Control of Gene Expression Prokaryotes and Operons Copyright








- Slides: 30

Control of Gene Expression Prokaryotes and Operons Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

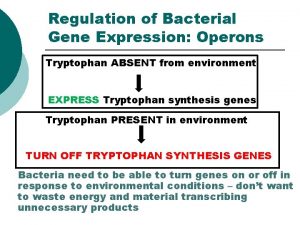
Regulated Gene Expression: an advantage • Lactose metabolism – disaccharide) - made of glucose & galactose – its oxidation provides cell with intermediates & energy – lactose absent, then no B-galactosidase – lactose present, enzyme levels rise ~1000 -fold • Tryptophan - essential amino acid; if not there, must be produced by bacterium at energy cost; needed for protein synthesis – if absent, cells make tryptophan – if present, genes repressed within minutes Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

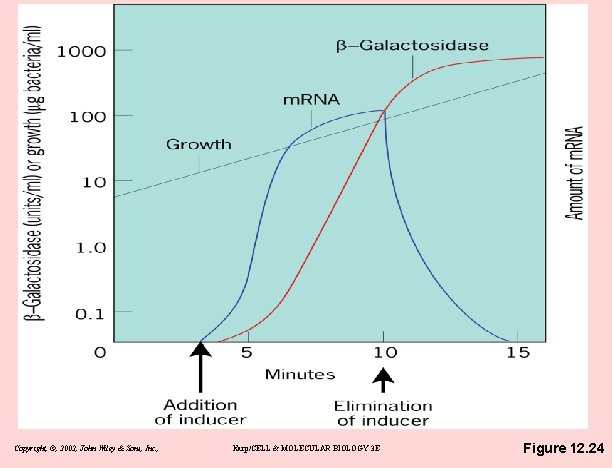
Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 12. 24

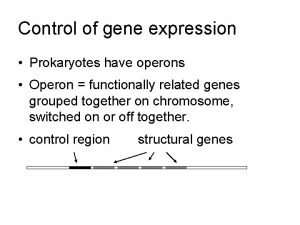
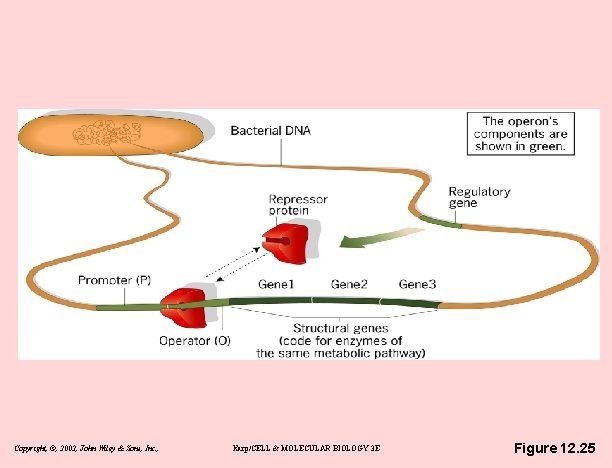
Bacterial operon - Jacob & Monod (Pasteur Inst. , 1961) • Components of operon (single m. RNA) – Structural genes - code for operon enzymes – Promoter – Operator - between promoter & genes – Repressor – binds to operator – Regulatory gene - codes for repressor protein • Repressor is key – it binds to operator, shielding promoter – Repressor regulated allosterically – presence or absence of inducer (lactose or tryptophan) Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 12. 25

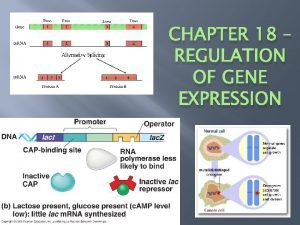
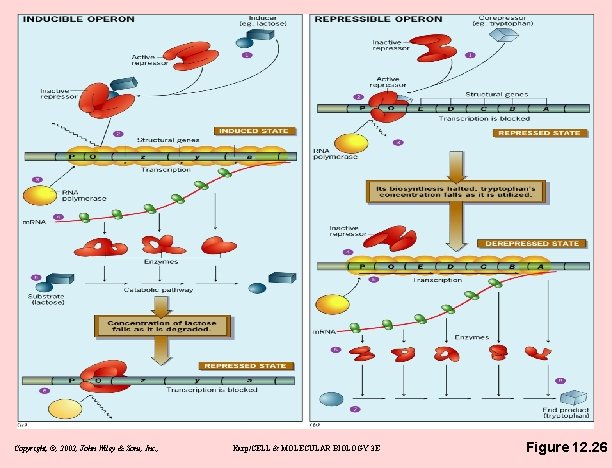
The lac operon - inducible operon • What are the structural genes in the lac operon? – z gene - encodes B-galactosidase – y gene - encodes galactoside permease; promotes lactose entry into cell – a gene - encodes thiogalactoside acetyltransferase; role is unclear • Inducible operon – If lactose present, binds repressor, changing its shape – Repressor binds promoter only in absence of inducer Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

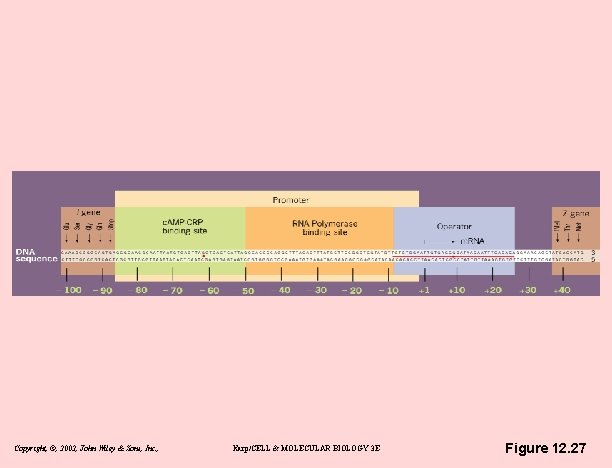
The lac operon - inducible operon • Positive control by cyclic AMP – Glucose inhibits lac expression – c. AMP inversely related to amount of glucose in medium – c. AMP activates lac – c. AMP binds to c. AMP receptor protein (CRP) – CRP binds DNA only if c. AMP bound – CRP-c. AMP complex allows RNA polymerase to transcribe – c. AMP-CRP complex is necessary for lac operon transcription Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 12. 27

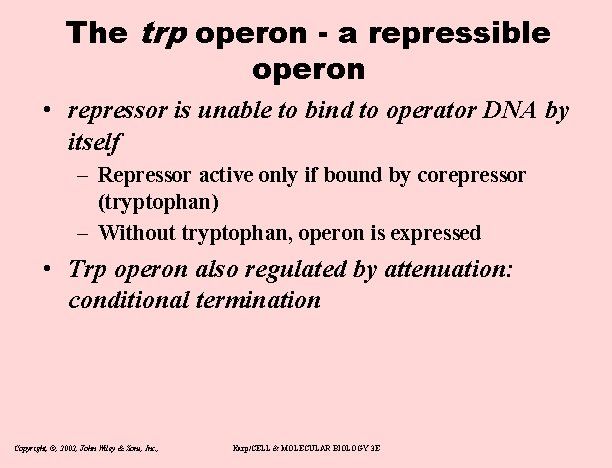
The trp operon - a repressible operon • repressor is unable to bind to operator DNA by itself – Repressor active only if bound by corepressor (tryptophan) – Without tryptophan, operon is expressed • Trp operon also regulated by attenuation: conditional termination Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 12. 26

Gene Structure and Gene Regulation in Eukaryotes Drosophila Genome Organization Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

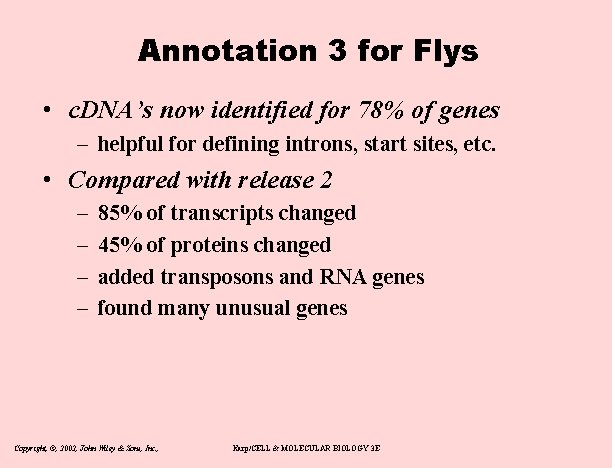
Annotation 3 for Flys • c. DNA’s now identified for 78% of genes – helpful for defining introns, start sites, etc. • Compared with release 2 – – 85% of transcripts changed 45% of proteins changed added transposons and RNA genes found many unusual genes Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

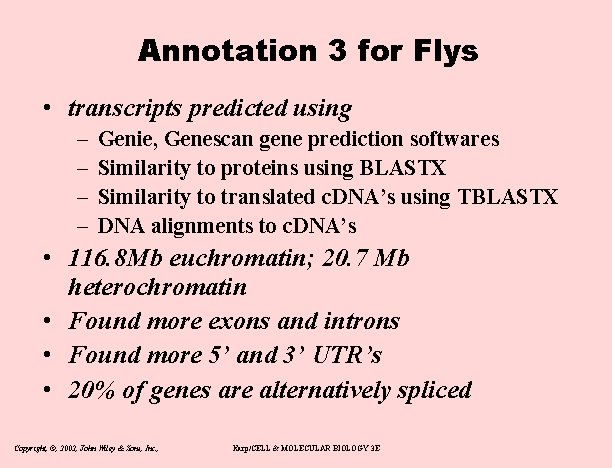
Annotation 3 for Flys • transcripts predicted using – – Genie, Genescan gene prediction softwares Similarity to proteins using BLASTX Similarity to translated c. DNA’s using TBLASTX DNA alignments to c. DNA’s • 116. 8 Mb euchromatin; 20. 7 Mb heterochromatin • Found more exons and introns • Found more 5’ and 3’ UTR’s • 20% of genes are alternatively spliced Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

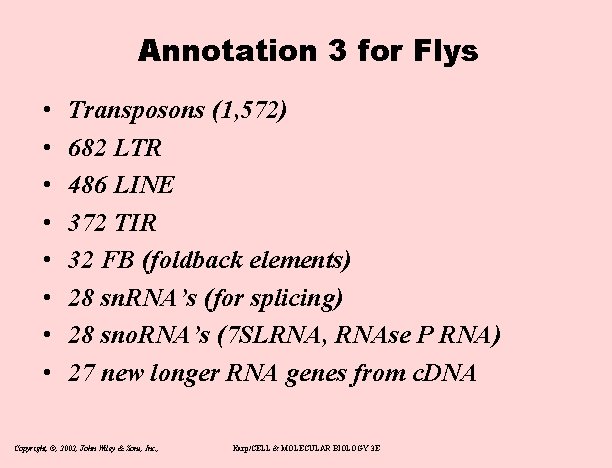
Annotation 3 for Flys • • Transposons (1, 572) 682 LTR 486 LINE 372 TIR 32 FB (foldback elements) 28 sn. RNA’s (for splicing) 28 sno. RNA’s (7 SLRNA, RNAse P RNA) 27 new longer RNA genes from c. DNA Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Annotation 3 for Flys • 17 pseudogenes (15 simple recombination, 1 is processed, 1 is very diverged) • 802 new protein coding genes • Resolved some repeated genes (Trypsin) • 345 genes from release 2 rejected (<50 aa’s, predicted only) Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

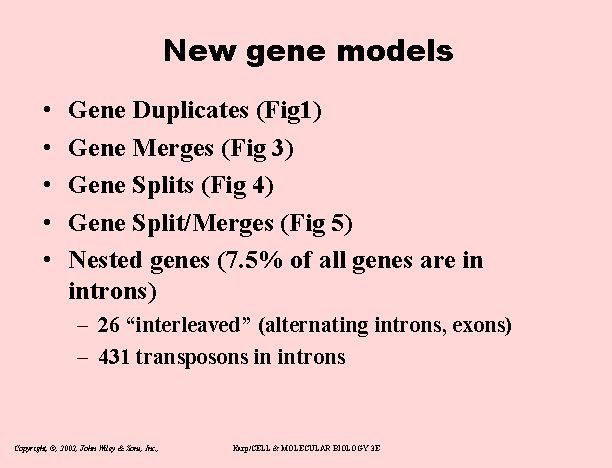
New gene models • • • Gene Duplicates (Fig 1) Gene Merges (Fig 3) Gene Splits (Fig 4) Gene Split/Merges (Fig 5) Nested genes (7. 5% of all genes are in introns) – 26 “interleaved” (alternating introns, exons) – 431 transposons in introns Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

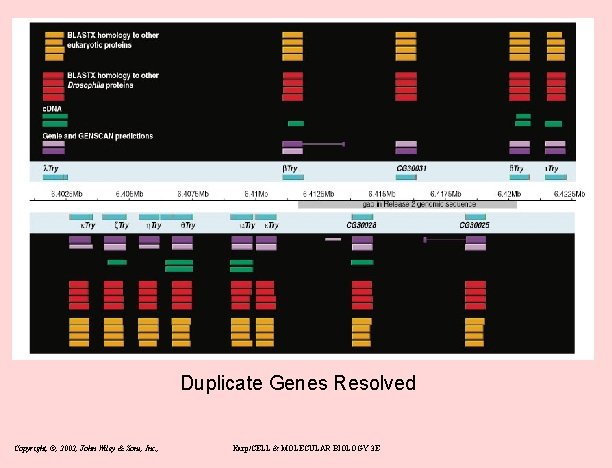
Duplicate Genes Resolved Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

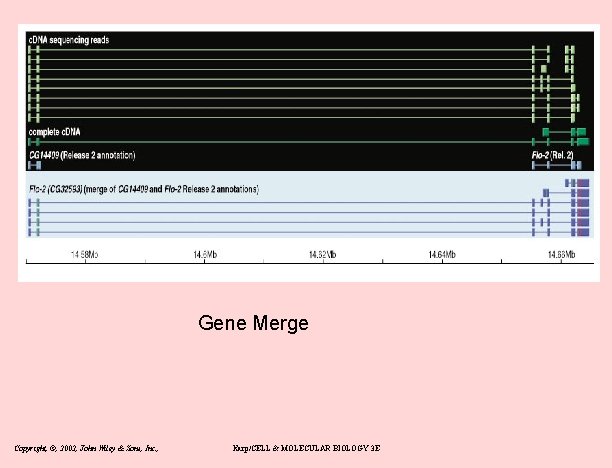
Gene Merge Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

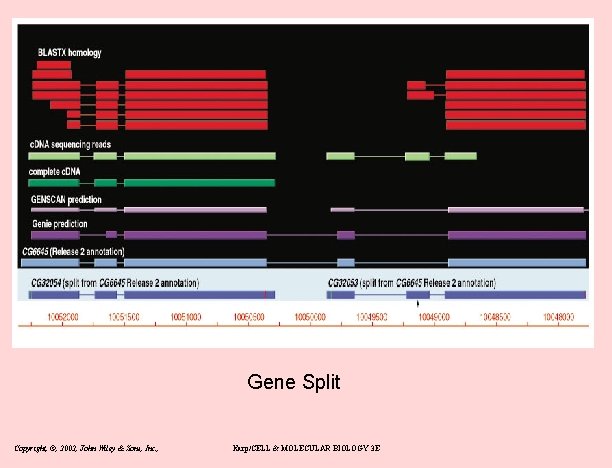
Gene Split Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

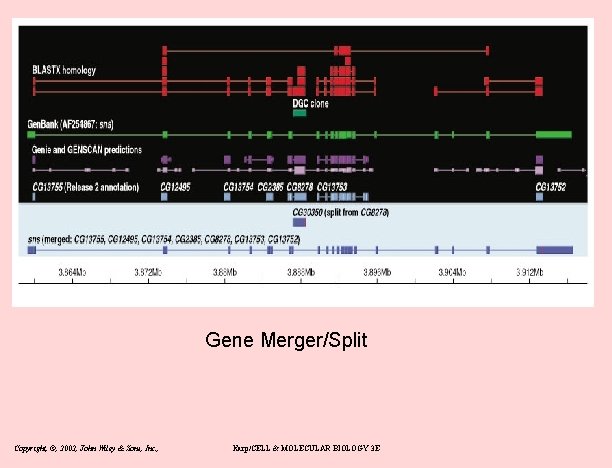
Gene Merger/Split Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

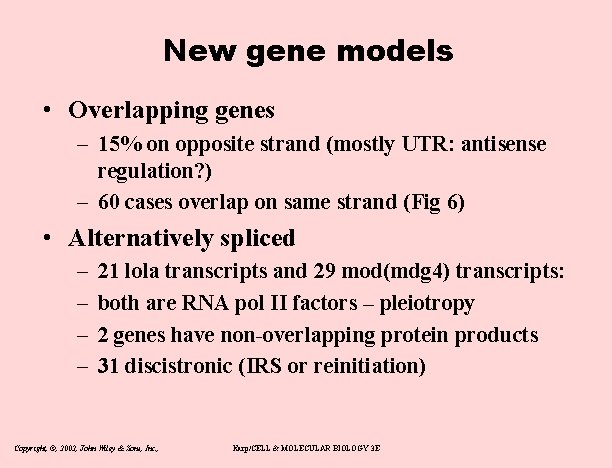
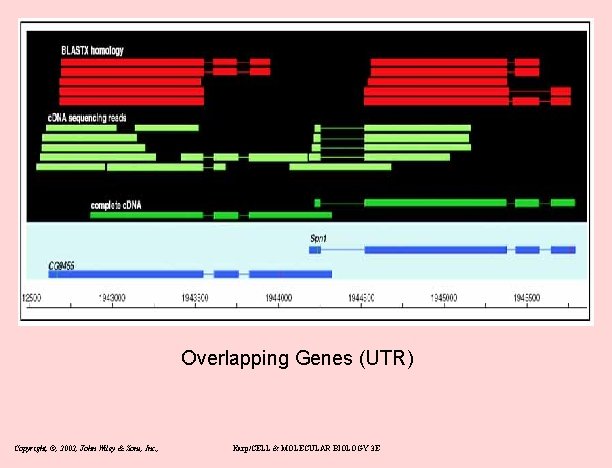
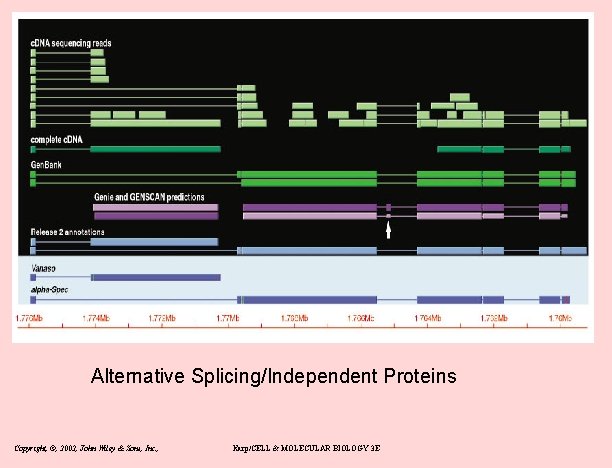
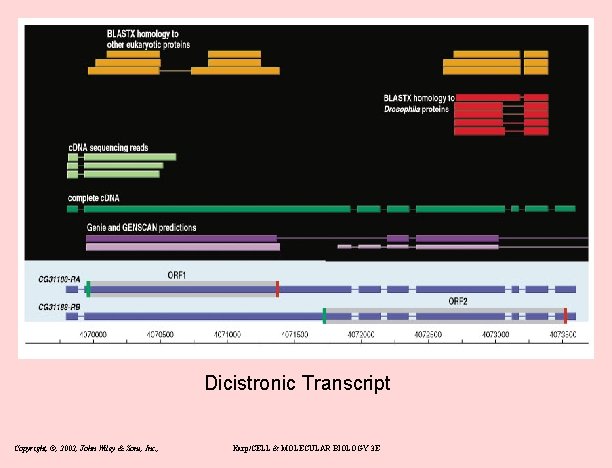
New gene models • Overlapping genes – 15% on opposite strand (mostly UTR: antisense regulation? ) – 60 cases overlap on same strand (Fig 6) • Alternatively spliced – – 21 lola transcripts and 29 mod(mdg 4) transcripts: both are RNA pol II factors – pleiotropy 2 genes have non-overlapping protein products 31 discistronic (IRS or reinitiation) Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Overlapping Genes (UTR) Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Alternative Splicing/Independent Proteins Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Dicistronic Transcript Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

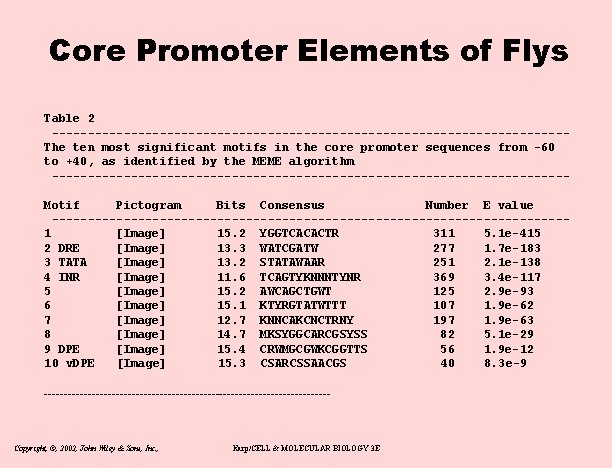
Core Promoters in Drosophila • • • Cap-trapped c. DNA 5’ ends TATA, INITIATOR, DPE, v. DPE. DRE Used to retrain Mac. Promoter 1, 941 TSS’s (11 base window) Covers 14% of all genes About 550 promotors already well described – Only 18% of new promoters matched old promoters – Only 30 seemed to have different TSS Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

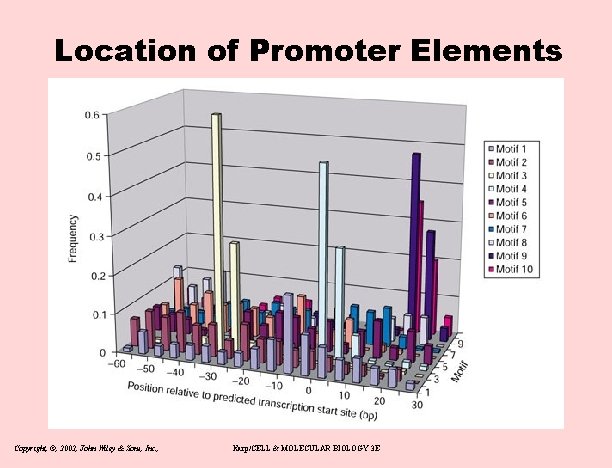
Core Promoter Elements of Flys Table 2 ------------------------------------The ten most significant motifs in the core promoter sequences from -60 to +40, as identified by the MEME algorithm ------------------------------------Motif Pictogram Bits Consensus Number E value ------------------------------------1 [Image] 15. 2 YGGTCACACTR 311 5. 1 e-415 2 DRE [Image] 13. 3 WATCGATW 277 1. 7 e-183 3 TATA [Image] 13. 2 STATAWAAR 251 2. 1 e-138 4 INR [Image] 11. 6 TCAGTYKNNNTYNR 369 3. 4 e-117 5 [Image] 15. 2 AWCAGCTGWT 125 2. 9 e-93 6 [Image] 15. 1 KTYRGTATWTTT 107 1. 9 e-62 7 [Image] 12. 7 KNNCAKCNCTRNY 197 1. 9 e-63 8 [Image] 14. 7 MKSYGGCARCGSYSS 82 5. 1 e-29 9 DPE [Image] 15. 4 CRWMGCGWKCGGTTS 56 1. 9 e-12 10 v. DPE [Image] 15. 3 CSARCSSAACGS 40 8. 3 e-9 ------------------------------------ Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

#1 -? ? , #2 -DRE Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

#3 -TATA, #4 -INR Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

#9 -DPE, #10 -v. DPE Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Location of Promoter Elements Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E
 Gene expression in prokaryotes and eukaryotes
Gene expression in prokaryotes and eukaryotes What is a positive and negative control
What is a positive and negative control Chapter 17 gene expression from gene to protein
Chapter 17 gene expression from gene to protein Dna pool noodles
Dna pool noodles Gene prediction in prokaryotes and eukaryotes
Gene prediction in prokaryotes and eukaryotes Prokaryotes vs eukaryotes gene regulation
Prokaryotes vs eukaryotes gene regulation At dna
At dna Gene by gene test results
Gene by gene test results Poltrp
Poltrp Dr kevin ahern
Dr kevin ahern Regulation of gene expression in bacteria
Regulation of gene expression in bacteria Chapter 18 regulation of gene expression
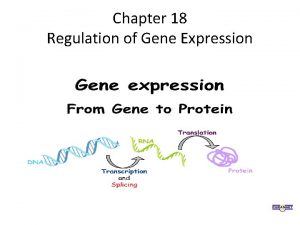
Chapter 18 regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Regulation of gene expression
Regulation of gene expression What is the first step in gene expression
What is the first step in gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Genetic effects on gene expression across human tissues
Genetic effects on gene expression across human tissues Ch 18+
Ch 18+ Rt pcr primer design tool
Rt pcr primer design tool Gene expression omnibus tutorial
Gene expression omnibus tutorial Gene expression
Gene expression Gene expression
Gene expression Gene expression
Gene expression Gene expression
Gene expression Lyonization of gene expression
Lyonization of gene expression Quadratic equation examples
Quadratic equation examples Chapter 20 viruses and prokaryotes
Chapter 20 viruses and prokaryotes Diff between prokaryotes and eukaryotes
Diff between prokaryotes and eukaryotes Chapter 20 viruses and prokaryotes
Chapter 20 viruses and prokaryotes Cytoskeleton nickname
Cytoskeleton nickname Prokaryotes and eukaryotes
Prokaryotes and eukaryotes