Complex and Dynamic Analysis of Nascent Transcription Introduction













- Slides: 34

Complex and Dynamic Analysis of Nascent Transcription Introduction and overview

Robin Dowell Bio. Frontiers Institute University of Colorado Boulder Robin. Dowell@colorado. edu Margaret. Gruca@colorado. edu Jacob. Stanley@colorado. edu Margaret Gruca Nascent analysis Jacob Stanley RNA-seq

Goals of this workshop 1. Understand the distinction between nascent transcription and steady state RNA (RNA-seq) analysis. 2. Work through a comparative transcriptome data analysis example. 3. Discuss basic statistical concepts that are paramount to understanding your data.

Schedule: Today • Session 1 (1: 30 pm - 2: 45 pm): Studying Transcription -- an Overview • Coffee Break (2: 45 -3: 15 pm) • Session 2 (3: 15 -4: 30 pm): Garbage In, Garbage Out: Quality control on sequence data • Session 3 (4: 30 -5: 15 pm): Maximize read usage through mapping strategies

Schedule: Tuesday Morning • Session 4 (8: 30 -9: 30 am): Learning to count: quantifying signal • Coffee Break (9: 30 -10: 00) • Session 5 (10 -11: 15) : Assessing changes in data (part I: resorting to statistics) • Session 6 (11: 15 -12: 30): Assessing changes in data (part II: differential expression)

Schedule: Tuesday Afternoon • Session 7 (1: 30 -2: 45): Annotation agnostic analysis: finding new things in the data • Coffee Break (2: 45 -3: 00 pm) • Session 8 (3: 15 -4: 30): Getting the most from your data: Integration with relevant other data

Key concepts of workshop • Understanding the experiment is key to its analysis. • A fundamental principle in data science is understanding your expectation and then evaluating your reality. • Reproducibility requires keeping good records. You *MUST* report on your software choices, options, order, etc.

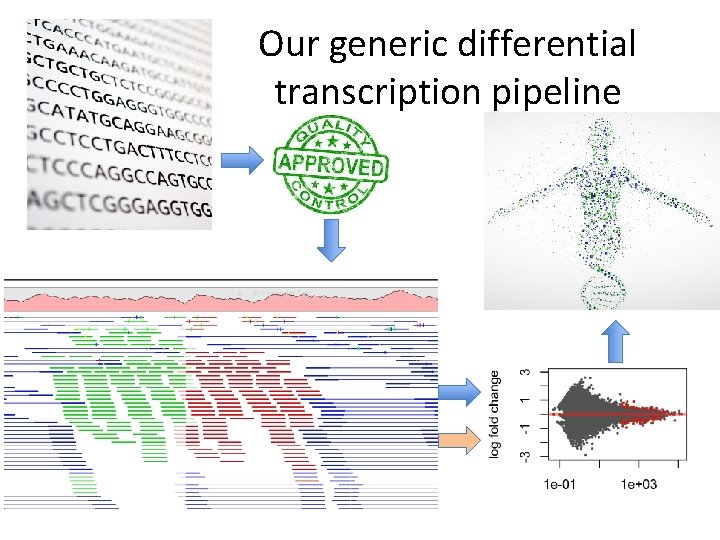
Our generic differential transcription pipeline

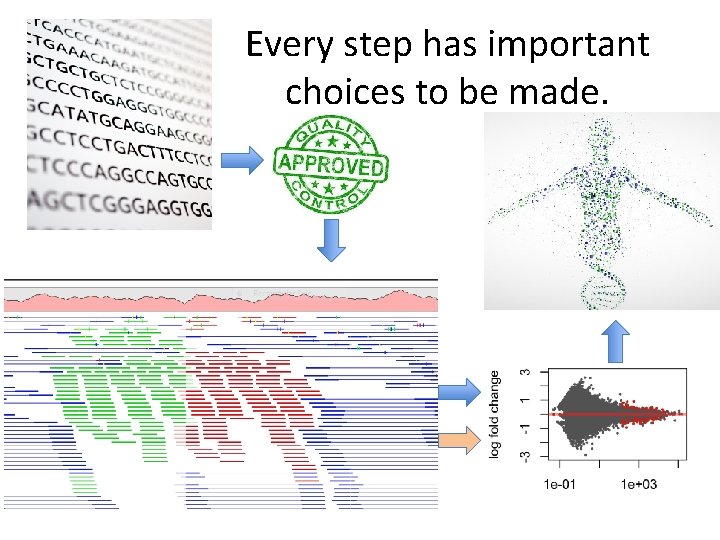
Every step has important choices to be made.

There is no ONE RIGHT answer. • Every algorithm/pipeline is different • Different assumptions • Order of processing can sometimes have a significant effect • Different options can dramatically change the result • MOST projects require customization and personalized attention to get useful data

Important consideration for bioinformatics (in general) Keep a digital notebook • • • Program and version Options and variables utilized Order of usage Specifics of compute environment Avoid “manual edits” whenever possible Backup your work!! You must be able to report on these elements of your analysis in order for your work to be REPRODUCIBLE!!!!

https: //rmghc. slack. com/

Our compute environment Amazon Web Services (https: //aws. amazon. com) graciously provided resources for the workshops and hackathon. We are using an EC 2 compute instance setup by Bio. Frontiers IT staff. https: //docs. google. com/spreadsheets/d/14 v. Stf 4 em. JO 6 i. REK 8 Vsy. SLa 7 O 45 vb 8 y. Rf. LOFm 3 NQIZuo/edit#gid=0

Assumptions of this workshop 1) You start with a bag of reads off the sequencer, BUT … MANY decisions pre-sequencing influence your analysis. So we MUST understand how the sample was handled BEFORE it was sequenced. 2) You already know how to use a Unix/Linux command line

http: //dowell. colorado. edu/Hack. Con/pages/hackcon. html

To view on your laptop … X 2 Go http: //dowell. colorado. edu/Hack. Con/pages/hackcon. html

What is IGV A desktop/server application for integrated visualization of multiple data types and annotations in the context of the genome Microarrays Epigenomics RNA-Seq NGS alignments Comparative genomics

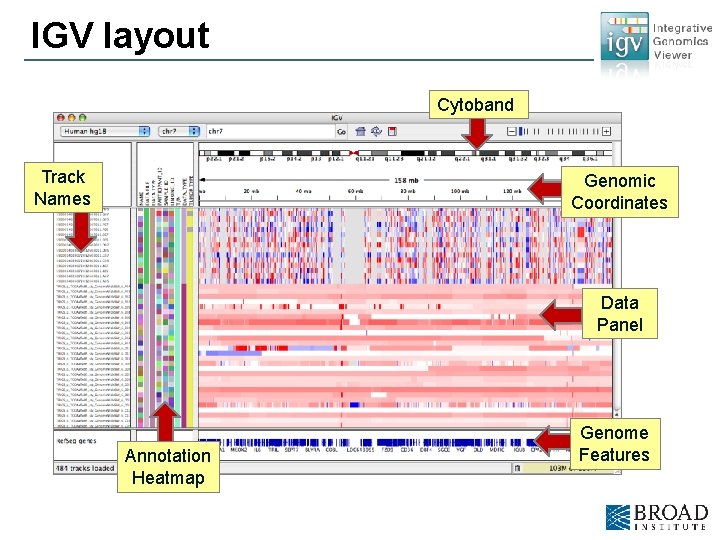
IGV layout Cytoband Track Names Genomic Coordinates Data Panel Annotation Heatmap Genome Features


Every cell contains essentially the same DNA, but is distinct


But how? Transcriptional Regulation

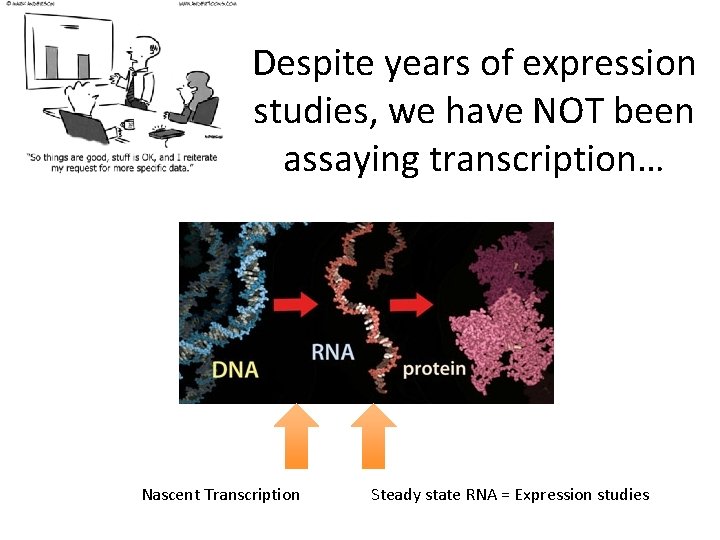
Despite years of expression studies, we have NOT been assaying transcription… Nascent Transcription Steady state RNA = Expression studies

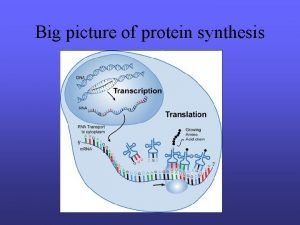
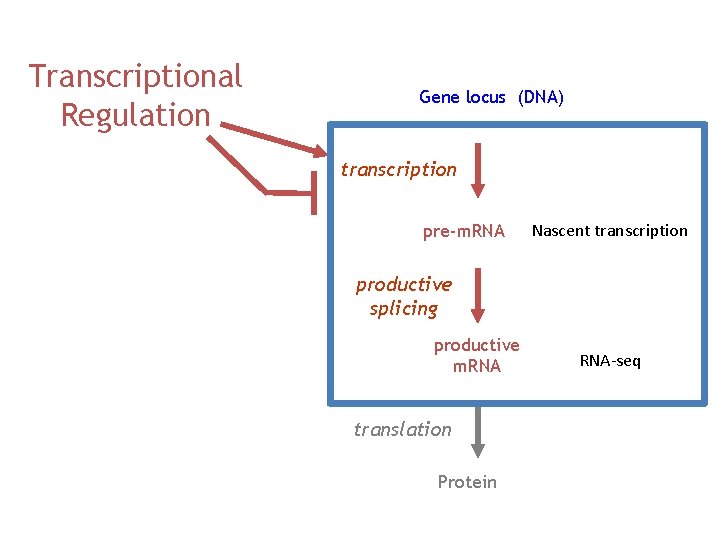
Transcriptional Regulation Gene locus (DNA) transcription pre-m. RNA Nascent transcription productive splicing productive m. RNA translation Protein RNA-seq

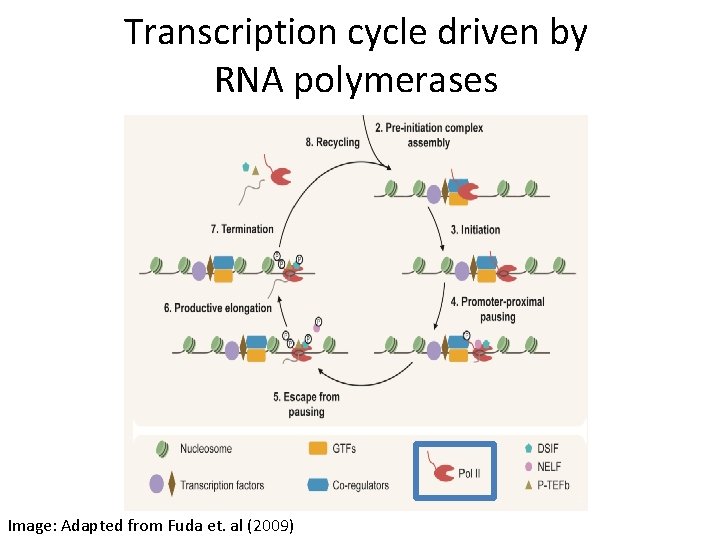
Transcription cycle driven by RNA polymerases Image: Adapted from Fuda et. al (2009)

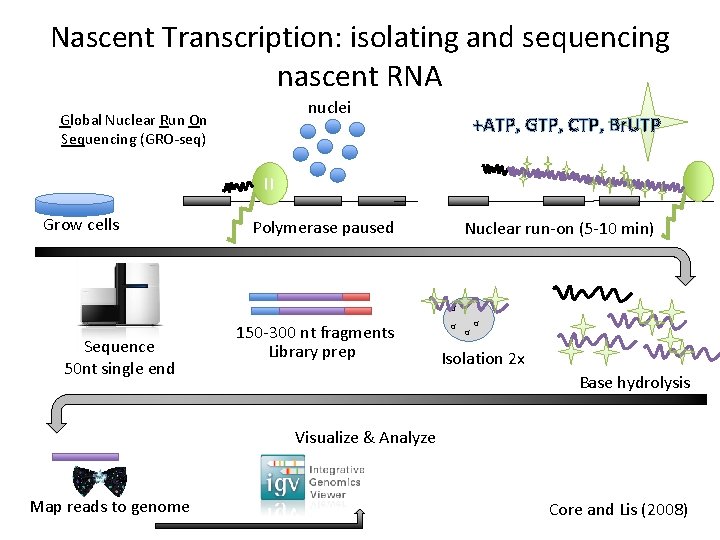
Nascent Transcription: isolating and sequencing nascent RNA nuclei Global Nuclear Run On Sequencing (GRO-seq) +ATP, GTP, CTP, Br. UTP = Grow cells Polymerase paused Nuclear run-on (5 -10 min) α Sequence 50 nt single end 150 -300 nt fragments Library prep α α α Isolation 2 x Base hydrolysis Visualize & Analyze Map reads to genome Core and Lis (2008)

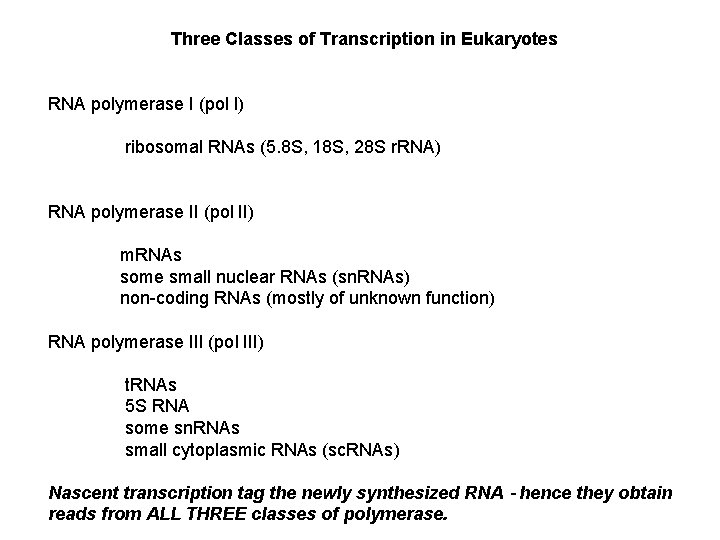
Three Classes of Transcription in Eukaryotes RNA polymerase I (pol I) ribosomal RNAs (5. 8 S, 18 S, 28 S r. RNA) RNA polymerase II (pol II) m. RNAs some small nuclear RNAs (sn. RNAs) non-coding RNAs (mostly of unknown function) RNA polymerase III (pol III) t. RNAs 5 S RNA some sn. RNAs small cytoplasmic RNAs (sc. RNAs) Nascent transcription tag the newly synthesized RNA – hence they obtain reads from ALL THREE classes of polymerase.

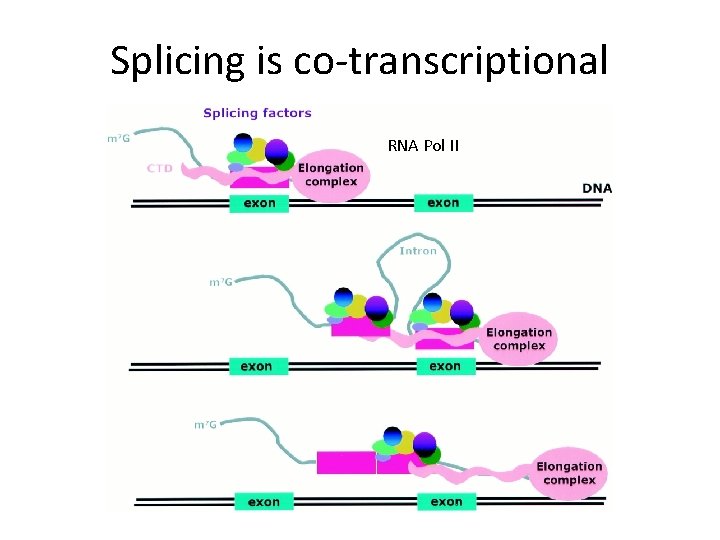
Splicing is co-transcriptional RNA Pol II

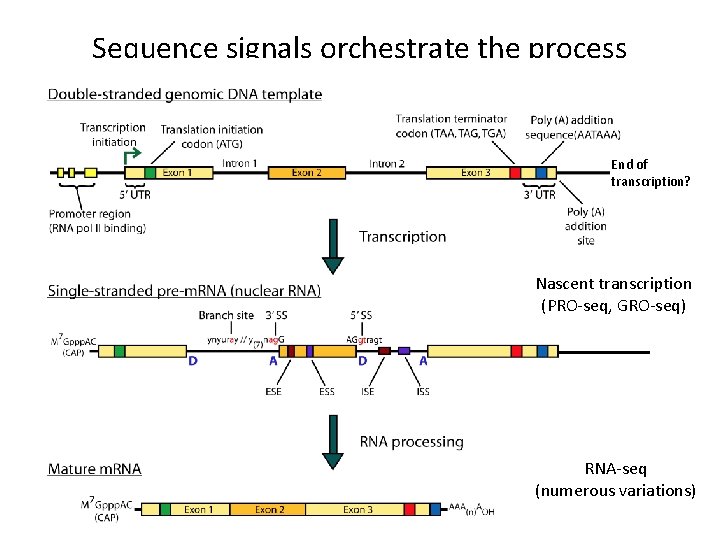
Sequence signals orchestrate the process End of transcription? Nascent transcription (PRO-seq, GRO-seq) RNA-seq (numerous variations)

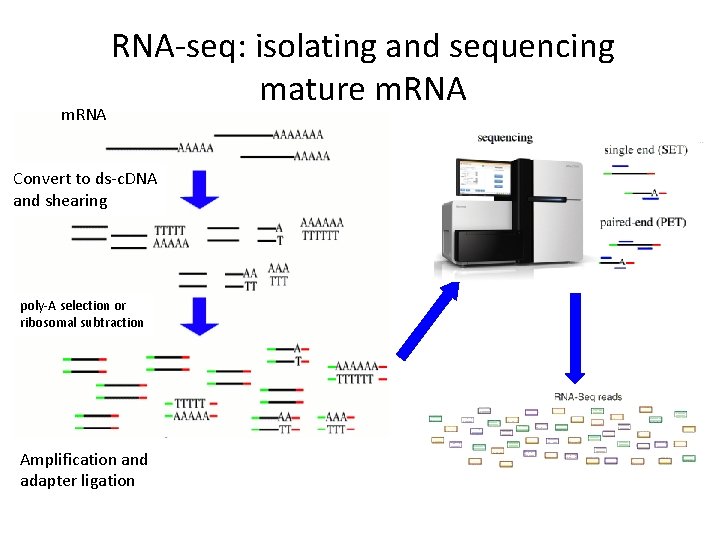
m. RNA-seq: isolating and sequencing mature m. RNA Convert to ds-c. DNA and shearing poly-A selection or ribosomal subtraction Amplification and adapter ligation

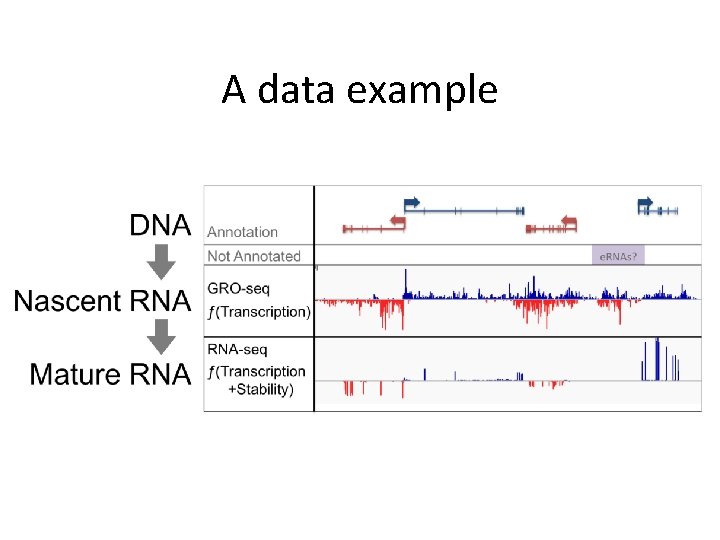
A data example

So what kinds of problems are most appropriate to each protocol?

But regardless of which transcription protocol (nascent or steady state), there are important consistent considerations! This is the single largest goal of this workshop!!

Schedule: Today • Session 1 (1: 30 pm - 2: 45 pm): Studying Transcription -- an Overview • Coffee Break (2: 45 -3: 15 pm) • Session 2 (3: 15 -4: 30 pm): Garbage In, Garbage Out: Quality control on sequence data • Session 3 (4: 30 -5: 15 pm): Maximize read usage through mapping strategies
 Nascent soap method
Nascent soap method Auxiliary method of emulsion
Auxiliary method of emulsion Dry gum method of emulsion
Dry gum method of emulsion Bottle method of emulsion
Bottle method of emulsion Simple compound complex and compound-complex sentences quiz
Simple compound complex and compound-complex sentences quiz Ghon complex
Ghon complex Simple, compound complex rules
Simple, compound complex rules Freud complexes
Freud complexes Psychodynamic and psychoanalytic theory difference
Psychodynamic and psychoanalytic theory difference Latent fixation
Latent fixation Cuckoo sandbox vm
Cuckoo sandbox vm Dynamic dynamic - bloom
Dynamic dynamic - bloom Transcription end result
Transcription end result Virusmax
Virusmax Dna to rna transcription
Dna to rna transcription Read the transcriptions and write the words
Read the transcriptions and write the words Protein synthesis bbc bitesize
Protein synthesis bbc bitesize Dna transcription and translation
Dna transcription and translation Dna replication transcription and translation
Dna replication transcription and translation Narrow and broad transcription
Narrow and broad transcription Dna and transcription tutorial
Dna and transcription tutorial Transcription and translation
Transcription and translation Transcription and translation picture
Transcription and translation picture Translation or transcription
Translation or transcription Biology transcription and translation
Biology transcription and translation Rna matching
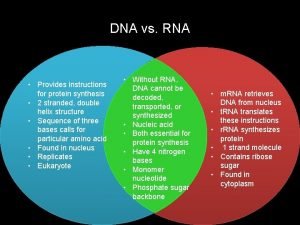
Rna matching Venn diagram to compare dna and rna
Venn diagram to compare dna and rna Transcription and translation
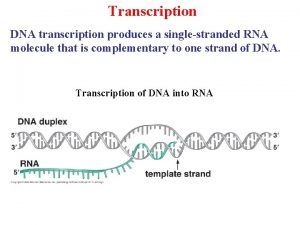
Transcription and translation Dna transcription
Dna transcription The sounds of language
The sounds of language Hiv reverse transcription
Hiv reverse transcription Speech phonetic transcription
Speech phonetic transcription Transcription translation replication
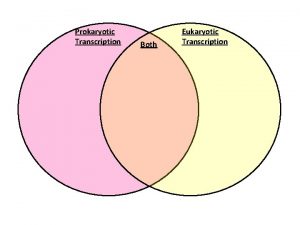
Transcription translation replication Eukaryotic vs prokaryotic transcription
Eukaryotic vs prokaryotic transcription Regressive assimilation
Regressive assimilation