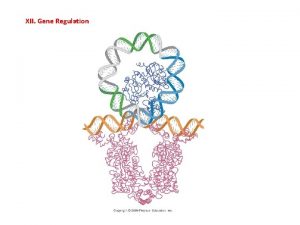
REGULATION OF GENE EXPRESSION Prokaryotes vs Eukaryotes Gene




- Slides: 29

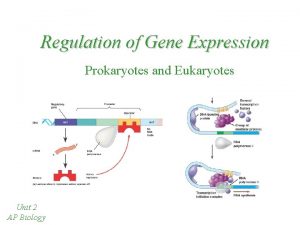
REGULATION OF GENE EXPRESSION Prokaryotes vs. Eukaryotes

Gene Expression is the process by which information from a gene is used in the synthesis of a functional protein or other gene product such as t. RNA or r. RNA. As you can imagine, not all genes are expressed at the same time in an organism.

Regulation of Gene Expression Both prokaryotic and eukaryotic cells, have the ability to turn gene expression on or off depending on what the cell or organism needs at any given time. Prokaryotic cells have evolved to be able to regulate gene expression in response to different environmental conditions. Eukaryotes regulate gene expression to maintain different cell types, in embryonic development, etc.

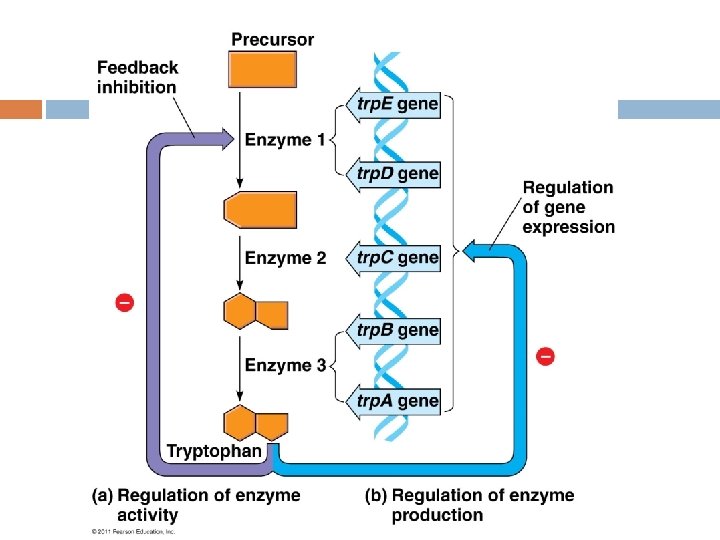
Why Regulate Gene Expression? Bacterial cells that can conserve energy have the selective advantage over cells that are unable to do so. Natural selection has favored bacteria who express genes only when their products are needed by the cell.

Why regulate gene expression? E. coli can adjust their metabolism to changing environments i. e. � They can adjust the activity of enzymes already present in the cell by Feedback inhibition where the product inhibits the first enzyme in a metabolic pathway. � Cells can adjust the production level of the enzyme and only produce it as needed.


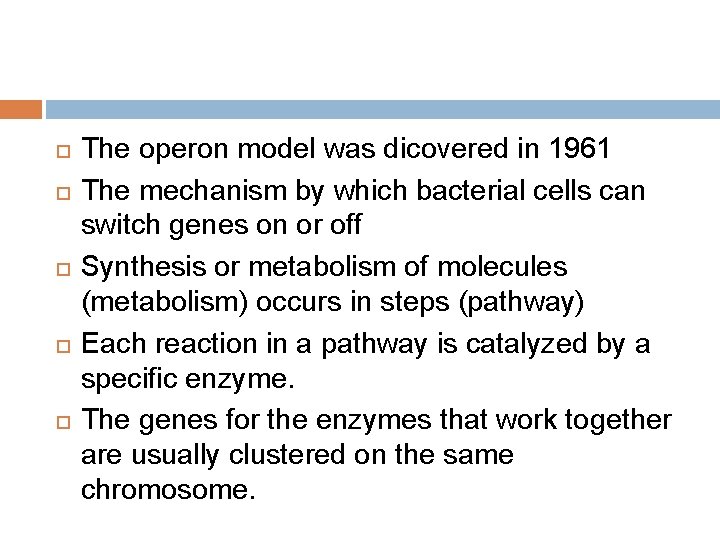
The operon model was dicovered in 1961 The mechanism by which bacterial cells can switch genes on or off Synthesis or metabolism of molecules (metabolism) occurs in steps (pathway) Each reaction in a pathway is catalyzed by a specific enzyme. The genes for the enzymes that work together are usually clustered on the same chromosome.

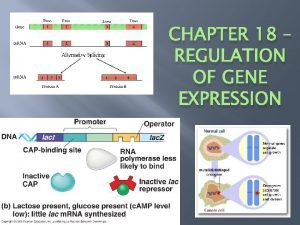
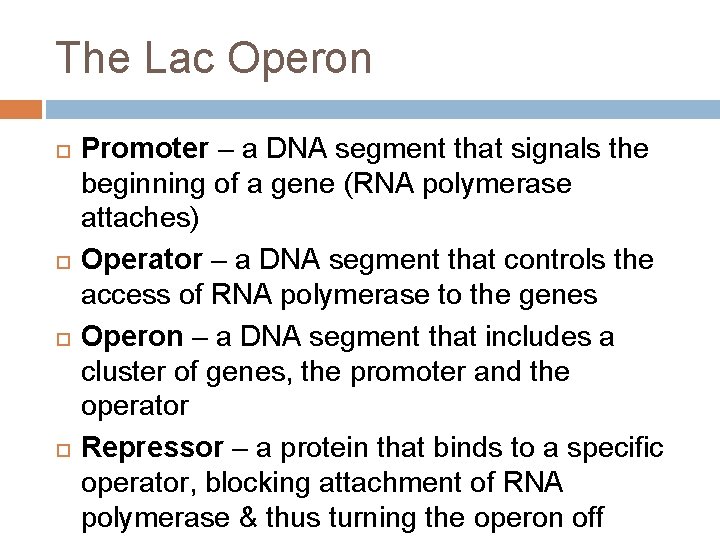
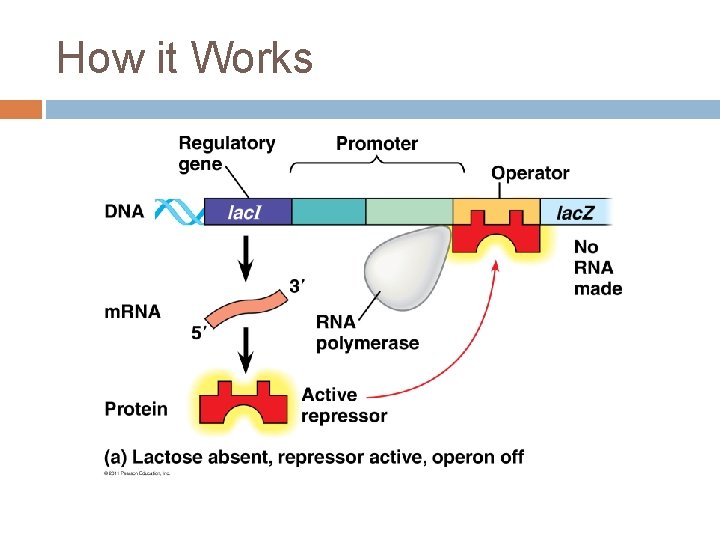
The Lac Operon Promoter – a DNA segment that signals the beginning of a gene (RNA polymerase attaches) Operator – a DNA segment that controls the access of RNA polymerase to the genes Operon – a DNA segment that includes a cluster of genes, the promoter and the operator Repressor – a protein that binds to a specific operator, blocking attachment of RNA polymerase & thus turning the operon off

The Lac Operon Regulatory genes – code for repressor proteins

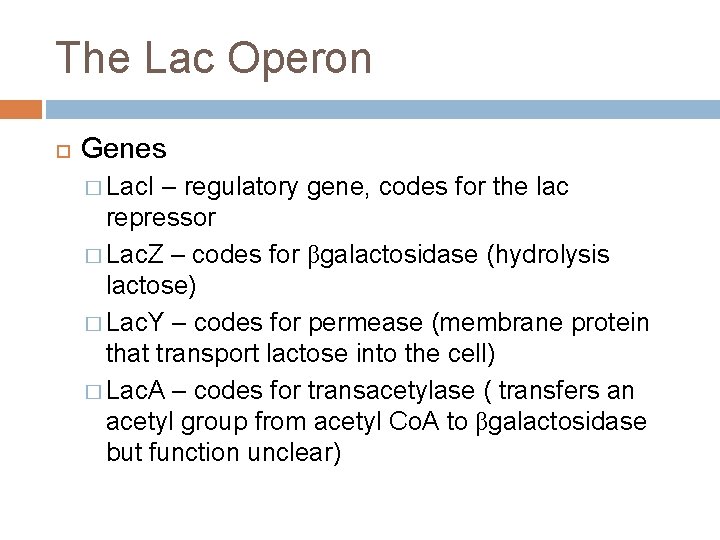
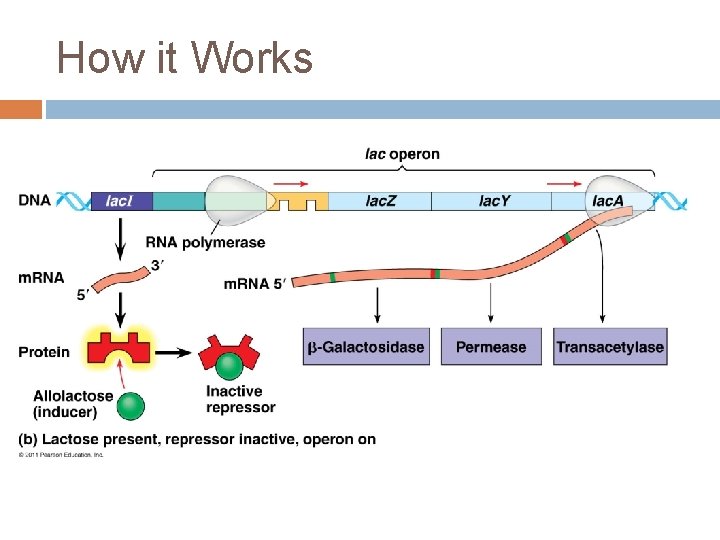
The Lac Operon Genes � Lac. I – regulatory gene, codes for the lac repressor � Lac. Z – codes for βgalactosidase (hydrolysis lactose) � Lac. Y – codes for permease (membrane protein that transport lactose into the cell) � Lac. A – codes for transacetylase ( transfers an acetyl group from acetyl Co. A to βgalactosidase but function unclear)

How it works The dissacharide lacose is available to E. coli in the human colon if the host drinks milk. Lactose metabolism begins with hydrolysis into ___ and _____. This reaction is catalyzed by β-galactosidase. In the absence of glucose, only a few molecules are present in the E. coli cell. If lactose is added to the environment, the number of molecules of enzyme increase a thousandfold

E. coli uses 3 enzymes to metabolize lactose. The genes for these enzymes are all clustered together on a molecule of DNA in the lac operon.

How it Works

How it Works

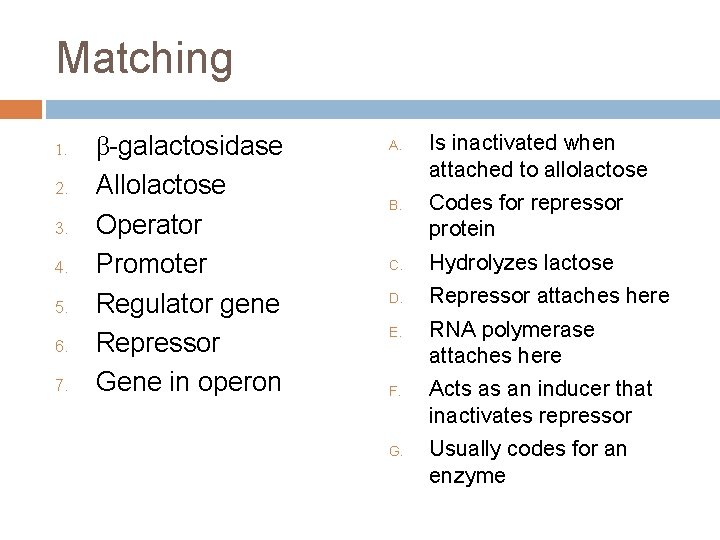
Matching 1. 2. 3. 4. 5. 6. 7. β-galactosidase Allolactose Operator Promoter Regulator gene Repressor Gene in operon A. B. Is inactivated when attached to allolactose Codes for repressor protein C. Hydrolyzes lactose D. Repressor attaches here E. F. G. RNA polymerase attaches here Acts as an inducer that inactivates repressor Usually codes for an enzyme

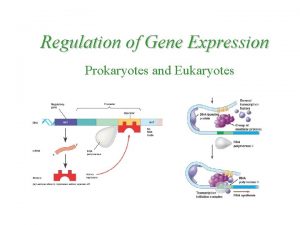
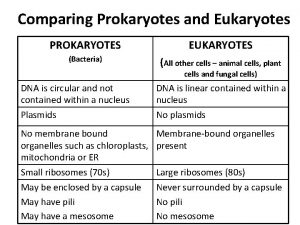
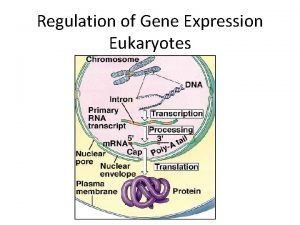
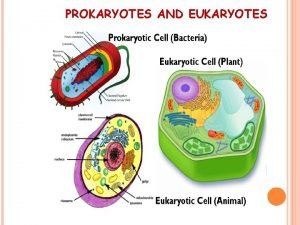
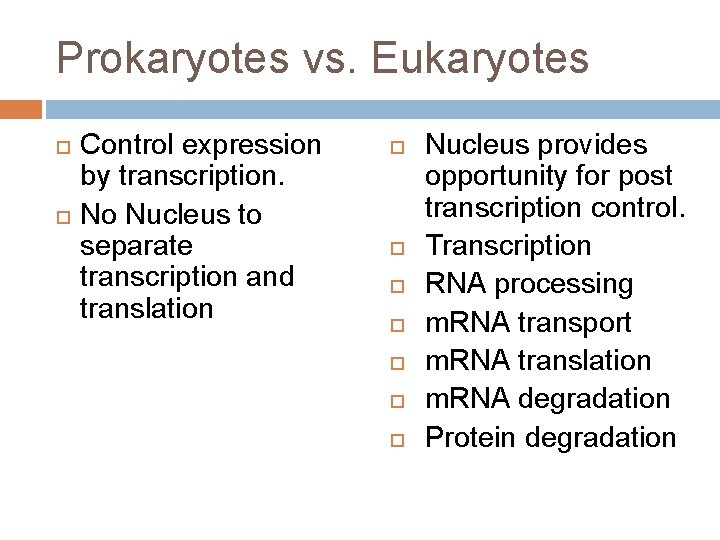
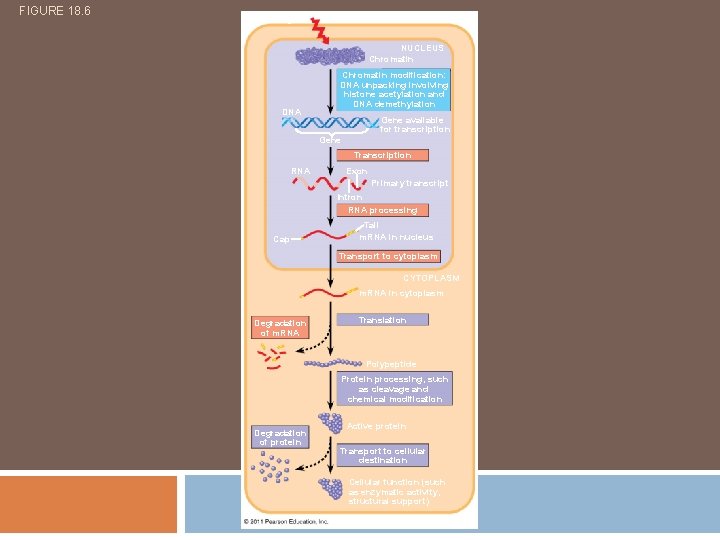
Prokaryotes vs. Eukaryotes Control expression by transcription. No Nucleus to separate transcription and translation Nucleus provides opportunity for post transcription control. Transcription RNA processing m. RNA transport m. RNA translation m. RNA degradation Protein degradation

FIGURE 18. 6 Signal NUCLEUS Chromatin DNA Chromatin modification: DNA unpacking involving histone acetylation and DNA demethylation Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Cap Tail m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing, such as cleavage and chemical modification Degradation of protein Active protein Transport to cellular destination Cellular function (such as enzymatic activity, structural support)

Control Points of Gene Expression Chromatin Modification Transcription RNA Processing (Post transcriptional) m. RNA degradation (Post transcriptional) Translation Protein Degradation (Post translational)

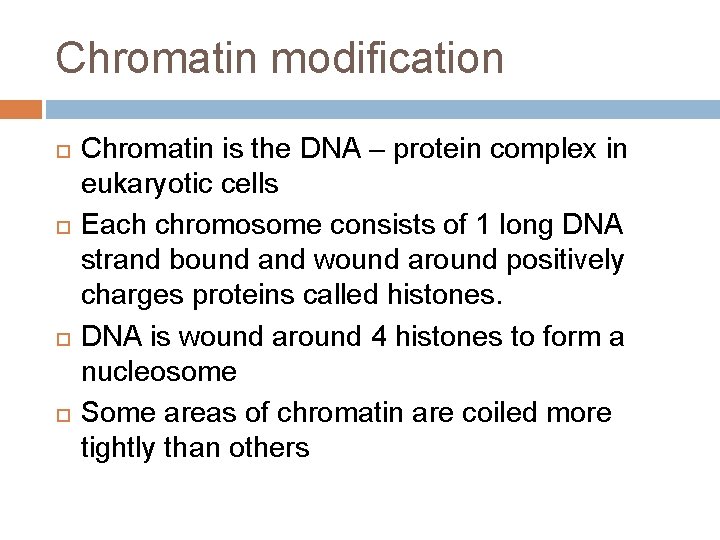
Chromatin modification Chromatin is the DNA – protein complex in eukaryotic cells Each chromosome consists of 1 long DNA strand bound and wound around positively charges proteins called histones. DNA is wound around 4 histones to form a nucleosome Some areas of chromatin are coiled more tightly than others

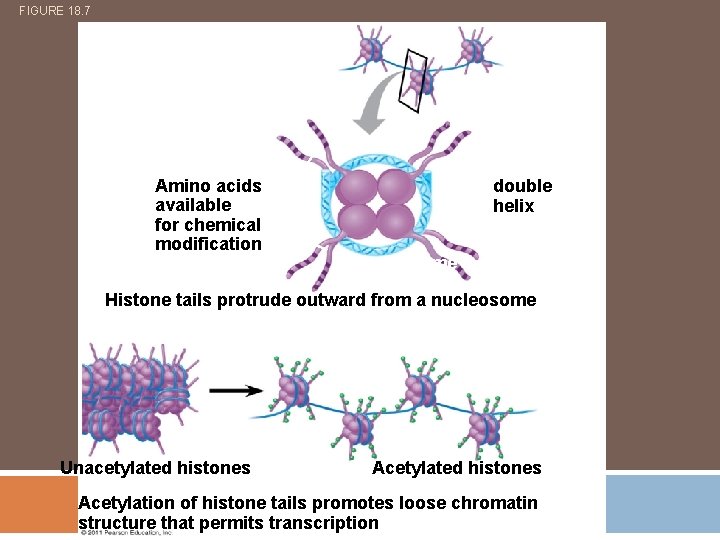
FIGURE 18. 7 Histone tails Amino acids available for chemical modification DNA double helix Nucleosome (end view) (a) Histone tails protrude outward from a nucleosome Unacetylated histones Acetylation of histone tails promotes loose chromatin structure that permits transcription

Genes within regions where chromatin is tightly coiled (heterochromatin) are usually not transcribed or expressed. Histones can be modified to regulate gene expression DNA can be modified to regulate gene expression

Histone tails protrude from the nucleosome and are accessible to enzymes that catalyse the addition or removal of certain chemical groups. Acetyl, methyl, phosphate groups Acetylation – promotes transcription (loosens chromatin) Methylation – prevents transcription (tightens chromatin)

Epigenetic inheritance Once methylated, genes usually stay that way through successive generations Enzymes methylate daughter strands according to the template strand in each round of replication. Chromatin modification such as methylation can be passed from generation to generation. Epigenetics is the inheritance of traits transmitted by mechanisms not involving the nucleotide sequence.

Changes in nucleotide sequences (mutations)are permanent. Changes in chromatin (methylation, acetylation) is reversible. Alterations in normal patterns of DNA methylation are seen in some cancers (inappropriate gene expression)

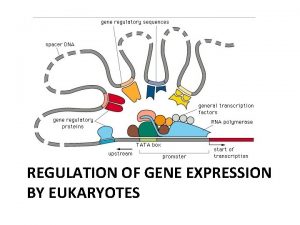
Transcriptional Control Includes transcription factors Enhancers Promoters Hormones

A Typical Eukaryotic Gene Consists of a promoter sequence – a sequence where RNA polymerase binds and starts transcription Introns Exons

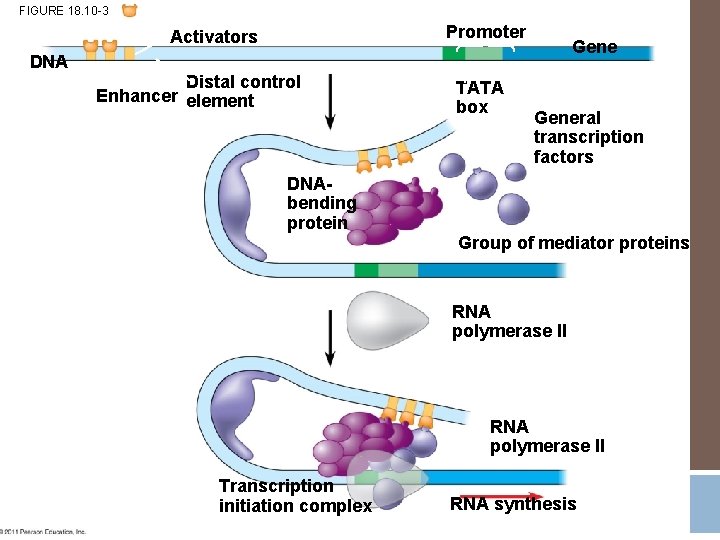
Upstream from the promoter sequence are: � Enhancer (distal control elements) � Proximal Control Elements

RNA polymerase attaches to the promoter sequence

FIGURE 18. 10 -3 Promoter Activators DNA Distal control Enhancer element TATA box General transcription factors DNAbending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis
 Prokaryotes vs eukaryotes gene regulation
Prokaryotes vs eukaryotes gene regulation Gene regulation
Gene regulation Gene prediction in prokaryotes and eukaryotes
Gene prediction in prokaryotes and eukaryotes Prokaryotes vs eukaryotes chart
Prokaryotes vs eukaryotes chart Types of organelles
Types of organelles Prokaryotic vs eukaryotic
Prokaryotic vs eukaryotic Polimerase
Polimerase Diff between prokaryotes and eukaryotes
Diff between prokaryotes and eukaryotes Prokaryotes vs eukaryotes
Prokaryotes vs eukaryotes Prokaryotic vs eukaryotic cells venn diagram
Prokaryotic vs eukaryotic cells venn diagram Prokaryotes and eukaryotes
Prokaryotes and eukaryotes Multiple choice questions on prokaryotes and eukaryotes
Multiple choice questions on prokaryotes and eukaryotes Prokaryotic vs eukaryotic transcription
Prokaryotic vs eukaryotic transcription Anatomy of prokaryotes and eukaryotes
Anatomy of prokaryotes and eukaryotes Prokaryotic cells vs eukaryotic cells
Prokaryotic cells vs eukaryotic cells Prokaryotes and eukaryotes
Prokaryotes and eukaryotes Regulation of gene expression
Regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18
Chapter 18 Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Regulation of gene expression
Regulation of gene expression Chapter 17 from gene to protein
Chapter 17 from gene to protein Section 4 gene regulation and mutation
Section 4 gene regulation and mutation Difference between positive and negative control
Difference between positive and negative control What is gene regulation
What is gene regulation Section 12-5 gene regulation answer key
Section 12-5 gene regulation answer key Gene regulation
Gene regulation Section 4 gene regulation and mutations
Section 4 gene regulation and mutations Differential gene regulation
Differential gene regulation