan motifs found in the E. coli regulatory network

Summary v Many complex networks in nature and technology have common features. v They differ considerably from random networks of the same size v By studying network structure and dynamics, and by using comparative network analysis, one can get answers of important biological questions.

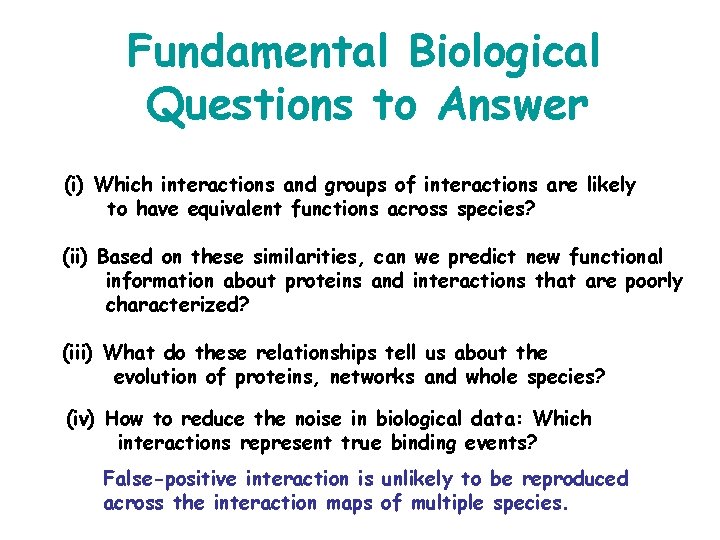
Fundamental Biological Questions to Answer (i) Which interactions and groups of interactions are likely to have equivalent functions across species? (ii) Based on these similarities, can we predict new functional information about proteins and interactions that are poorly characterized? (iii) What do these relationships tell us about the evolution of proteins, networks and whole species? (iv) How to reduce the noise in biological data: Which interactions represent true binding events? False-positive interaction is unlikely to be reproduced across the interaction maps of multiple species.

Barabasi and Oltvai (2004) Network Biology: understanding the cell’s functional organization. Nature Reviews Genetics 5: 101 -113 Stogatz (2001) Exploring complex networks. Nature 410: 268 -276 Hayes (2000) Graph theory in practice. American Scientist 88: 9 -13/104 -109 Mason and Verwoerd (2006) Graph theory and networks in Biology Keller (2005) Revisiting scale-free networks. Bio. Essays 27. 10: 1060 -1068
 Perché vediamo cose nelle nuvole
Perché vediamo cose nelle nuvole Sistemi elettronici integrati
Sistemi elettronici integrati Sistemi elettronici integrati
Sistemi elettronici integrati Circuiti integrati logici
Circuiti integrati logici Irigem bassano del grappa
Irigem bassano del grappa Cescot rimini corsi
Cescot rimini corsi Codici classroom unife medicina corsi a scelta
Codici classroom unife medicina corsi a scelta Massaggio tantra nudi
Massaggio tantra nudi Corsi team formation
Corsi team formation Scuola dello sport campania
Scuola dello sport campania Válvula ileocecal
Válvula ileocecal Mato grosso plateau physical map
Mato grosso plateau physical map A grosso has a group of soloists rather than just one
A grosso has a group of soloists rather than just one Massacre da baía do garcez
Massacre da baía do garcez Qual a função do intestino delgado
Qual a função do intestino delgado Bandeira do mato grosso do sul
Bandeira do mato grosso do sul Cabo coaxial fino e grosso
Cabo coaxial fino e grosso Miositis por cuerpos de inclusion
Miositis por cuerpos de inclusion Concerto grosso definicion
Concerto grosso definicion Rondone codacuta
Rondone codacuta Vsg
Vsg Definition concerto grosso
Definition concerto grosso Graciela grosso
Graciela grosso Quais são as categorias da narrativa
Quais são as categorias da narrativa Presente imperfecto
Presente imperfecto Recuerda es un verbo
Recuerda es un verbo Modo de transmissão
Modo de transmissão Indicar subjuntivo
Indicar subjuntivo Subjuntivo ejemplos
Subjuntivo ejemplos Modo de transmissão
Modo de transmissão Weirdo subjunctive
Weirdo subjunctive Preterito imperfecto subjuntivo
Preterito imperfecto subjuntivo Macroestructura del texto expositivo
Macroestructura del texto expositivo Condizionale semplice
Condizionale semplice Kazi modo
Kazi modo Ojalá es adverbio
Ojalá es adverbio Modo indicativo e subjuntivo
Modo indicativo e subjuntivo Chegamos muito tarde para a aula
Chegamos muito tarde para a aula Modo condiciones y estilo de vida
Modo condiciones y estilo de vida Modo geocentrico
Modo geocentrico Tutorial de pseint
Tutorial de pseint Concordancia de los tiempos
Concordancia de los tiempos Concepto de un verbo
Concepto de un verbo Modo de transmissão
Modo de transmissão Estilo narrativo
Estilo narrativo Modo de rezar el rosario
Modo de rezar el rosario Formas nominais do verbo
Formas nominais do verbo Que son los modos de discurso
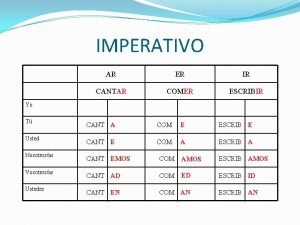
Que son los modos de discurso Imperativo cantar
Imperativo cantar Ejemplos de un monologo
Ejemplos de un monologo Modi verbali
Modi verbali Libros de fon?tica ac?stica inglesa
Libros de fon?tica ac?stica inglesa El capitalismo cuando surge
El capitalismo cuando surge Verbo gerundio
Verbo gerundio Clases sociales del modo de producción esclavista
Clases sociales del modo de producción esclavista Elementos de la función exponencial
Elementos de la función exponencial Tiempo condicional
Tiempo condicional Verbos en presente
Verbos en presente Vertical
Vertical Modo de organización discursiva
Modo de organización discursiva Adverbio de negación
Adverbio de negación Frasi al passato
Frasi al passato Modo de produção primitivo
Modo de produção primitivo Dialogico concepto
Dialogico concepto Locuzioni
Locuzioni Campo tenor y modo halliday
Campo tenor y modo halliday Legat damnacyjny
Legat damnacyjny