DNA structure and replication Hershey and Chase experiments











- Slides: 50

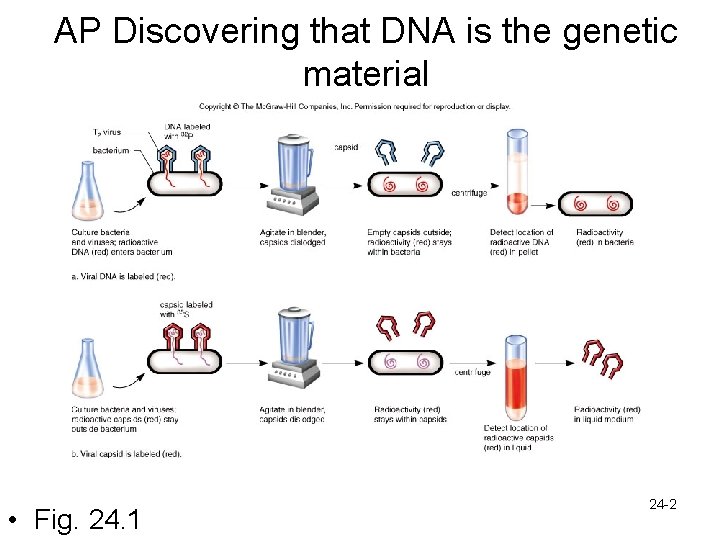
DNA structure and replication • Hershey and Chase experiments – Determined that DNA is the genetic material – Experiment 1 • Viruses with DNA labeled with 32 P were incubated with E. coli – Mixed in a blender to remove virus particles attached to cells – Centrifuged so bacteria formed a pellet • Results- viral DNA was inside the bacteria – Experiment 2 • Viral proteins in capsids were labeled with 35 S and viruses were incubated with E. coli – Mixed in blender and centrifuged • Results- labeled proteins were washed off with the capsids and were not inside the bacteria 24 -1

AP Discovering that DNA is the genetic material • Fig. 24. 1 24 -2

Biology 12 : DNA Structure, Function & Replication 24 -3

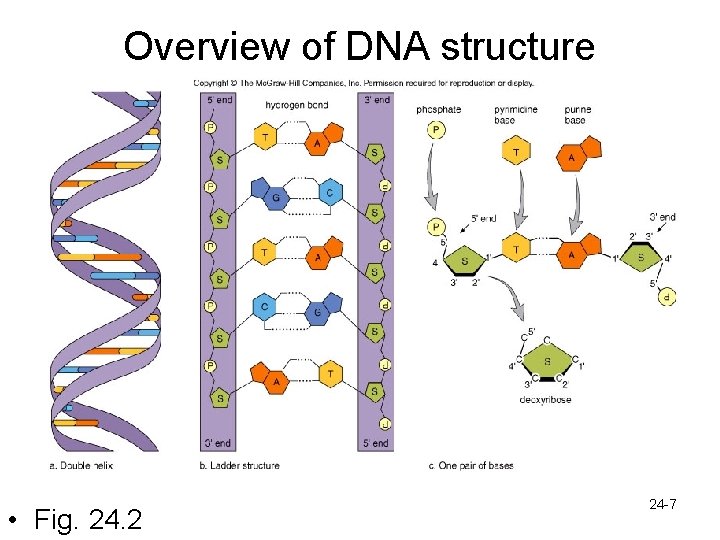
Structure of DNA – Determined by Watson and Crick • Double helix • Composed of monomers called nucleotides • Each nucleotide has 3 parts – phosphoric acid (phosphate) – deoxyribose sugar – Nitrogen base- 4 possible » Adenine and guanine: purine bases- double ring structure » Cytosine and thymine: pyrimidine bases-single ring structure 24 -4

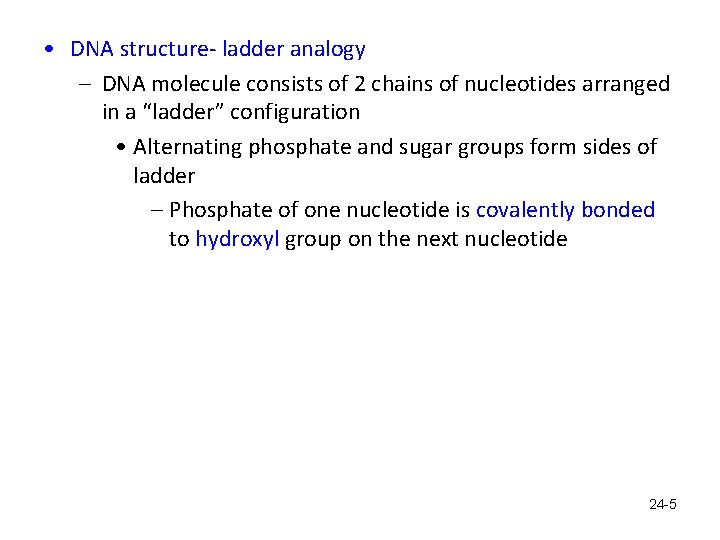
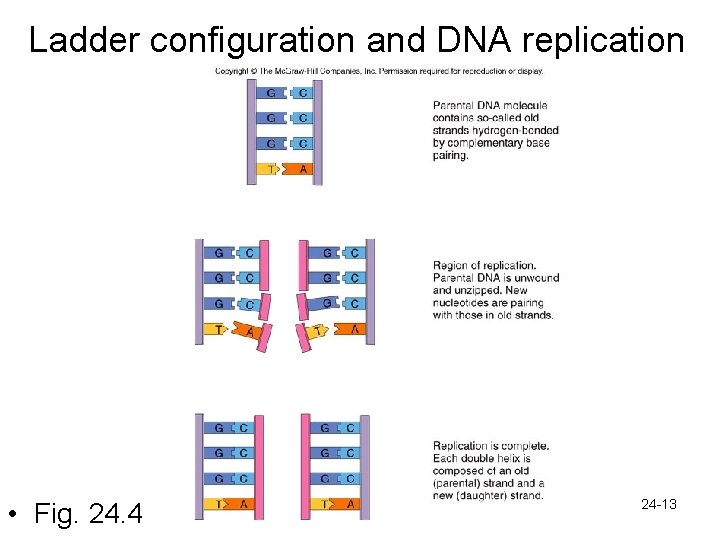
• DNA structure- ladder analogy – DNA molecule consists of 2 chains of nucleotides arranged in a “ladder” configuration • Alternating phosphate and sugar groups form sides of ladder – Phosphate of one nucleotide is covalently bonded to hydroxyl group on the next nucleotide 24 -5

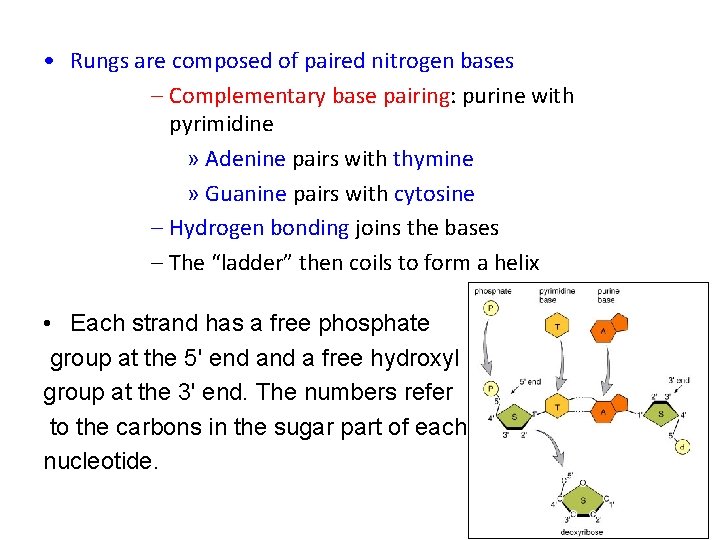
• Rungs are composed of paired nitrogen bases – Complementary base pairing: purine with pyrimidine » Adenine pairs with thymine » Guanine pairs with cytosine – Hydrogen bonding joins the bases – The “ladder” then coils to form a helix • Each strand has a free phosphate group at the 5' end a free hydroxyl group at the 3' end. The numbers refer to the carbons in the sugar part of each nucleotide. 24 -6

Overview of DNA structure • Fig. 24. 2 24 -7

Stop and Check 1. Which two nitrogenous bases are purines? 2. Which of the following describes the base pairing in a DNA molecule? a) b) c) d) A binds with T by forming two H-bonds C binds with G by forming two H-bonds A binds with G by forming 3 H-bonds C binds with T by forming 3 H-bonds 24 -8

Answers 1. Adenine and Guanine 2. A 24 -9

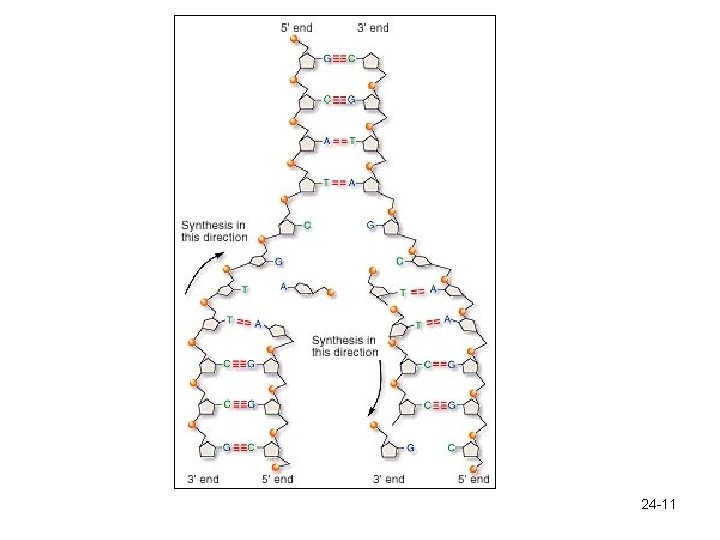
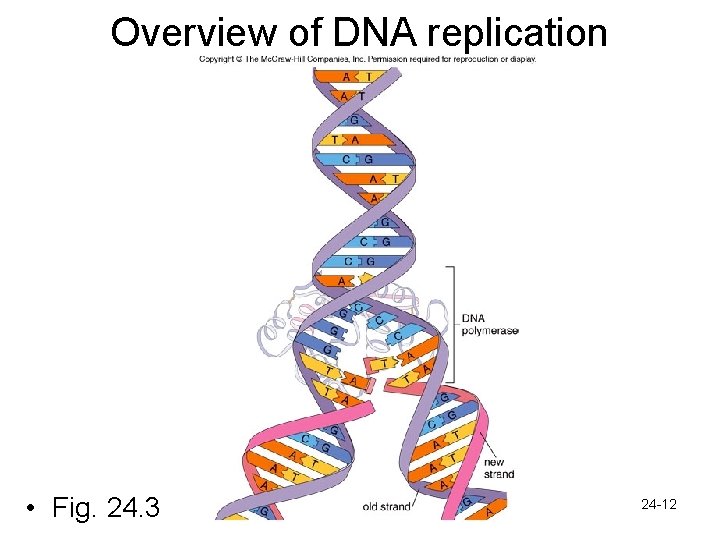
DNA Replication – Each strand acts as a template for a new strand. – DNA strands separate • Helicase unwinds and unzips DNA - separates the hydrogen bonds – Free, unattached nucleotides pair with their complementary bases through Hydrogen bonding. – DNA polymerase (-ase = enzyme) adds nucleotides to the newly forming strand from the 3’ end. – Two DNA strands are formed, each containing one strand from the old DNA and one from the newly formed strand. – Semi-conservative replication 24 -10

24 -11

Overview of DNA replication • Fig. 24. 3 24 -12

Ladder configuration and DNA replication • Fig. 24. 4 24 -13

Homework 1. Read pages 114 -116. You NEED to know this like you know your name in order to grasp the rest of the unit. 2. DNA structure and Replication Worksheet. Questions 46 -49 p. 151 24 -14

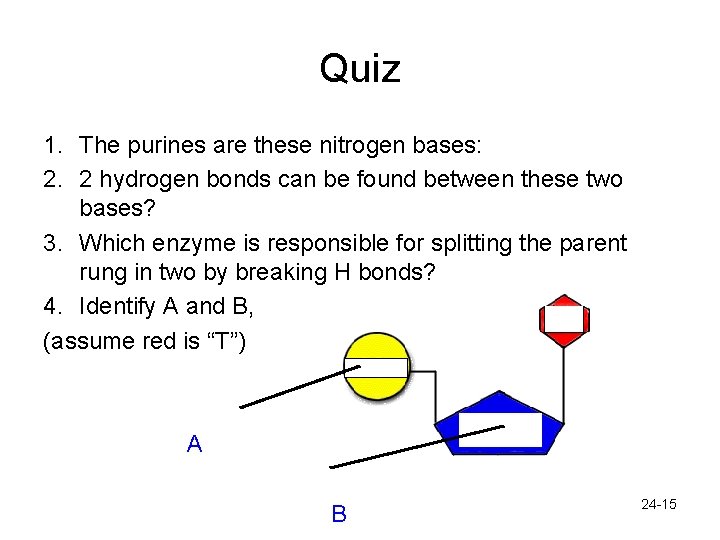
Quiz 1. The purines are these nitrogen bases: 2. 2 hydrogen bonds can be found between these two bases? 3. Which enzyme is responsible for splitting the parent rung in two by breaking H bonds? 4. Identify A and B, (assume red is “T”) A B 24 -15

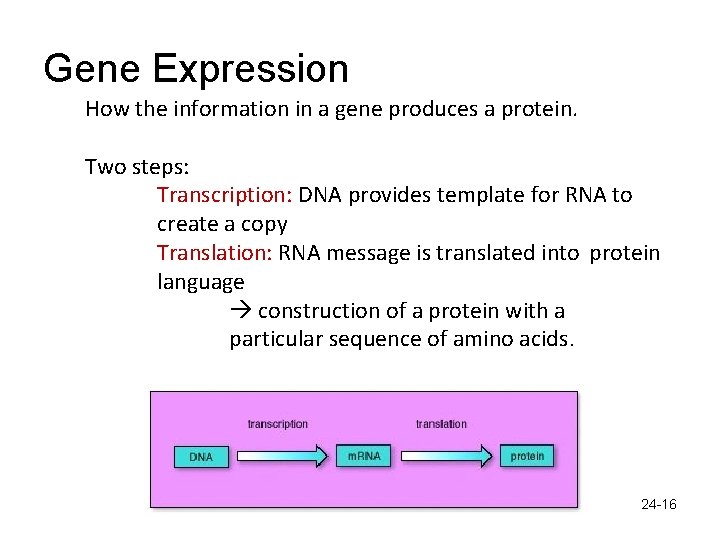
Gene Expression How the information in a gene produces a protein. Two steps: Transcription: DNA provides template for RNA to create a copy Translation: RNA message is translated into protein language construction of a protein with a particular sequence of amino acids. 24 -16

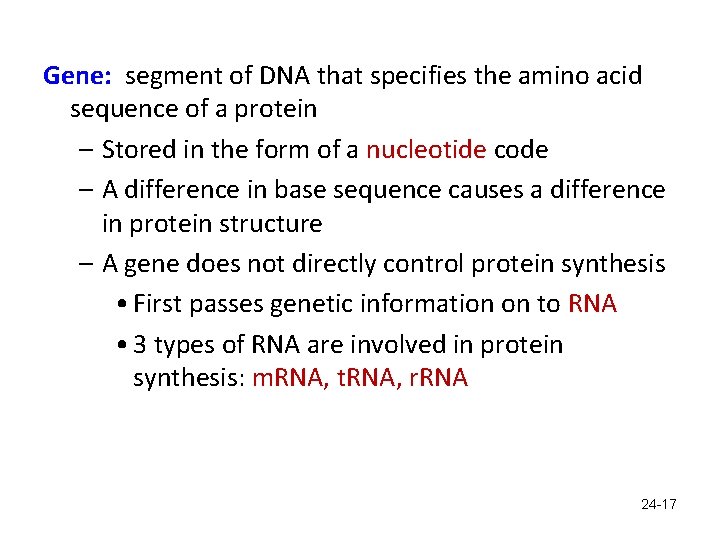
Gene: segment of DNA that specifies the amino acid sequence of a protein – Stored in the form of a nucleotide code – A difference in base sequence causes a difference in protein structure – A gene does not directly control protein synthesis • First passes genetic information on to RNA • 3 types of RNA are involved in protein synthesis: m. RNA, t. RNA, r. RNA 24 -17

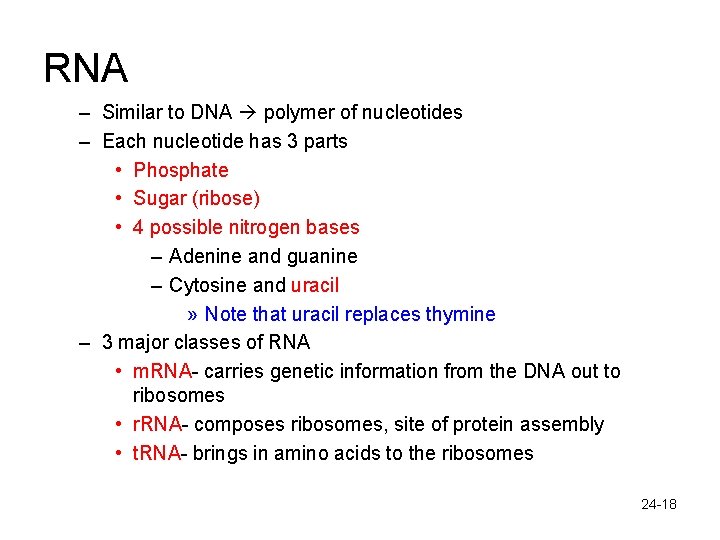
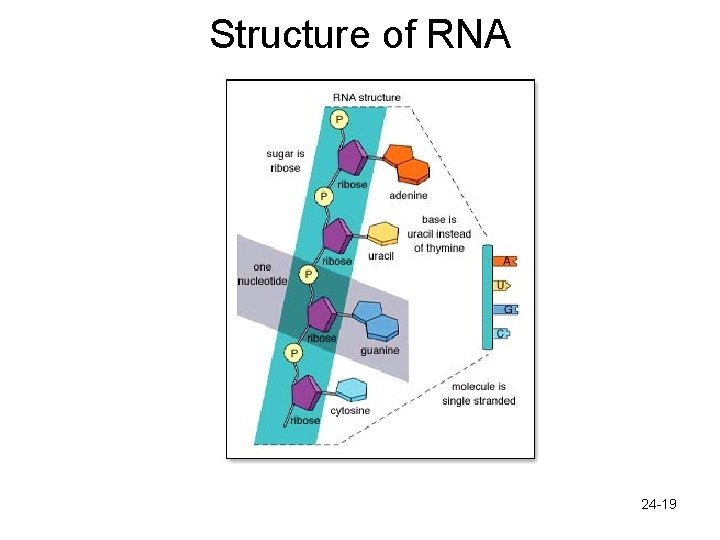
RNA – Similar to DNA polymer of nucleotides – Each nucleotide has 3 parts • Phosphate • Sugar (ribose) • 4 possible nitrogen bases – Adenine and guanine – Cytosine and uracil » Note that uracil replaces thymine – 3 major classes of RNA • m. RNA- carries genetic information from the DNA out to ribosomes • r. RNA- composes ribosomes, site of protein assembly • t. RNA- brings in amino acids to the ribosomes 24 -18

Structure of RNA 24 -19

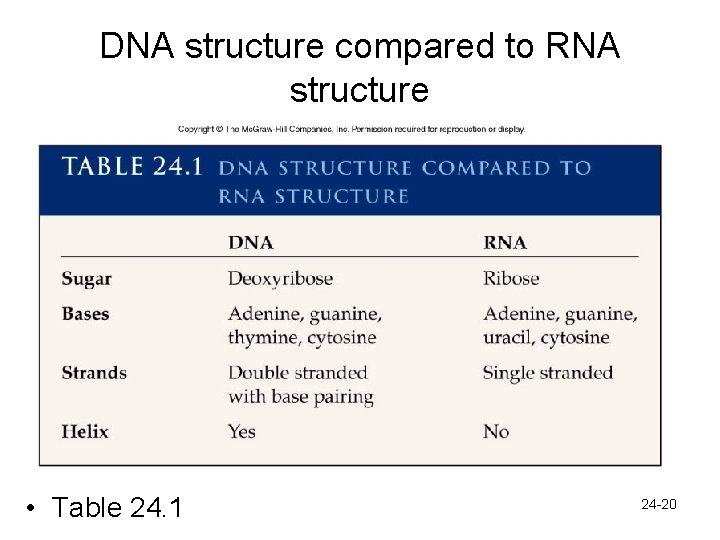
DNA structure compared to RNA structure • Table 24. 1 24 -20

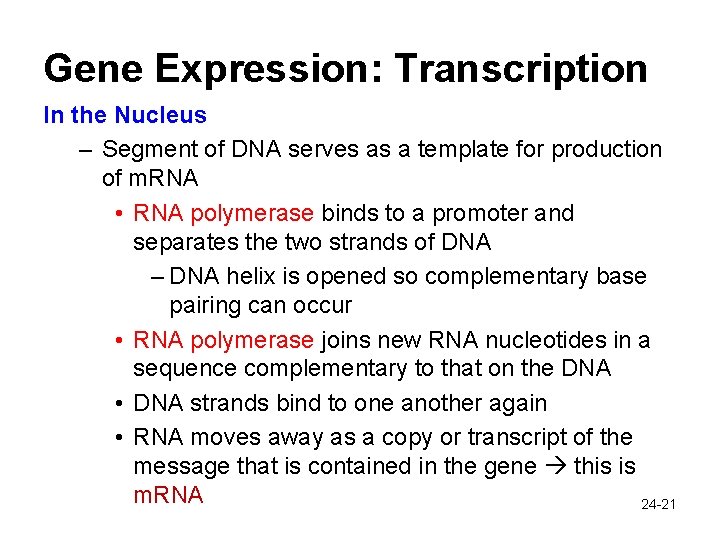
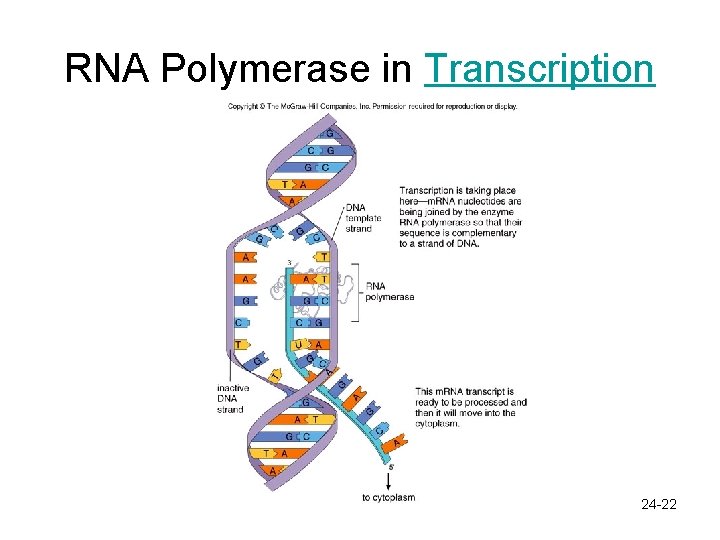
Gene Expression: Transcription In the Nucleus – Segment of DNA serves as a template for production of m. RNA • RNA polymerase binds to a promoter and separates the two strands of DNA – DNA helix is opened so complementary base pairing can occur • RNA polymerase joins new RNA nucleotides in a sequence complementary to that on the DNA • DNA strands bind to one another again • RNA moves away as a copy or transcript of the message that is contained in the gene this is m. RNA 24 -21

RNA Polymerase in Transcription 24 -22

Homework: 1. Read pages 117 -118, “Gene Expression” and “Transcription” 2. Complete Transcription worksheet. 3. Questions page 148 # 1 -6, 10, 22, 45 24 -23

Warm up Quiz 1. What is the base that is unique to RNA? 2. What would be the m. RNA code for GATCCT? 3. What is the end product of the process of transcription? 4. Why do cells need to undergo transcription? (why doesn’t DNA directly result in a particular protein). 24 -24

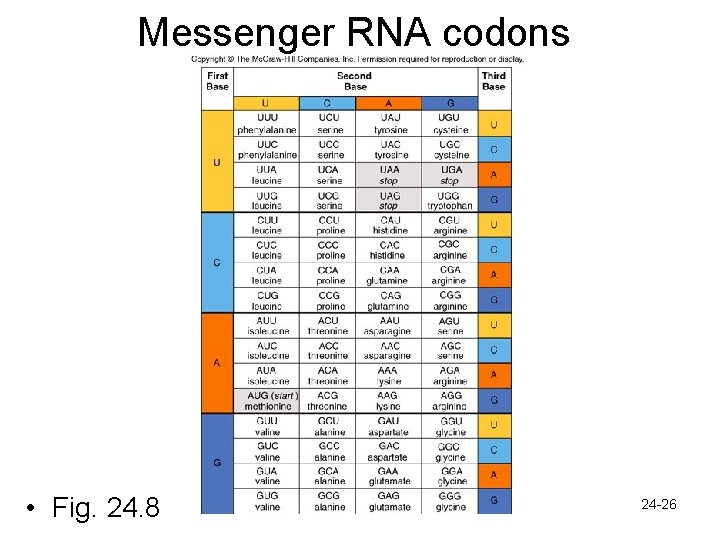
Gene Expression: Genetic Code • Translating RNA to proteins requires the genetic code • Triplet code- each 3 -nucleotide unit of a m. RNA molecule is called a codon • There are 64 different m. RNA codons – 61 code for particular amino acids » Redundant code-some amino acids have numerous code words » Provides some protection against mutations – 3 are stop codons signal polypeptide termination – One codon stands for methionine - signals polypeptide initiation (start codon) 24 -25

Messenger RNA codons • Fig. 24. 8 24 -26

Gene Expression: from m. RNA to Protein Synthesis • Translation – m. RNA carries genetic message from the DNA in the nucleus, through the cytoplasm to the ribosome. – Process where information carried in a sequence of bases on m. RNA is used to direct synthesis of a polypeptide 24 -27

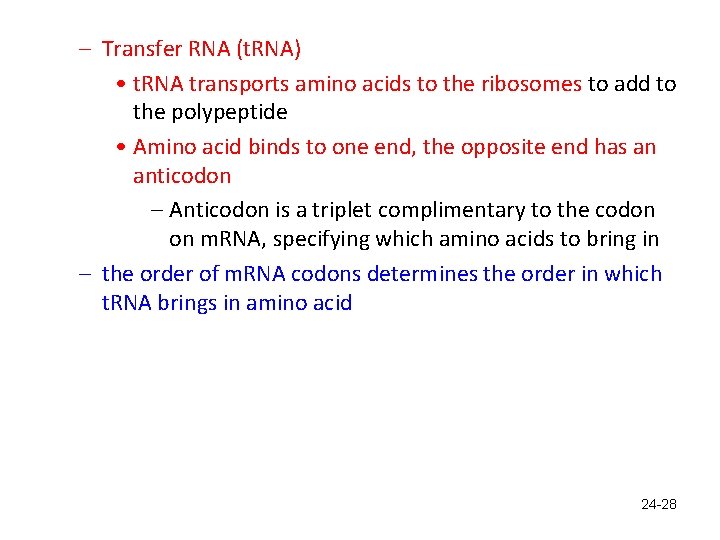
– Transfer RNA (t. RNA) • t. RNA transports amino acids to the ribosomes to add to the polypeptide • Amino acid binds to one end, the opposite end has an anticodon – Anticodon is a triplet complimentary to the codon on m. RNA, specifying which amino acids to bring in – the order of m. RNA codons determines the order in which t. RNA brings in amino acid 24 -28

Transfer RNA; amino acid carrier • Fig. 24. 9 24 -29

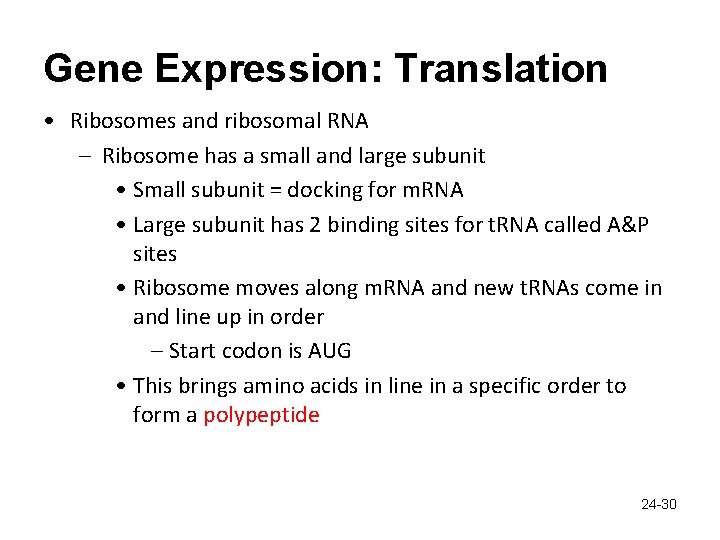
Gene Expression: Translation • Ribosomes and ribosomal RNA – Ribosome has a small and large subunit • Small subunit = docking for m. RNA • Large subunit has 2 binding sites for t. RNA called A&P sites • Ribosome moves along m. RNA and new t. RNAs come in and line up in order – Start codon is AUG • This brings amino acids in line in a specific order to form a polypeptide 24 -30

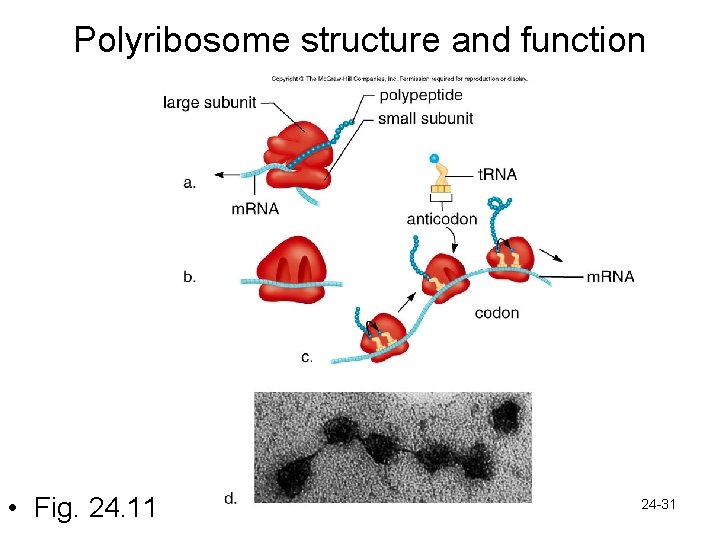
Polyribosome structure and function • Fig. 24. 11 24 -31

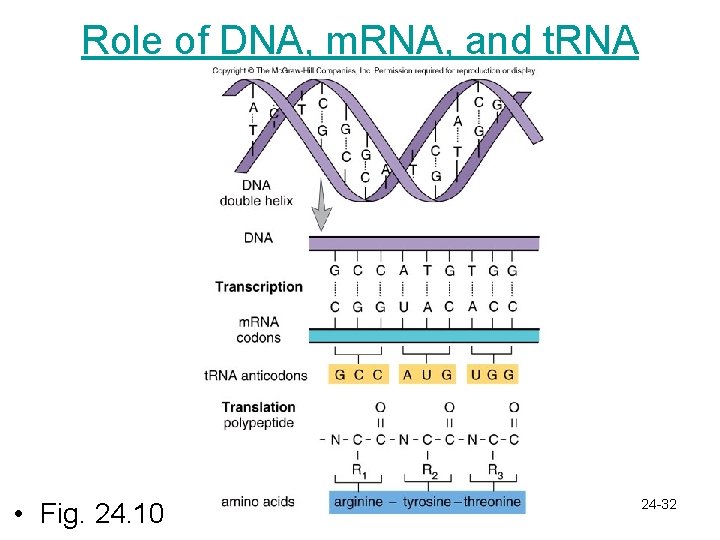
Role of DNA, m. RNA, and t. RNA • Fig. 24. 10 24 -32

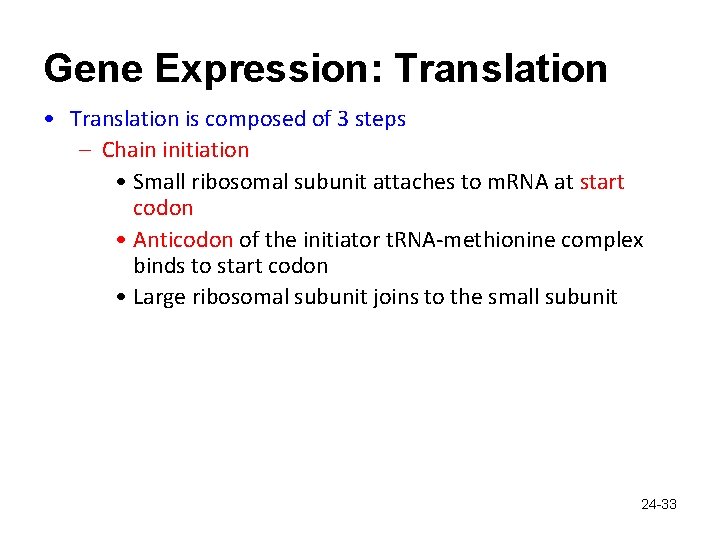
Gene Expression: Translation • Translation is composed of 3 steps – Chain initiation • Small ribosomal subunit attaches to m. RNA at start codon • Anticodon of the initiator t. RNA-methionine complex binds to start codon • Large ribosomal subunit joins to the small subunit 24 -33

– Chain elongation • Amino acids are added one at a time • Each new t. RNA-amino acid complex at the second binding site receives a peptide from a t. RNA at the first binding site • This t. RNA breaks away, and the ribosome moves forward one codon • The t. RNA at the second binging site now occupies the first site • Movement of the ribosome is called translocation 24 -34

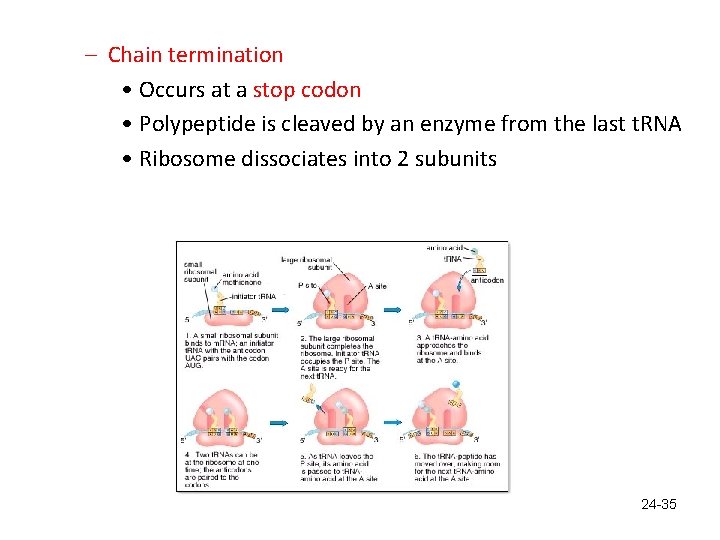
– Chain termination • Occurs at a stop codon • Polypeptide is cleaved by an enzyme from the last t. RNA • Ribosome dissociates into 2 subunits 24 -35

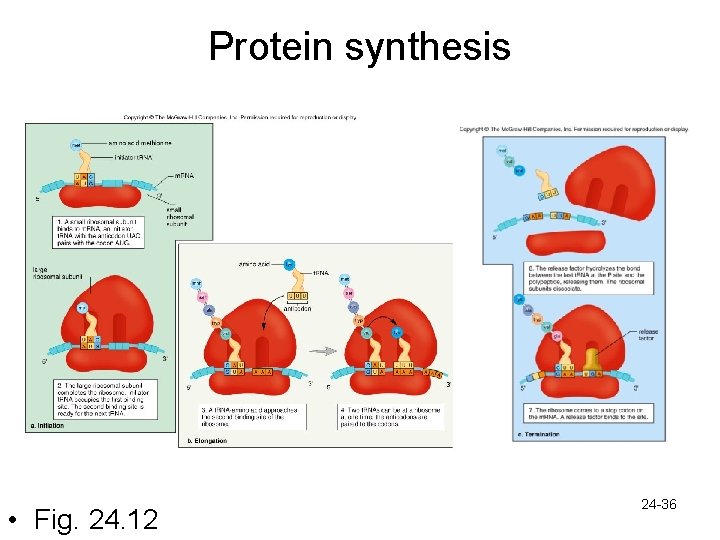
Protein synthesis • Fig. 24. 12 24 -36

Gene Expression: Review – DNA in nucleus contains a triplet base code • Each group of 3 bases stands for a specific amino acid – Transcription- complementary m. RNA is made from the template strand of DNA • Every 3 bases along m. RNA is called a codon • m. RNA is processed before it leaves the nucleus to go out to ribosomes – Translation • Initiation- initiator codon, 2 ribosomal subunits, and t. RNA-methionine • Chain elongation- anticodons of t. RNA line up along m. RNA codons • Chain termination- at the termination codon 24 -37 polypeptide is released

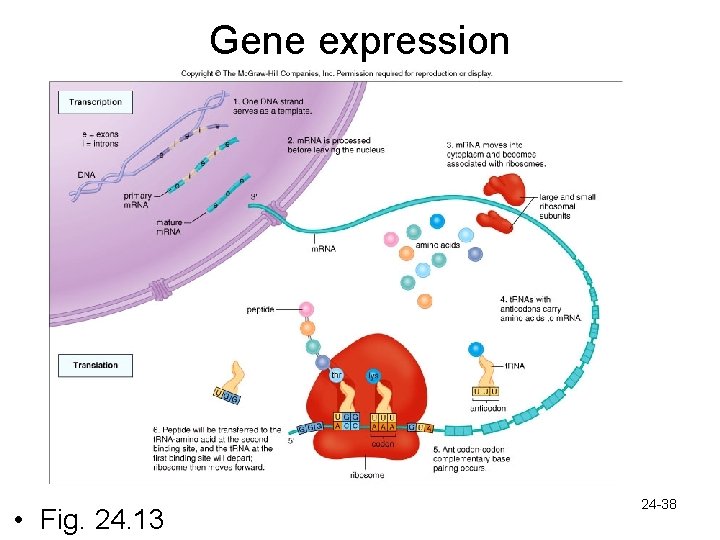
Gene expression • Fig. 24. 13 24 -38

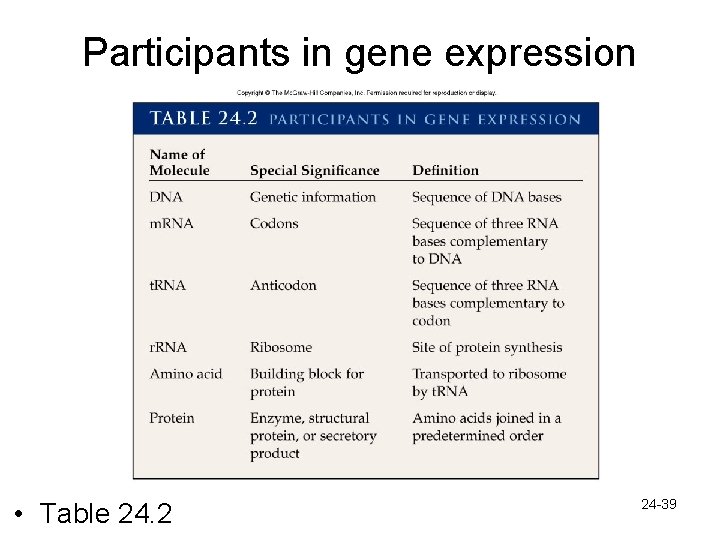
Participants in gene expression • Table 24. 2 24 -39

Gene Expression: Mutations • Genes and gene mutations – Causes of gene mutations • Gene mutation is a change in base code sequence • Errors in replication – Rare – DNA polymerase “proof reads” new strands and errors are cleaved out • Mutagens – Environmental influences – Radiation, UV light, chemicals – Rate is low because DNA repair enzymes monitor and repair DNA 24 -40

24 -41

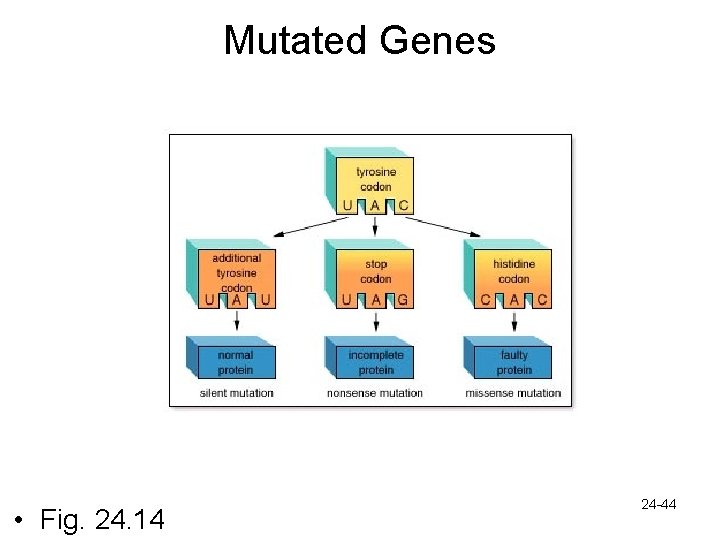
Mutations Point Mutation - substitute one base for another e. x Sickle Cell anemia Original: A T A C Mutant : T T A C Frameshift Mutation - a base is either added or removed which causes a shift in the reading frame. Many genes affects Original: A T A C A A G C C A Mutant: A T T A C A A G C C A Silent Mutation - a base is changed but the resulting amino acid is the same as in the non mutant DNA. No outward changes. Original: A A A C A G Mutant: A A G C A G Nonsense Mutation - a codon is changed to a STOP codon Original: A T A C C C A A A Mutant: A T T C C C A A A 24 -42

24 -43

Mutated Genes • Fig. 24. 14 24 -44

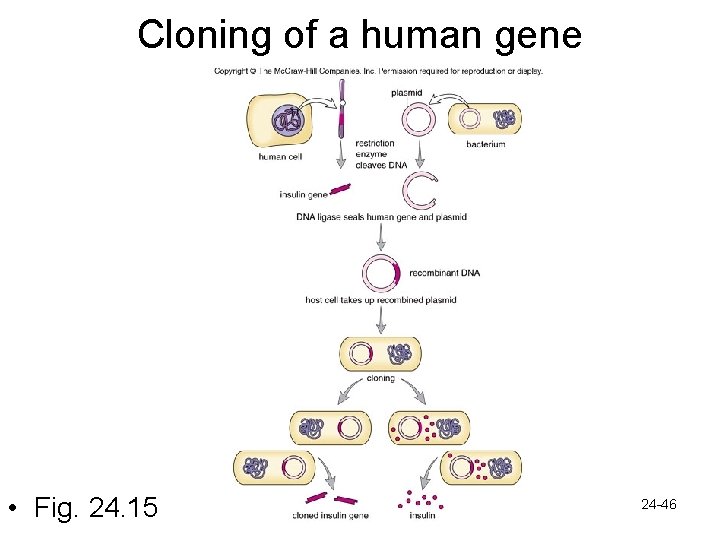
24. 3 DNA technology • The cloning of a gene – Production of identical copies by asexual means • Gene cloning-many copies of a gene – Gene cloning by recombinant DNA technology • r. DNA-contains DNA from 2 or more sources • Restriction enzyme- breaks open vector DNA – Vector is a plasmid-when plasmid replicates, inserted genes will be cloned – Restriction enzyme breaks it open at specific sequence of bases-”sticky ends” – Foreign DNA to be inserted also cleaved with same restriction enzyme so ends match • Foreign DNA is inserted into plasmid DNA and “sticky ends” pair up • DNA ligase seals them together 24 -45

Cloning of a human gene • Fig. 24. 15 24 -46

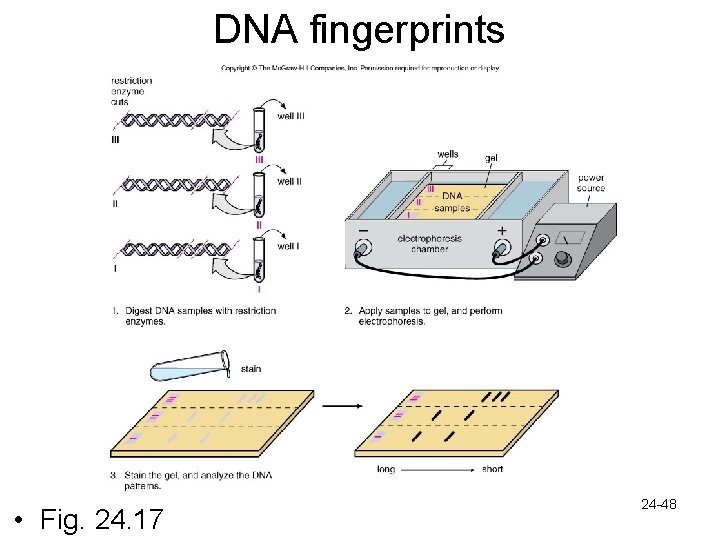
DNA technology cont’d. • DNA fingerprinting – Permits identification of individuals and their relatives – Based on differences between sequences in nucleotides between individuals – Detection of the number of repeating segments (called repeats) are present at specific locations in DNA • Different numbers in different people • PCR amplifies only particular portions of the DNA • The greater the number of repeats, the greater the amount of DNA that is amplified • The quantity of DNA that results at completion of PCR tells the number of repeats 24 -47

DNA fingerprints • Fig. 24. 17 24 -48

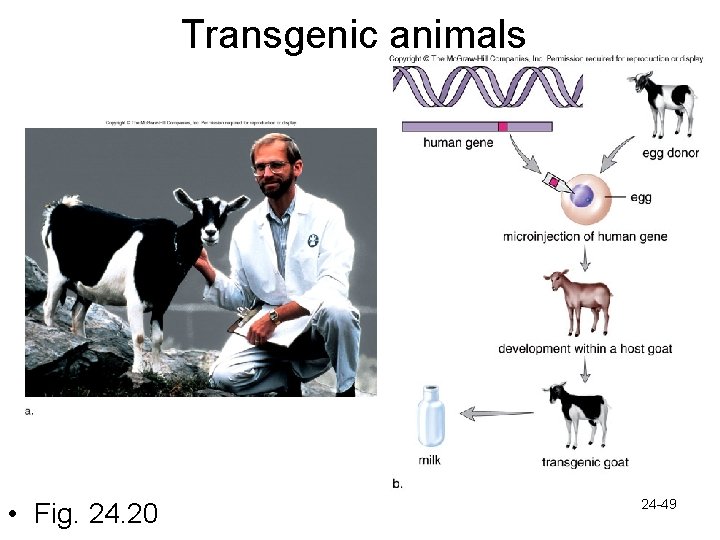
Transgenic animals • Fig. 24. 20 24 -49

24 -50