DNA Structure Replication Overview DNA Structure Structure of



















- Slides: 79

DNA Structure & Replication

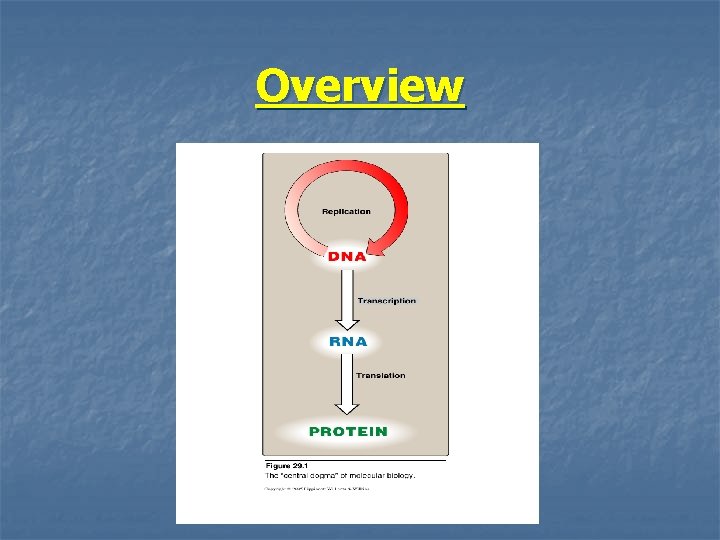
Overview

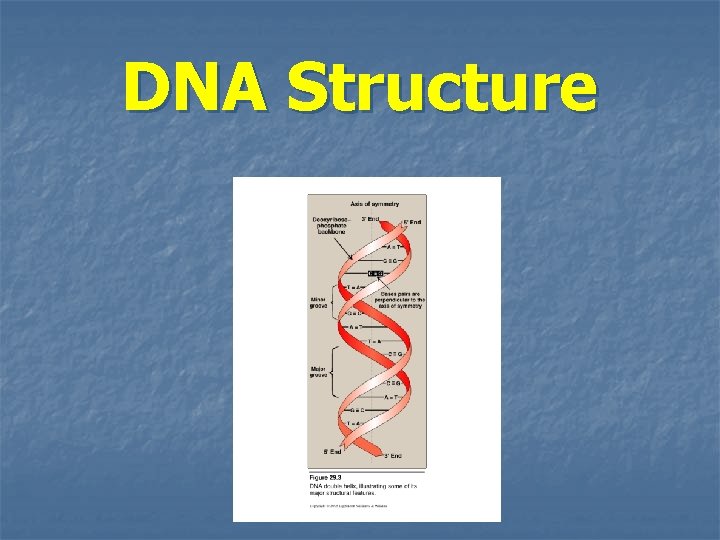
DNA Structure

Structure of DNA n n DNA is a double-stranded molecule (two strands) The two strands wind around each other forming a double helix Each strand of DNA molecule is a polydeoxyribonucleotide: (i. e. many monodeoxyribouncleotides) base (A, G, C, T) + sugar (deoxyribose) + phosphoric acid Monodeoxyribonucleotides: linked by 3`- 5`phosphodiester bonds (PDE) to form polydeoxyribonucleotide n In eukaryotic cells (with a nucleus): DNA is associated with various types of proteins (nucleoproteins) present in the nucleus n In prokaryotes, the protein-DNA complex is present in the nucleoid

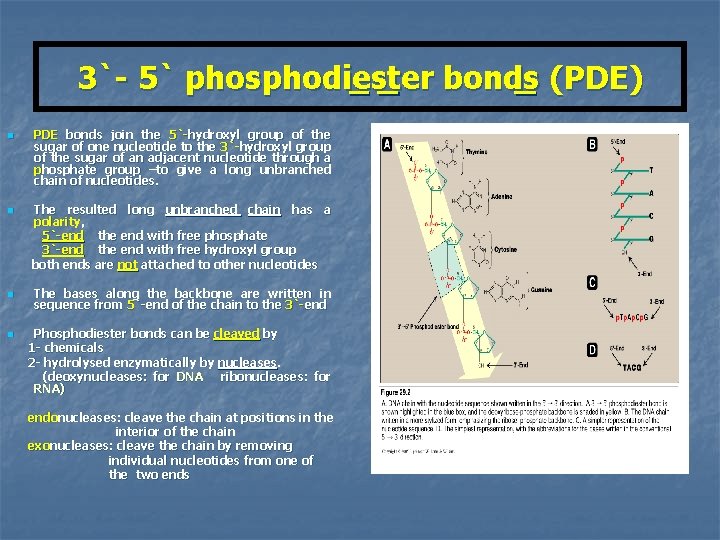
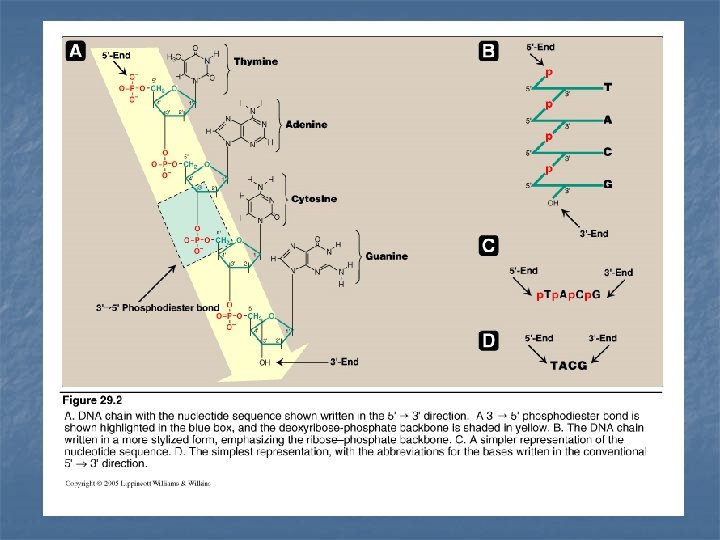
3`- 5` phosphodiester bonds (PDE) n n PDE bonds join the 5`-hydroxyl group of the sugar of one nucleotide to the 3`-hydroxyl group of the sugar of an adjacent nucleotide through a phosphate group –to give a long unbranched chain of nucleotides. The resulted long unbranched chain has a polarity, 5`-end the end with free phosphate 3`-end the end with free hydroxyl group both ends are not attached to other nucleotides The bases along the backbone are written in sequence from 5`-end of the chain to the 3`-end Phosphodiester bonds can be cleaved by 1 - chemicals 2 - hydrolysed enzymatically by nucleases. (deoxynucleases: for DNA ribonucleases: for RNA) endonucleases: cleave the chain at positions in the interior of the chain exonucleases: cleave the chain by removing individual nucleotides from one of the two ends


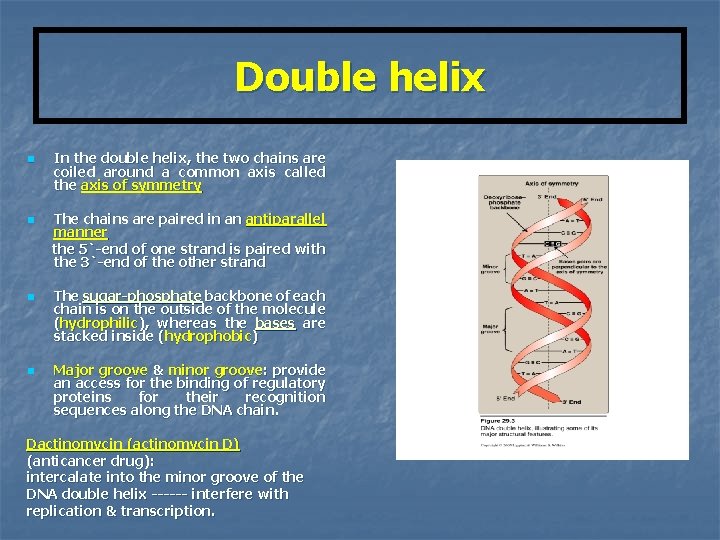
Double helix n n In the double helix, the two chains are coiled around a common axis called the axis of symmetry The chains are paired in an antiparallel manner the 5`-end of one strand is paired with the 3`-end of the other strand The sugar-phosphate backbone of each chain is on the outside of the molecule (hydrophilic), whereas the bases are stacked inside (hydrophobic) Major groove & minor groove: provide an access for the binding of regulatory proteins for their recognition sequences along the DNA chain. Dactinomycin (actinomycin D) (anticancer drug): intercalate into the minor groove of the DNA double helix ------ interfere with replication & transcription.


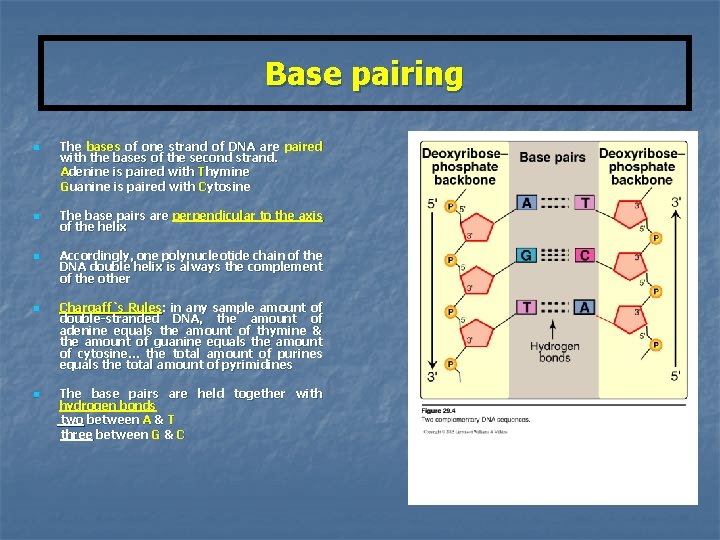
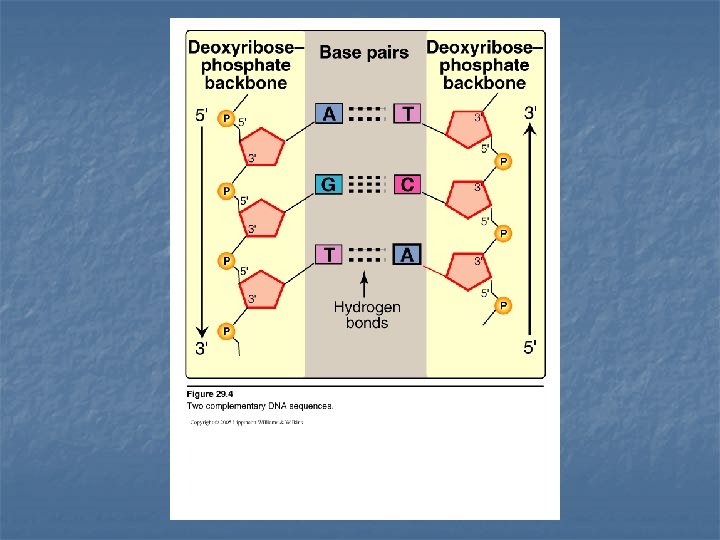
Base pairing n n n The bases of one strand of DNA are paired with the bases of the second strand. Adenine is paired with Thymine Guanine is paired with Cytosine The base pairs are perpendicular to the axis of the helix Accordingly, one polynucleotide chain of the DNA double helix is always the complement of the other Chargaff`s Rules: in any sample amount of double-stranded DNA, the amount of adenine equals the amount of thymine & the amount of guanine equals the amount of cytosine… the total amount of purines equals the total amount of pyrimidines The base pairs are held together with hydrogen bonds two between A & T three between G & C


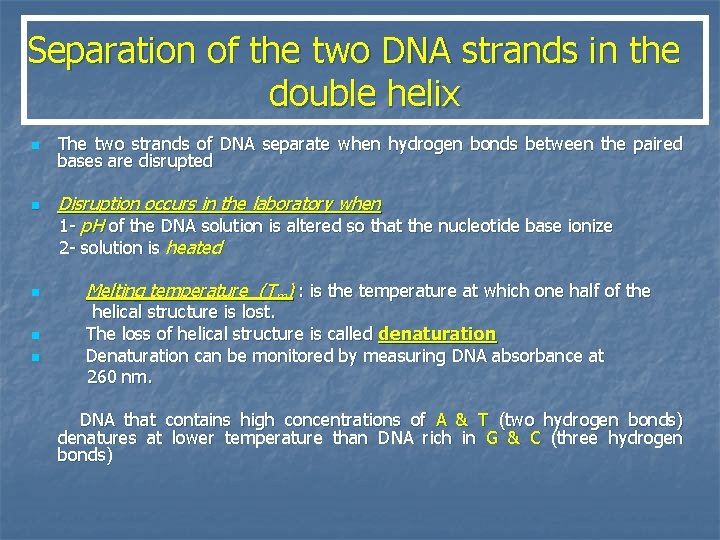
Separation of the two DNA strands in the double helix n n n The two strands of DNA separate when hydrogen bonds between the paired bases are disrupted Disruption occurs in the laboratory when 1 - p. H of the DNA solution is altered so that the nucleotide base ionize 2 - solution is heated Melting temperature (Tm) : is the temperature at which one half of the helical structure is lost. The loss of helical structure is called denaturation Denaturation can be monitored by measuring DNA absorbance at 260 nm. DNA that contains high concentrations of A & T (two hydrogen bonds) denatures at lower temperature than DNA rich in G & C (three hydrogen bonds)

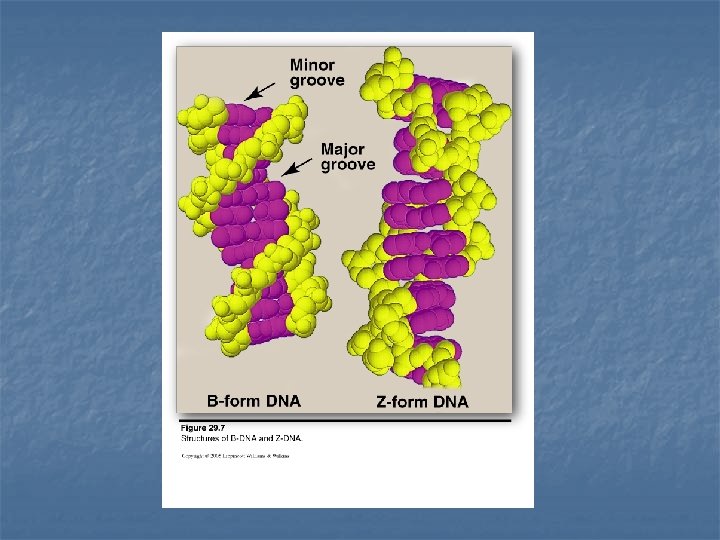
Structural forms of the double helix n B-DNA: Right-handed helix with 10 residues (base pairs) per 360 o turn of the helix The planes of the bases perpendicular to the helical axis. Chromosomal DNA consists primarily of B-DNA n A-DNA: Produced by moderately dehydrating the B-DNA form Right handed helix 11 residues per turn The planes of the bases are tilted 20 o away from the perpendicular to the helical axis. Found in DNA-RNA hybrid or RNA-RNA double stranded region. n Z-DNA left handed helix 12 base pairs per turn Zigzag sugar-phosphate backbone Occur in regions of DNA that have sequence of alternating purines and pyrimidines


Circular DNA molecules Eukaryote n Each chromosome in the nucleus of the eukaryote contains one long linear molecule of double-stranded DNA, bound to a complex mixture of proteins to form chromatin. -6+ n In addition, eukaryotes have closed circular DNA molecules in their mitochondria Prokaryotes n have single, double stranded, circular chromosome Each chromosome is associated with histone-like proteins and RNA that can condense the DNA to form a nucleoid. n In addition, most species of bacteria contain small, circular extrachromosomal DNA molecules called plasmids Plasmids carry genetic information and can undergo replication

Prokaryotic DNA synthesis (REPLICATION)

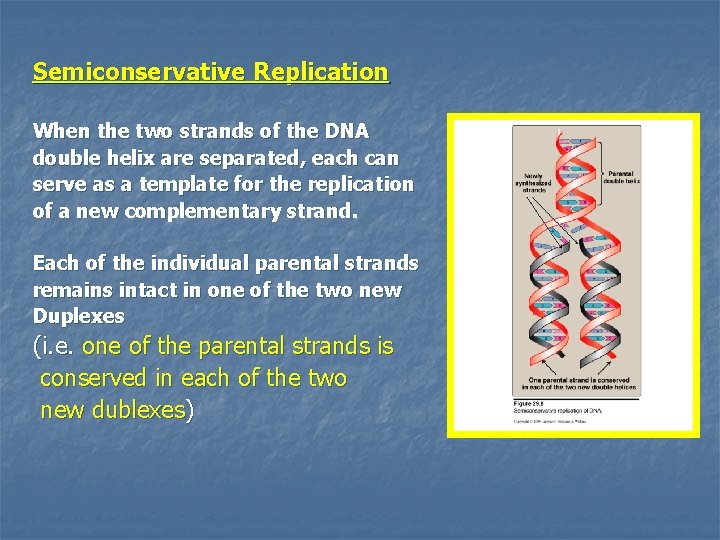
Semiconservative Replication When the two strands of the DNA double helix are separated, each can serve as a template for the replication of a new complementary strand. Each of the individual parental strands remains intact in one of the two new Duplexes (i. e. one of the parental strands is conserved in each of the two new dublexes)

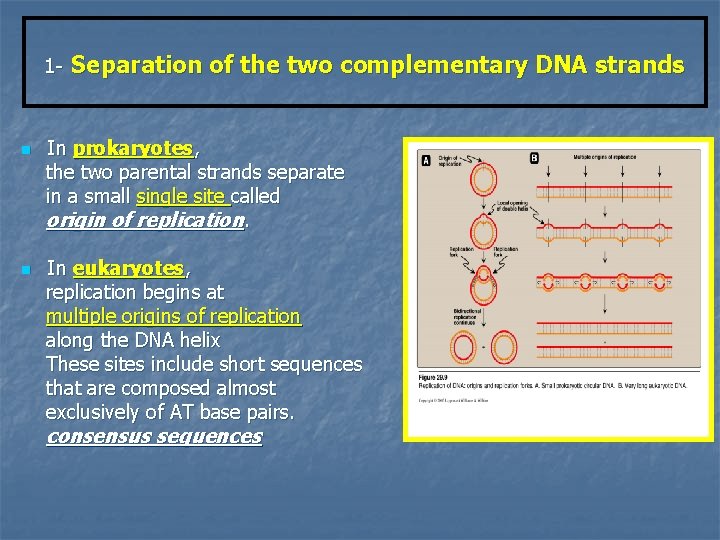
1 - n n Separation of the two complementary DNA strands In prokaryotes, the two parental strands separate in a small single site called origin of replication. In eukaryotes, replication begins at multiple origins of replication along the DNA helix These sites include short sequences that are composed almost exclusively of AT base pairs. consensus sequences

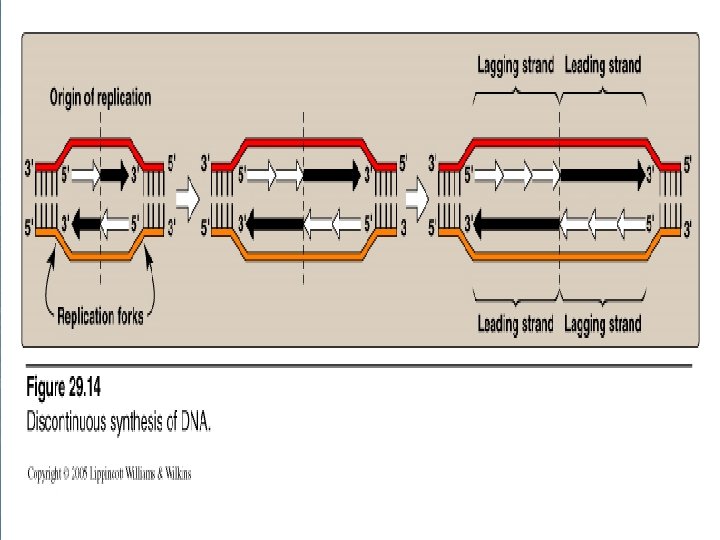
2 - Formation of the replication fork n n n As the two strands unwind and separate, they form a region called the replication fork, in which active synthesis occurs. Replication fork moves along the DNA molecule as synthesis occurs. Replication of double-stranded DNA is bidirectional (i. e. the replication forks moves in both directions away from the origin)

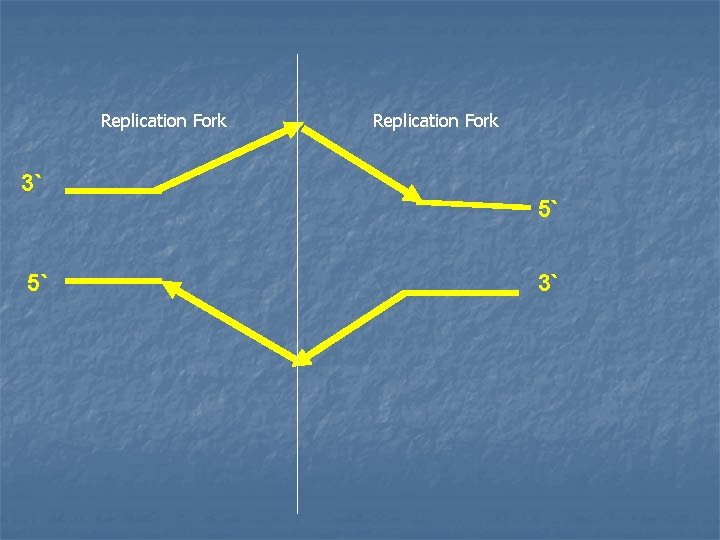
Replication Fork 3` 5` Replication Fork 5` 3`

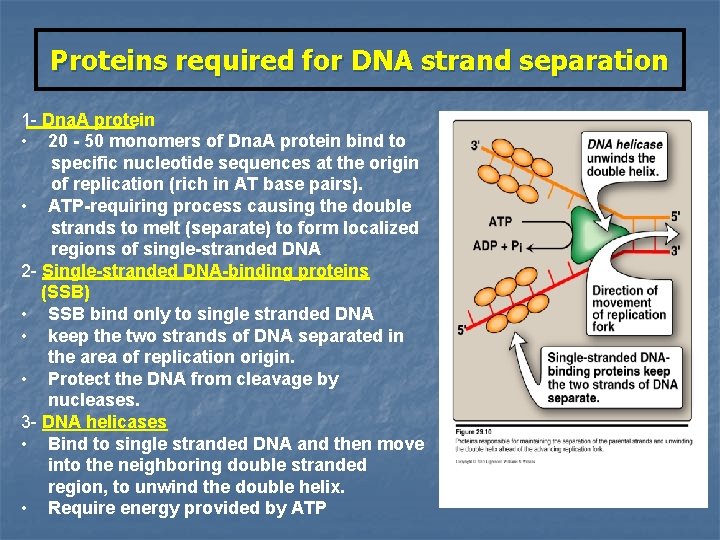
Proteins required for DNA strand separation Initiation of DNA replication requires the recognition of the origin of replication by a group of proteins that form the prepriming complex These proteins are responsible for: 1 - Maintaining the separation of the parental strands 2 - Unwinding the double helix ahead of the advancing replication fork.

Proteins required for DNA strand separation 1 - Dna. A protein • 20 - 50 monomers of Dna. A protein bind to specific nucleotide sequences at the origin of replication (rich in AT base pairs). • ATP-requiring process causing the double strands to melt (separate) to form localized regions of single-stranded DNA 2 - Single-stranded DNA-binding proteins (SSB) • SSB bind only to single stranded DNA • keep the two strands of DNA separated in the area of replication origin. • Protect the DNA from cleavage by nucleases. 3 - DNA helicases • Bind to single stranded DNA and then move into the neighboring double stranded region, to unwind the double helix. • Require energy provided by ATP

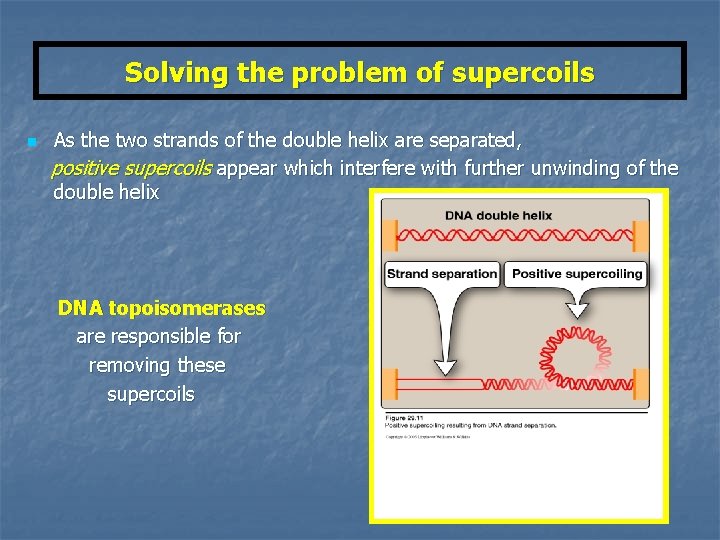
Solving the problem of supercoils n As the two strands of the double helix are separated, positive supercoils appear which interfere with further unwinding of the double helix DNA topoisomerases are responsible for removing these supercoils

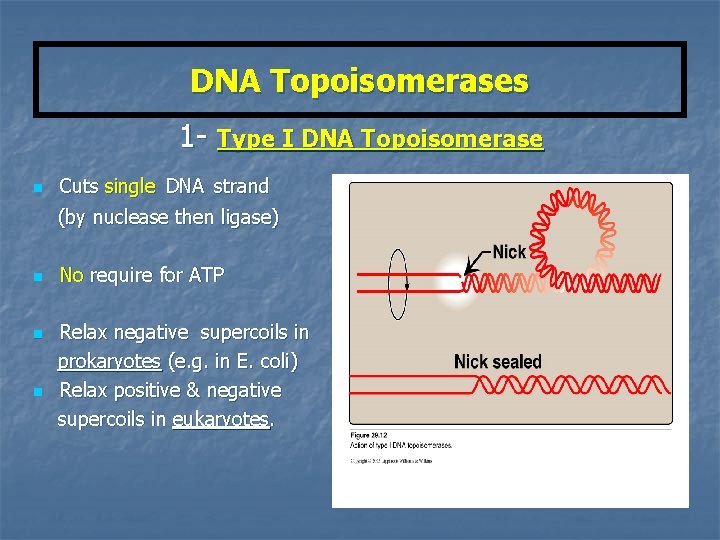
DNA Topoisomerases 1 - Type I DNA Topoisomerase n Cuts single DNA strand (by nuclease then ligase) n n n No require for ATP Relax negative supercoils in prokaryotes (e. g. in E. coli) Relax positive & negative supercoils in eukaryotes.

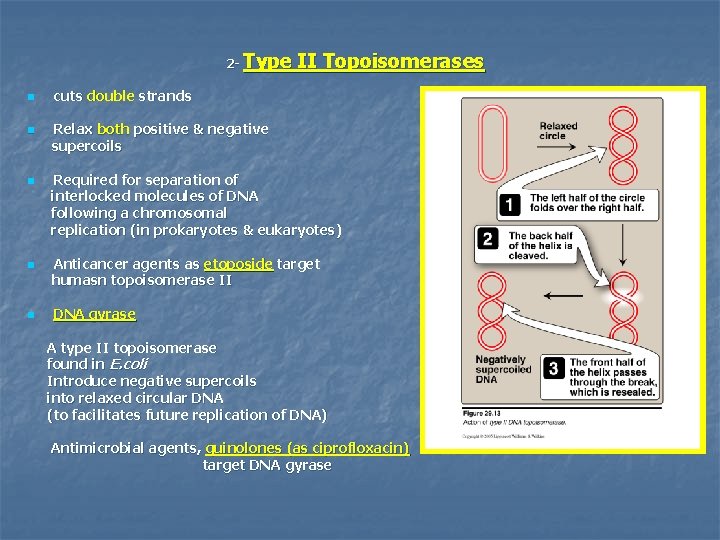
2 n n n Type II Topoisomerases cuts double strands Relax both positive & negative supercoils Required for separation of interlocked molecules of DNA following a chromosomal replication (in prokaryotes & eukaryotes) Anticancer agents as etoposide target humasn topoisomerase II DNA gyrase A type II topoisomerase found in E. coli Introduce negative supercoils into relaxed circular DNA (to facilitates future replication of DNA) Antimicrobial agents, quinolones (as ciprofloxacin) target DNA gyrase

3 - Direction of DNA replication n DNA polymerase is only able to read the parental nucleotide sequences in the 3`-5` direction n It synthesizes the new DNA strands in the 5`-3` direction. n So, beginning with one prenatal double helix, the two newly synthesized DNA must grow in opposite directions: 1 - One in 5`- 3` direction towards the replication fork 2 - One in 5`- 3` direction away from the replication fork.

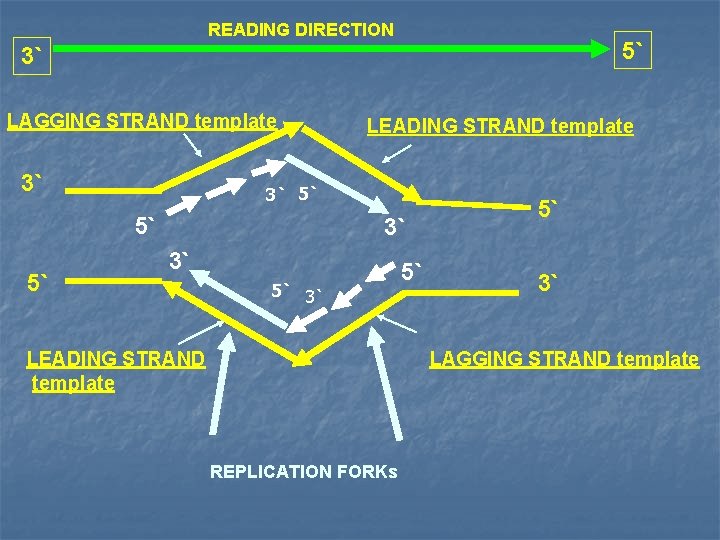
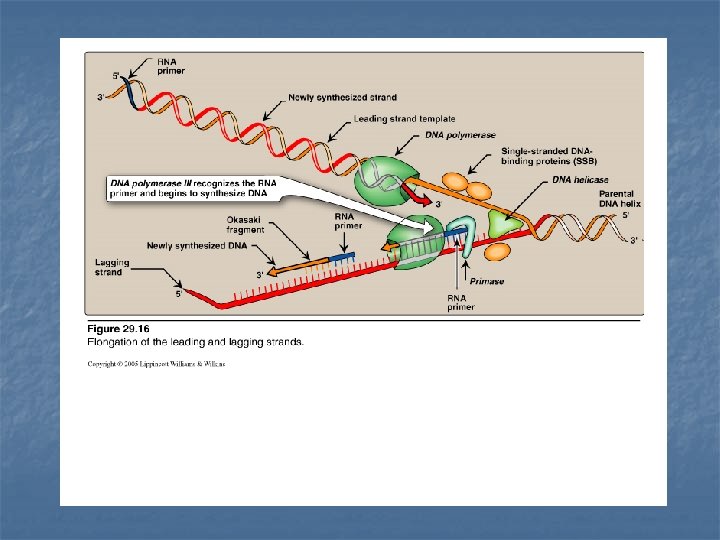
READING DIRECTION 5` 3` LAGGING STRAND template 3` 3` 5` 5` 5` LEADING STRAND template 3` 3` 5` 3` LEADING STRAND template 5` 5` 3` LAGGING STRAND template REPLICATION FORKs


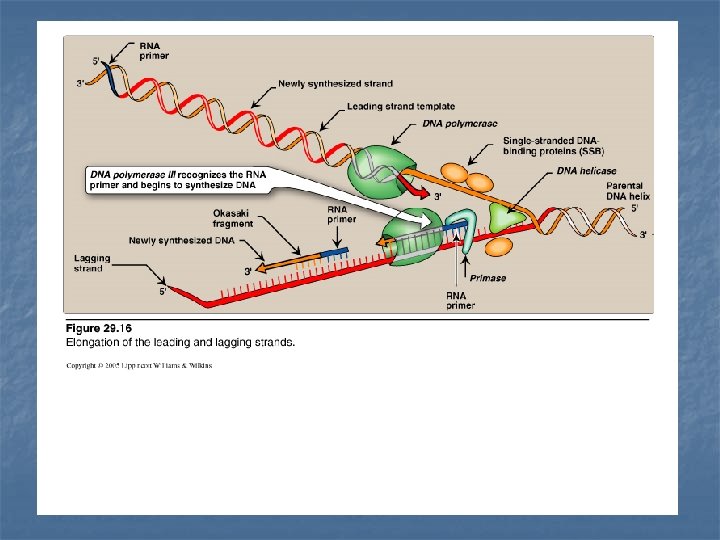
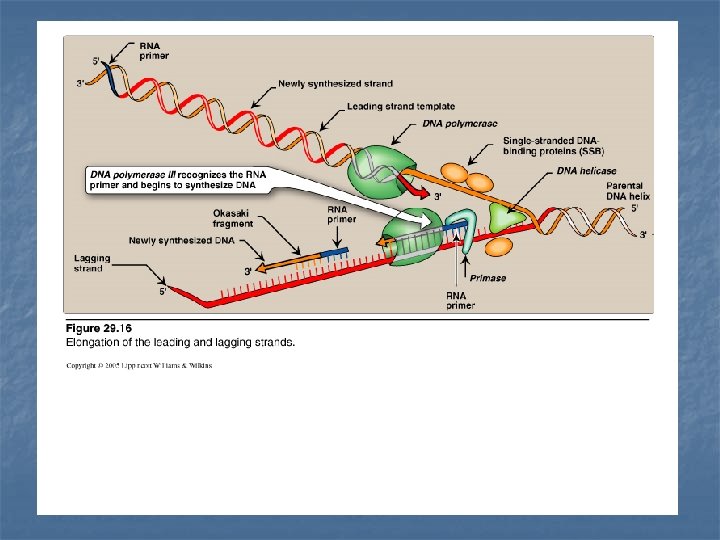
n - n Leading strand The strand that is being copied in the direction of the advancing replication fork Synthesized continuously. Lagging strand - The strand that is being copied in the direction away from the replication fork - Synthesized discontinuously with small fragments of DNA (Okazaki fragments) - They become eventually joined to become a single continuous DNA strand


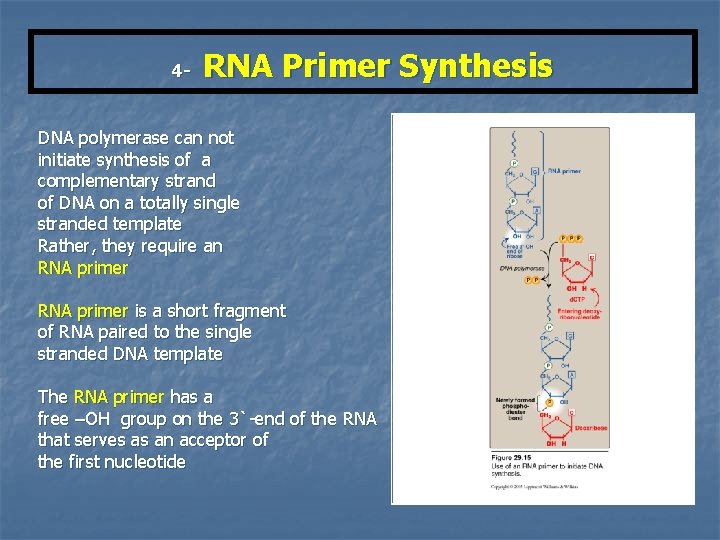
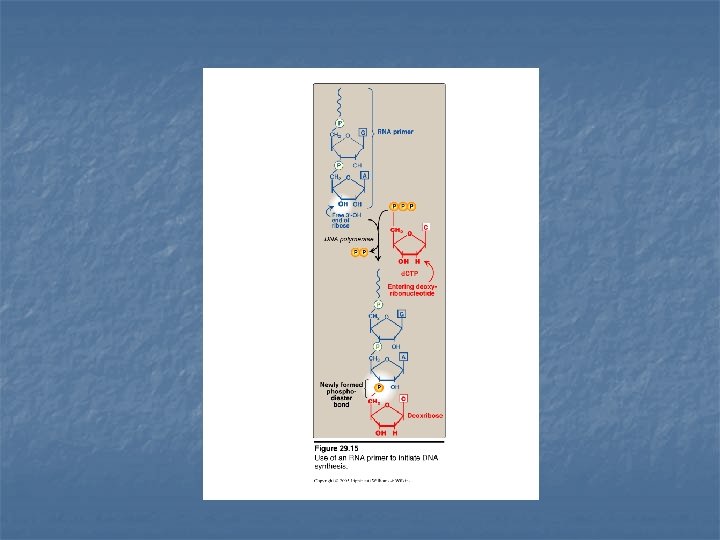
4 - RNA Primer Synthesis DNA polymerase can not initiate synthesis of a complementary strand of DNA on a totally single stranded template Rather, they require an RNA primer is a short fragment of RNA paired to the single stranded DNA template The RNA primer has a free –OH group on the 3`-end of the RNA that serves as an acceptor of the first nucleotide


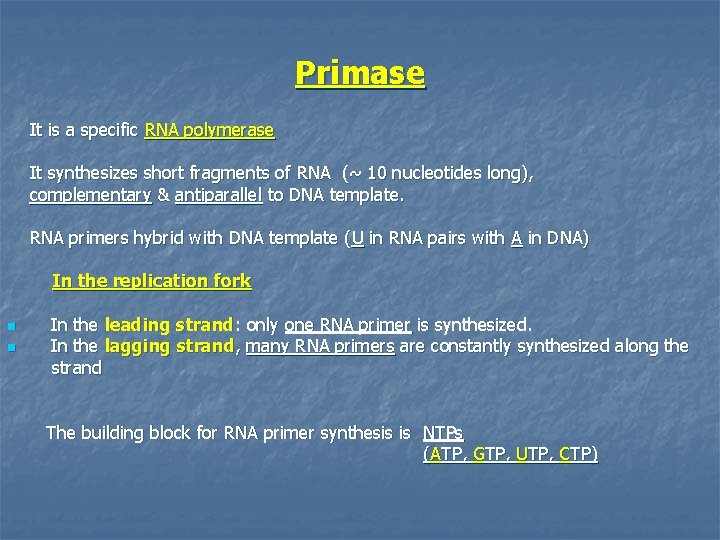
Primase It is a specific RNA polymerase It synthesizes short fragments of RNA (~ 10 nucleotides long), complementary & antiparallel to DNA template. RNA primers hybrid with DNA template (U in RNA pairs with A in DNA) In the replication fork n n In the leading strand: only one RNA primer is synthesized. In the lagging strand, many RNA primers are constantly synthesized along the strand The building block for RNA primer synthesis is NTPs (ATP, GTP, UTP, CTP)

Primosome (Primase + Prepriming complex) This complex initiates Okazaki fragment formation by moving along the template for the lagging strand in the 5`-3` direction periodically recognizing specific sequences of nucleotides that direct it to create an RNA primer that is synthesized in the 5`-3` direction.


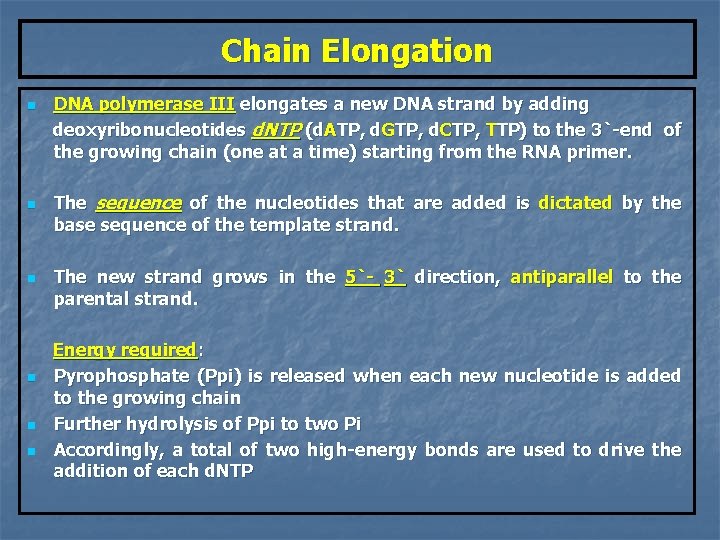
Chain Elongation n n n DNA polymerase III elongates a new DNA strand by adding deoxyribonucleotides d. NTP (d. ATP, d. GTP, d. CTP, TTP) to the 3`-end of the growing chain (one at a time) starting from the RNA primer. The sequence of the nucleotides that are added is dictated by the base sequence of the template strand. The new strand grows in the 5`- 3` direction, antiparallel to the parental strand. Energy required: Pyrophosphate (Ppi) is released when each new nucleotide is added to the growing chain Further hydrolysis of Ppi to two Pi Accordingly, a total of two high-energy bonds are used to drive the addition of each d. NTP

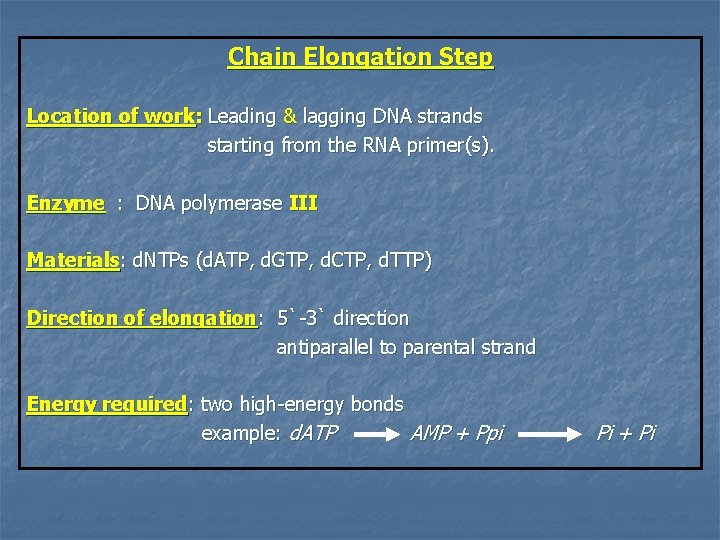
Chain Elongation Step Location of work: Leading & lagging DNA strands starting from the RNA primer(s). Enzyme : DNA polymerase III Materials: d. NTPs (d. ATP, d. GTP, d. CTP, d. TTP) Direction of elongation: 5`-3` direction antiparallel to parental strand Energy required: two high-energy bonds example: d. ATP AMP + Ppi Pi + Pi


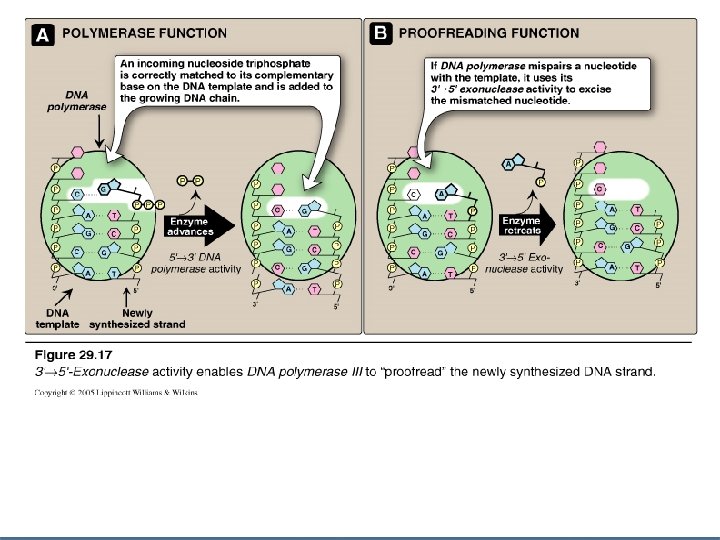
Proofreading of newly synthesized DNA n Misreading of the template sequence could result in mutations. n To ensure replication correctness of sequence, DNA polymerase III has a proofreading activity (3`- 5` exonuclease activity) n Steps : 1 - DNA polymerase III checks for the correctness of matching of added nucleotide to its complementary base on the template. 2 - If a wrong nucleotide is added, 3`-5` exonuclease activity edits the mistake


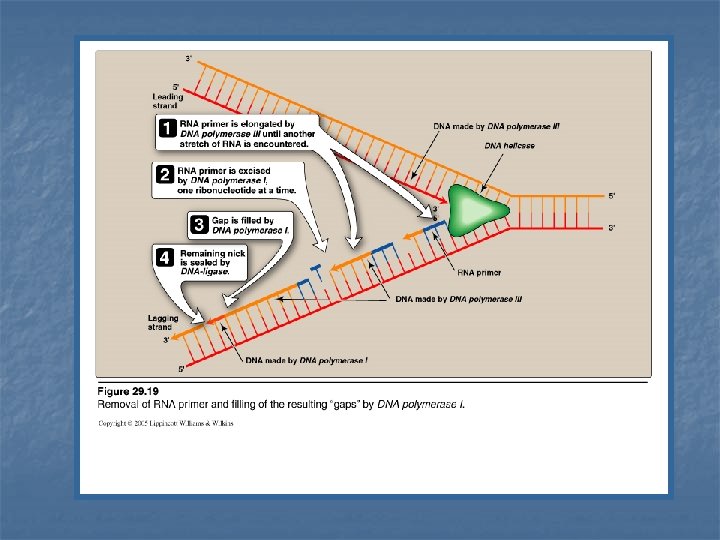
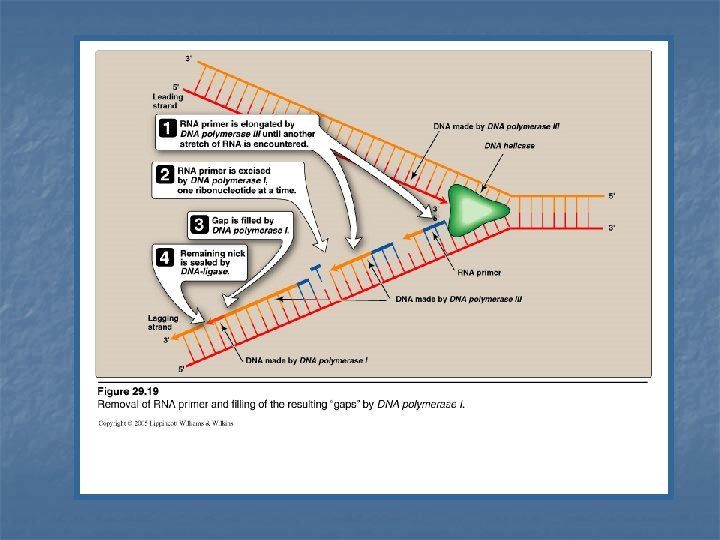
Excision of RNA primers & their replacement by DNA n n On the lagging strand DNA polymerase III continues to synthesize DNA until it is blocked by an RNA primer. At that stage, 1 - RNA primer is removed (excised) by DNA polymerase I (by 5`-3` exonuclease activity) 2 - Then, the gap is filled by DNA polymerase I (by 5`-3` polymerase activity) 3 - DNA polymerase I proofreads the filled gap DNA (by 3`- 5` exonuclease activity)


DNA Polymerase III DNA Polymerase I 5`-3`polymerase activity synthesis of new DNA strand 5`-3`polymerase activity synthesis (filling) of gap of removed RNA primer Step 2 3`-5` exonuclease activity proof reading of new strand 3`-5` exonuclease activity proof reading of the DNA filling the gap of removed RNA primer Step 3 5`-3` exonuclease activity removal (excision) of RNA primers. Step 1 Function Synthesis ---- Proofread Function Removal ----synthesis ----- Proofread

DNA ligase The final phosphodiester linkage between the 5`-phosphate group on the DNA chain synthesized by DNA polymerase III and the 3`-OH group on the chain made by DNA polymerase I is catalyzed by DNA Ligase. This process requires energy derived from the cleavage of ATP to AMP + Ppi

Eukaryotic DNA Replication n n The process of eukaryotic DNA replication closely follows that of prokaryotic DNA synthesis. Main Diffrerences: 1 - Multiple origins of replication in eukaryotes. Single origin of replication in prokaryotes. 2 - RNA primers are removed by RNase H in eukaryotes.

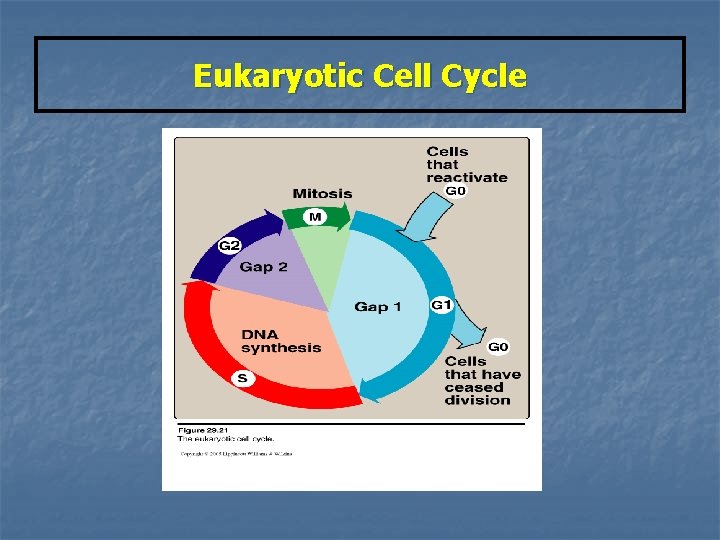
Eukaryotic Cell Cycle

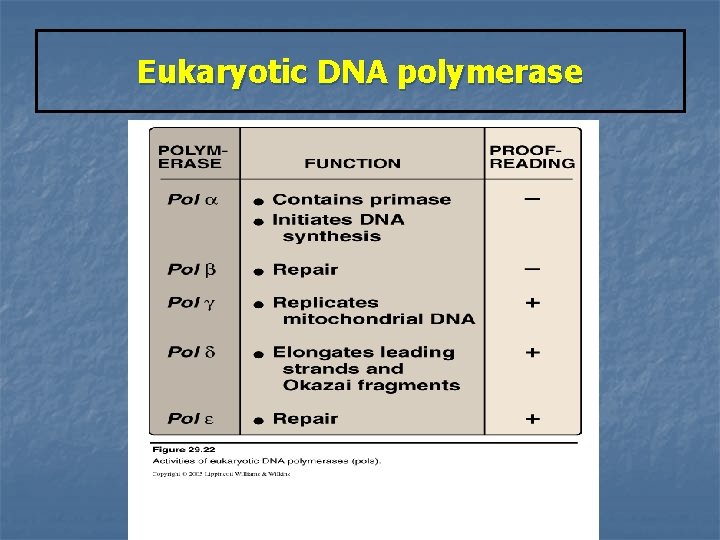
Eukaryotic DNA polymerase n Pol a : n Pol d : 1 - completes DNA synthesis on the leading strand & elongates multisubunit enzyme 1 - Primase activity Synthesizes a short RNA primer 2 - Initiates DNA synthesis by the 5`-3` polymerase activity. on leading & the beginning of each Okazaki fragment on the lagging strand ---- adding of a short piece of DNA. each Okazaki fragment 2 - Proofread newly synthesized DNA n Pol b pol e : involved in carrying out DNA repair. n Pol g : replicates mitochondrial DNA

Eukaryotic DNA polymerase


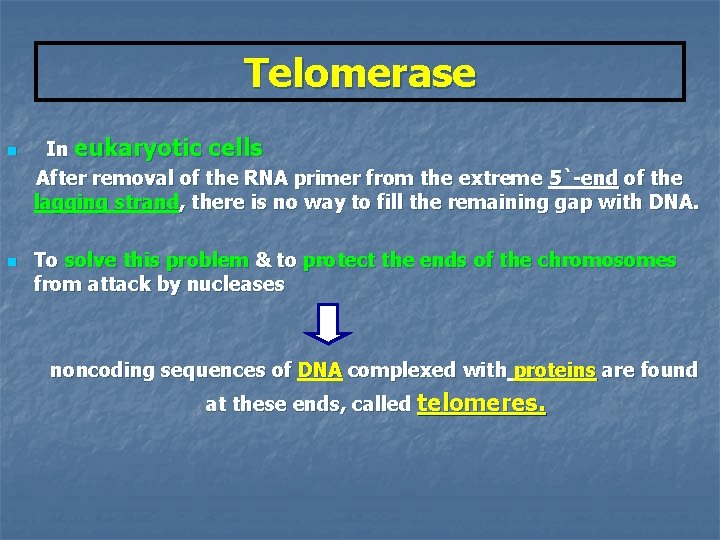
Telomerase n n In eukaryotic cells After removal of the RNA primer from the extreme 5`-end of the lagging strand, there is no way to fill the remaining gap with DNA. To solve this problem & to protect the ends of the chromosomes from attack by nucleases noncoding sequences of DNA complexed with proteins are found at these ends, called telomeres.

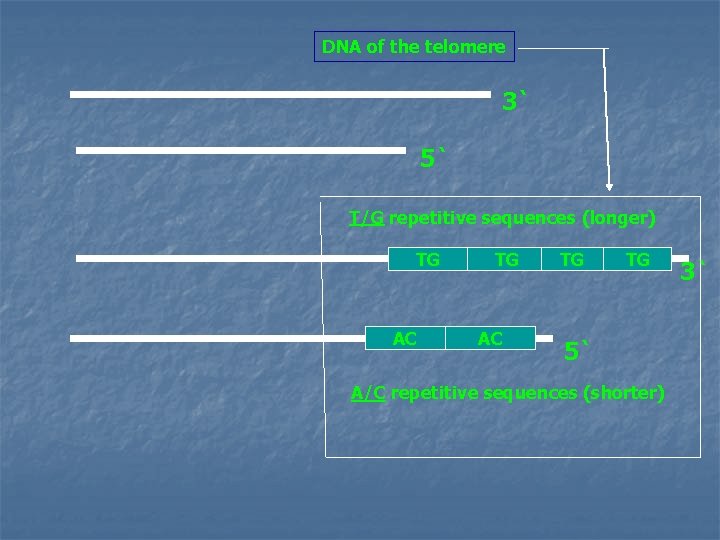
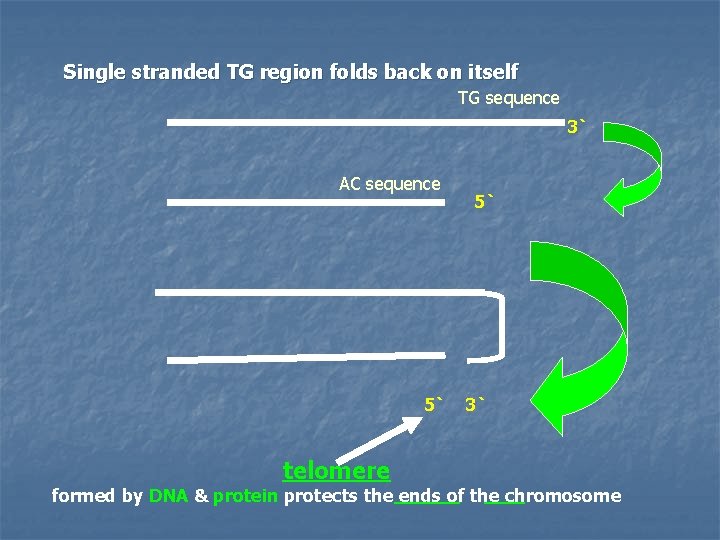
Telomeres n n DNA of telomeres consists of repetitive sequence of T`s & G`s, base paired to a complementary chain of A`s and C`s Tx Gy: where x and y are in the range of 1 : 4 The TG strand is longer than its complement leaving a region of single stranded DNA at the 3`-end of the double helix (few hundred nucleotides long). This single stranded region folds back on itself, forming a structure that is stabilized by protein to protect the ends of the chromosomes In aging cells, the ends of their chromosomes get slightly shorter with each cell division until the telomeres are gone and DNA essential for cell function is degraded. This phenomenon is related to cellular aging & death.

DNA of the telomere 3` 5` T/G repetitive sequences (longer) TG AC TG TG 5` A/C repetitive sequences (shorter) 3`

Single stranded TG region folds back on itself TG sequence 3` AC sequence 5` telomere 5` 3` formed by DNA & protein protects the ends of the chromosome

In aging cells, the ends of their chromosomes get slightly shorter with each cell division until the telomeres are gone and DNA essential for cell function is degraded. This phenomenon is related to cellular aging & death.

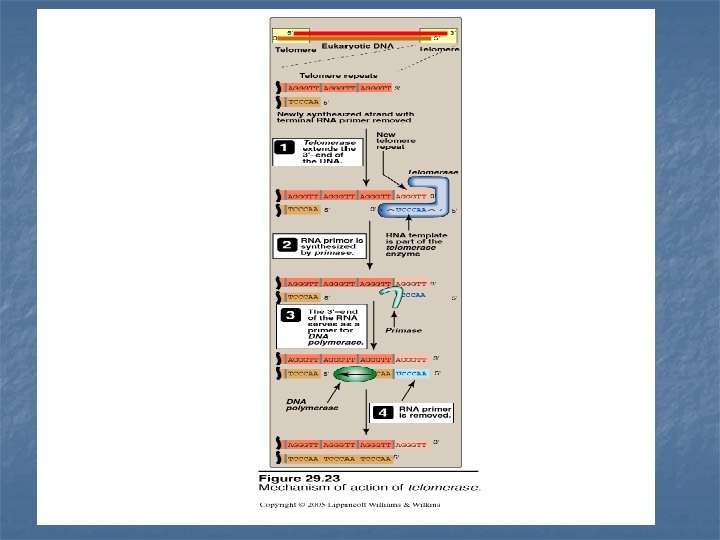
So, what is a telomerase ? n n Cells that do not age (as germ-line cells and cancer cells) contain an enzyme called telomerase responsible for replacing these lost ends of telomeres. Telomerase is a special kind of reverse transcriptase that carries its own RNA molecule of about 150 nucleotides long. This RNA contains copies of A/C sequence that is complementary to T/G repeat sequence Steps of telomere elongation 1 -The telomerase RNA serves as template for extending T/G DNA strand (longer strand) 2 -The 3`-end of the RNA serves as a primer for DNA polymerase to extend the A/C DNA strand (shorter strand) 3 -Once the next repeat sequence is complete, telomerase RNA is translocated to the newly synthesized end of the DNA and the process is repeated.


Reverse Transcriptase n n A retrovirus as HIV, carries the genome in the form of single-stranded RNA molecul. Following infection of a host cell, the viral enzyme, reverse transcriptase, uses the RNA as a template for the synthesis of viral DNA Then, DNA becomes integrated into host chromosomes. n n n Reverse transcriptase moves along the RNA template in the 3`-5` direction, synthesizing new DNA in the 5`-3` direction No proofreading in reverse transcriptase high mutation rate of such viruses As an attempt to prevent HIV infection from progressing to AIDS, patients are treated by inhibitors of reverse transcriptase (nucleosides or non-nucleosides inhib. ), in addition to protease inhibitors (which targets another HIV maturation enzyme.

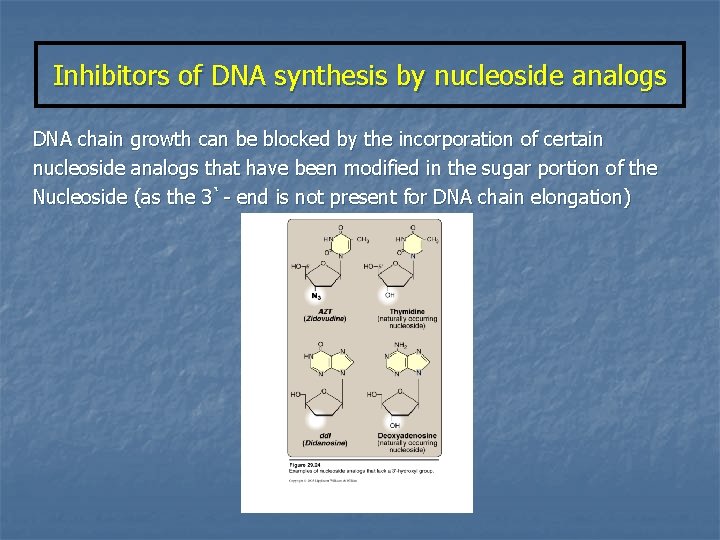
Inhibitors of DNA synthesis by nucleoside analogs DNA chain growth can be blocked by the incorporation of certain nucleoside analogs that have been modified in the sugar portion of the Nucleoside (as the 3`- end is not present for DNA chain elongation)

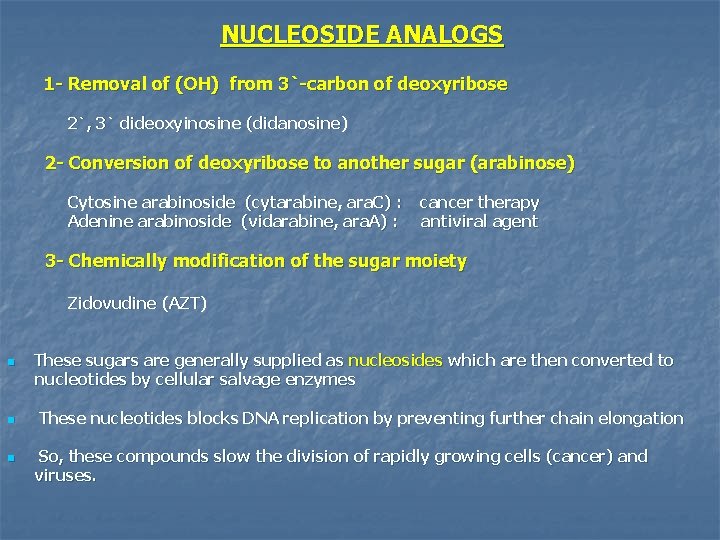
NUCLEOSIDE ANALOGS 1 - Removal of (OH) from 3`-carbon of deoxyribose 2`, 3` dideoxyinosine (didanosine) 2 - Conversion of deoxyribose to another sugar (arabinose) Cytosine arabinoside (cytarabine, ara. C) : Adenine arabinoside (vidarabine, ara. A) : cancer therapy antiviral agent 3 - Chemically modification of the sugar moiety Zidovudine (AZT) n n n These sugars are generally supplied as nucleosides which are then converted to nucleotides by cellular salvage enzymes These nucleotides blocks DNA replication by preventing further chain elongation So, these compounds slow the division of rapidly growing cells (cancer) and viruses.

n n n Cytosine arabinoside (cytarabine, ara. C) used as an anticancer chemotherapy Adenine arabinoside (vidarabine, ara. A) used as an antiviral Zidoviduline (AZT)

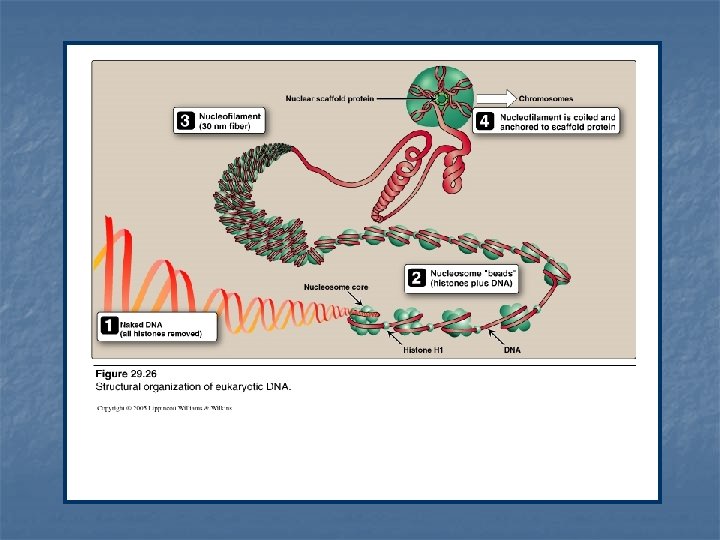
Organization of the eukaryotic DNA n n n Typical human DNA contains 46 chromosomes Total DNA is ~ one meter long It is important to effectively packaging such large amount of genetic material into a volume of the size of a cell nucleus for effective replication & gene expression. To fulfill so, DNA interacts with large number of proteins Eukaryotic DNA is associated with basic proteins called histones. These serve to order the DNA into basic structural units called nucleossomes (resemble beads on a string) Nucleosomes are further arranged into more complex structures that organize and condense the long DNA molecules into chromosomes

Histones and the formation of nucleosomes n n n 5 classes of histones H 1, H 2 A, H 2 B, H 3 & H 4 Histones are positively charged at physiologic p. H (high contents of lysine & argenine) They can form ionic bonds with negatively charged DNA (in addition with Mg 2+ can neutralize –ve charged DNA phosphate groups)

1 - Formation of the nucleosome n Two molecules each of H 2 A, H 2 B, H 3 and H 4 form the structural core of the individual nucleosome (bead). n Around this core, a segment of DNA double helix is wound twice forming a negatively supertwisted helix. n Neighboring nucleosomes are joined by linker DNA ~ 50 base pairs long. n H 1 binds to the linker DNA chain between the nucleosome beads. n H 1 is the most tissue-specific and species-specific of the histone.


2 - Higher levels of organization n Nucleosomes can be packed more tightly to form polyuncleosome (nucleofilament or 30 -nm fiber) The fiber is organized into loops that are anchored a nuclear scafold proteins. Additional levels of organization lead to the final chromosomal structure.


DNA Repair DNA repair is required in the following cases: 1 - Mismatches: which occur after DNA replication including proofreading n incorrect base-pairing n insertion of one to few extra nucleotides 2 - DNA damage due to environmental insults: leads to alteration or removal of nucleotide bases n chemical: nitrous oxide n radiation: UV light : (can fuse 2 pyrimidines adjacent to each other in DNA) High-energy radiation: (can cause double-stranded breaks) 3 - Spontaneous alteration or loss of bases from mammalian DNA (at a rate of thousands per cell per day)

Effect of unrepaired mismatch and DNA damage: n n A permanent mutation loss of control over the proliferation of the mutated cells CANCER

General Steps of Repair n Recognizing the lesion n Excision of the damaged section of the DNA strand n Fill the gap using the sister strand as a template

A. Strand-directed mismatch repair system in cases of replication errors escaping the proofreading function during DNA synthesis 1 - Identification of the mismatched strand Proteins which identify and remove the mispaired nucleotide(s) must be able to discriminate between the template strand the newly synthesized strand containing the mistake. GATC sequences occurring every 1000 nucleotides are methylated on the adenine The methylation is not done immediately after synthesis ---- so, the newly synthesized DNA is temporarily hemimethylated (i. e. the parental strand is methylated but, the newly synthesized strand is not)

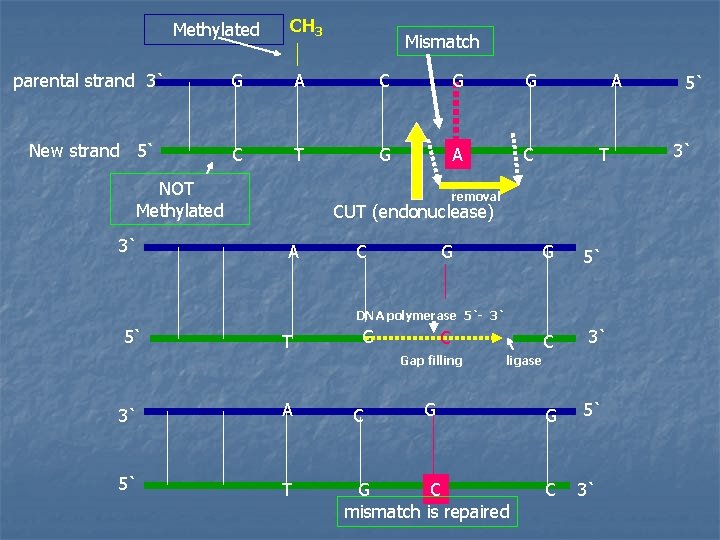
2 - Repair of damaged DNA i- endonuclease cuts the mismatched strand. ii- the mismatched base(S) are removed. iii- the gap left by removal of the bases is filled using the sister strand as a template by a 5`-3` DNA polymerase. ( DNA polymerase I in E. coli) iv- The cut ends of the DNA are ligated (spliced) by DNA ligase. A defect in mismatch repair in humans may cause herditary nonpolyposis colon cancer (HNPCC), an inherited cancer

Methylated CH 3 Mismatch parental strand 3` G A C G G New strand 5` C T G A C NOT Methylated 3` A T removal CUT (endonuclease) A C G G 5` DNA polymerase 5`- 3` 5` T G C Gap filling 3` A 5` T C ligase G G G C mismatch is repaired C C 3` 5` 3`

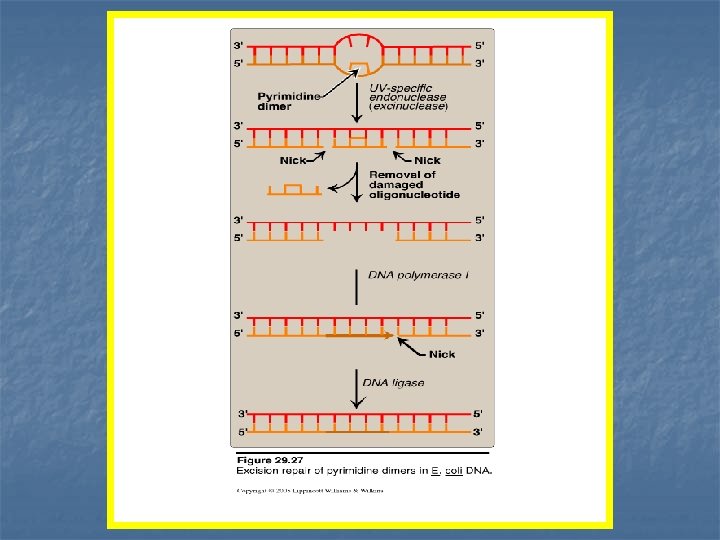
B-Repair of damage caused by ultraviolet light Exposure of a cell to ultraviolet light can result in the covalent joining of two adjacent pyrimidines (usually thymines), producing a dimer (thymine dimer) prevents DNA polymerase from replicating the DNA strand beyond the dimer formation. -----------------------------------------------n Thymine dimers has to be excised for the continuity of DNA replication by the following system: 1 - Recognition & excision of the dimer by UV-specific endonuclease. called: uvr. ABC excinuclease on both 5` & 3` sides of the dimer 2 - Damaged oligonucleotide is released, leaving a gap in the DNA strand. 3 - The gap is filled by nucleotides using the sister strand as a template.


Ultraviolet Radiation & Cancer n n Pyrimidine dimers can be formed in the skin of humans exposed to unfiltered sunlight. In xeroderma pigmentosum ( a rare genetic disease), the cells cannot repair the damaged DNA extensive accumulation of mutations Skin Cancers The most common form of this disease is caused by the absence of UV-specific excinucleases.

C- Correction of base alterations (base excision repair) I - The bases of DNA can be altered either : 1 - spontaneously (as the case of cytosine which is spontaneously deaminated slowly to uracil) 2 - by the action of deaminating or alkylating compounds as nitrous oxide is formed by the cell from precursor as nitrosamines, nitrites & nitrates. nitrous oxide deaminates cytosine, adenine & guanine II- Bases are lost spontaneously ~ 10, 000 purine bases are lost per cell per day.

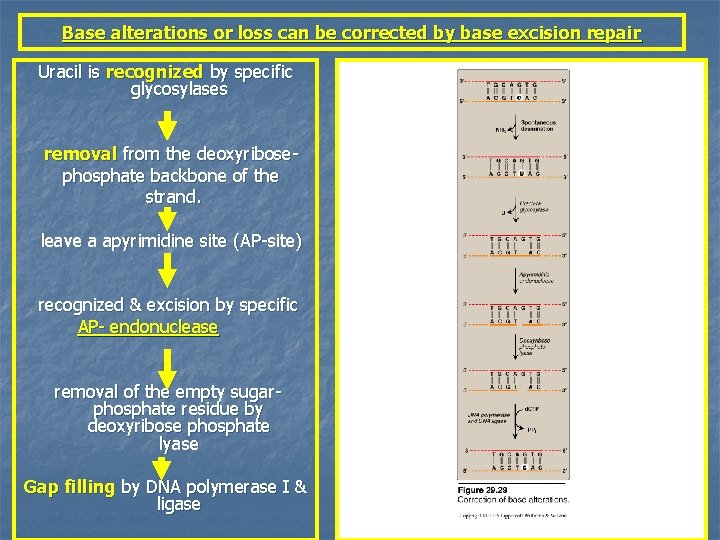
Base alterations or loss can be corrected by base excision repair Uracil is recognized by specific glycosylases removal from the deoxyribosephosphate backbone of the strand. leave a apyrimidine site (AP-site) recognized & excision by specific AP- endonuclease removal of the empty sugarphosphate residue by deoxyribose phosphate lyase Gap filling by DNA polymerase I & ligase

D- Repair of double-stranded breaks High energy radiation or oxidative free radicals can cause double stranded breaks in DNA --- lethal to cells. Double-stranded breaks in DNA can occur naturally during gene rearrangement

Repair of Double-stranded DNA breaks (cont. ) 1 n n nonhomologous end-joining repair The ends of two DNA fragments are brought together by a group of proteins that cause their religation. This does not require that the two DNA sequences have any sequence homology Error prone & mutagenic Defects in this repair system are associated with pdp to cancer & immunodeficiency syndrome.

2 - n n Homologous recombination repair Uses the enzymes that normally perform genetic recombination between homologous chromosomes during meiosis This system is used by the lower eukaryotes to repair double-stranded breaks.