The Signal Hypothesis and the Targeting of Nascent

- Slides: 35

The Signal Hypothesis and the Targeting of Nascent Polypeptides to the Secretory Pathway Tuesday 8/30/2018 Mike Mueckler mmueckler@wustl. edu

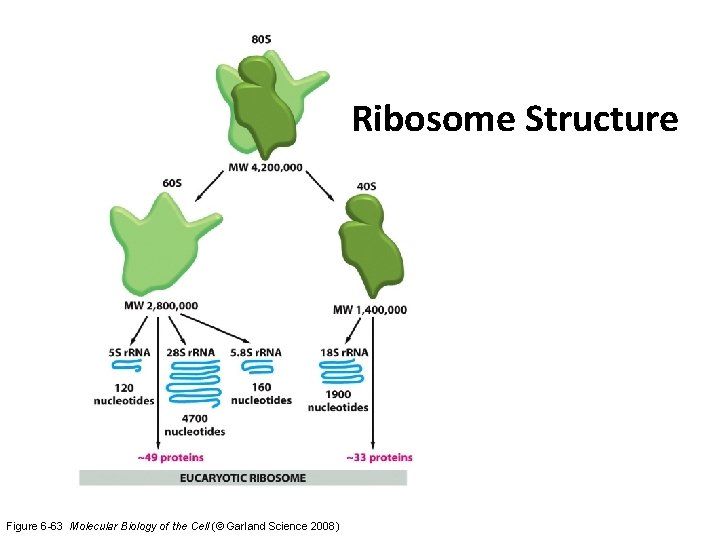
Ribosome Structure Figure 6 -63 Molecular Biology of the Cell (© Garland Science 2008)

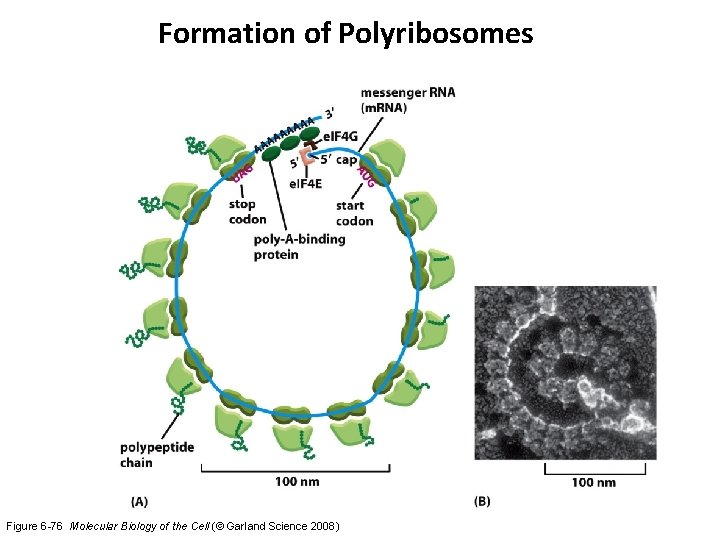
Formation of Polyribosomes Figure 6 -76 Molecular Biology of the Cell (© Garland Science 2008)

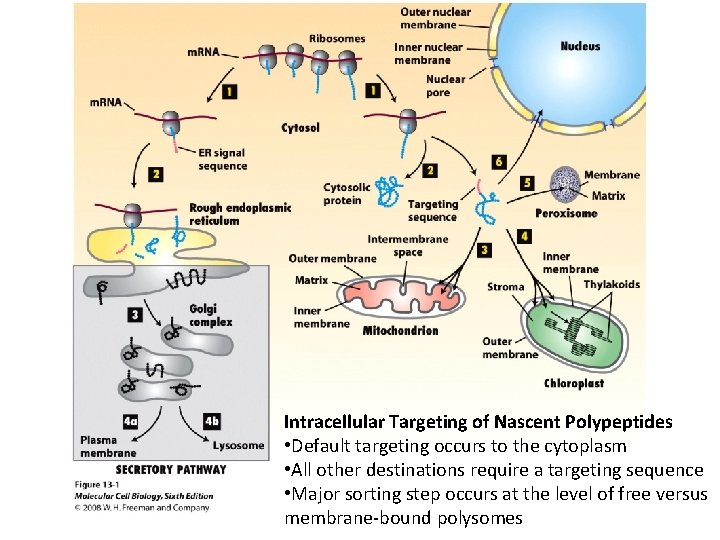
Intracellular Targeting of Nascent Polypeptides • Default targeting occurs to the cytoplasm • All other destinations require a targeting sequence • Major sorting step occurs at the level of free versus membrane-bound polysomes

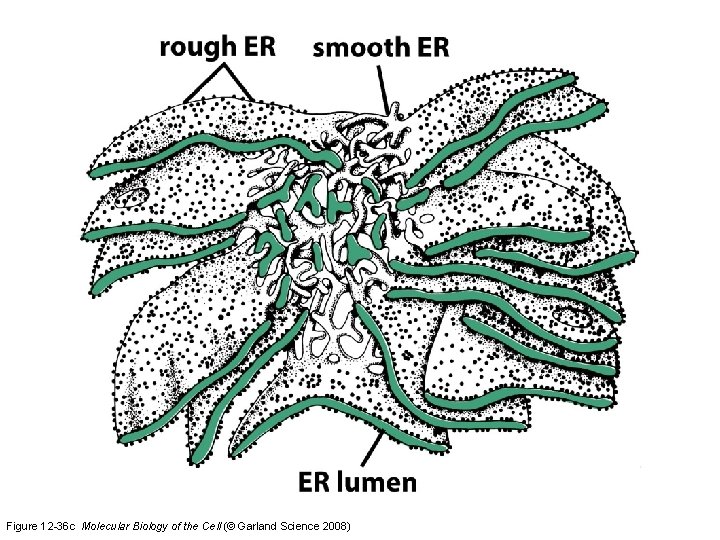
Figure 12 -36 c Molecular Biology of the Cell (© Garland Science 2008)

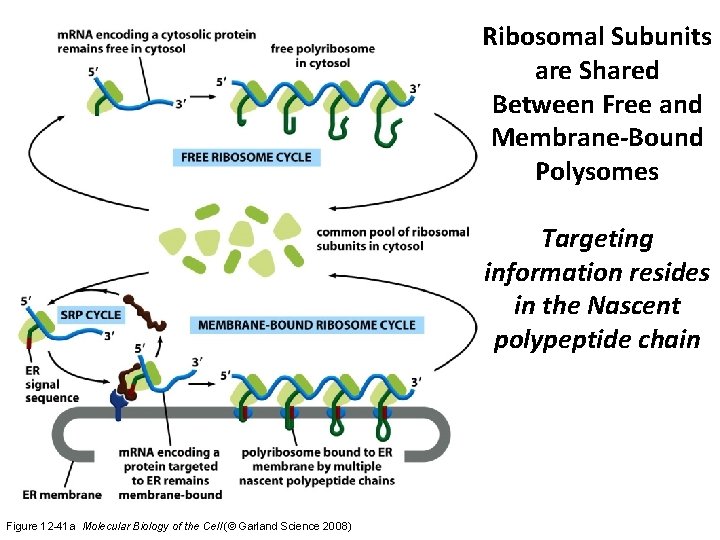
Ribosomal Subunits are Shared Between Free and Membrane-Bound Polysomes Targeting information resides in the Nascent polypeptide chain Figure 12 -41 a Molecular Biology of the Cell (© Garland Science 2008)

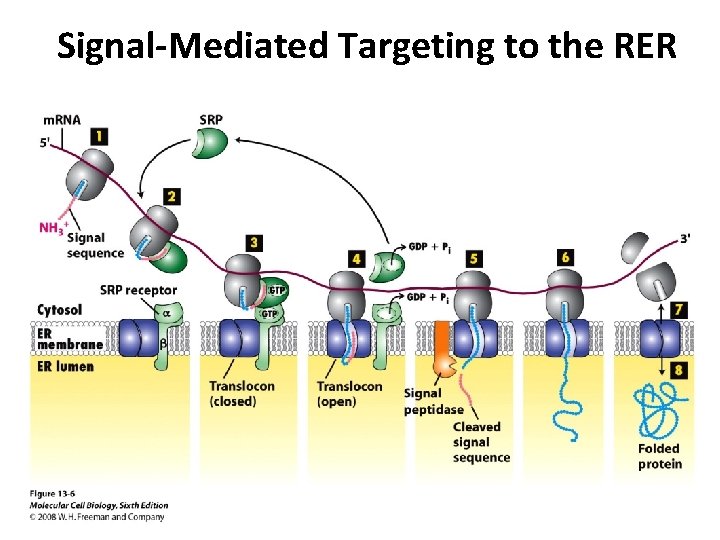
Signal-Mediated Targeting to the RER

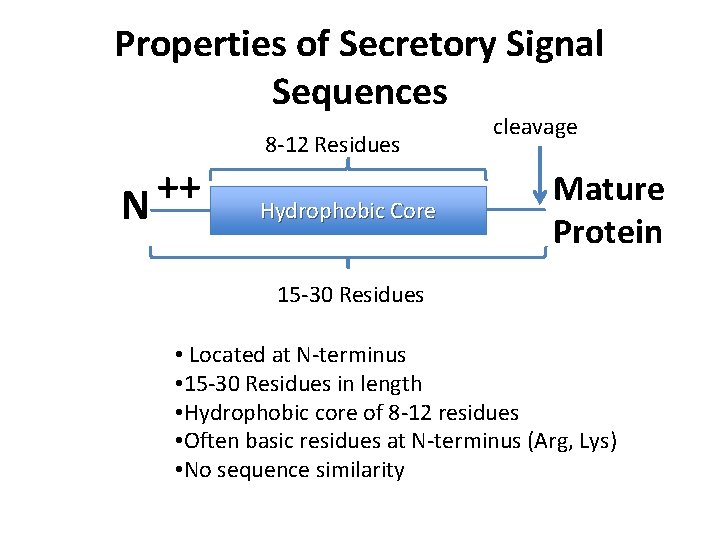
Properties of Secretory Signal Sequences ++ N 8 -12 Residues Hydrophobic Core cleavage Mature Protein 15 -30 Residues • Located at N-terminus • 15 -30 Residues in length • Hydrophobic core of 8 -12 residues • Often basic residues at N-terminus (Arg, Lys) • No sequence similarity

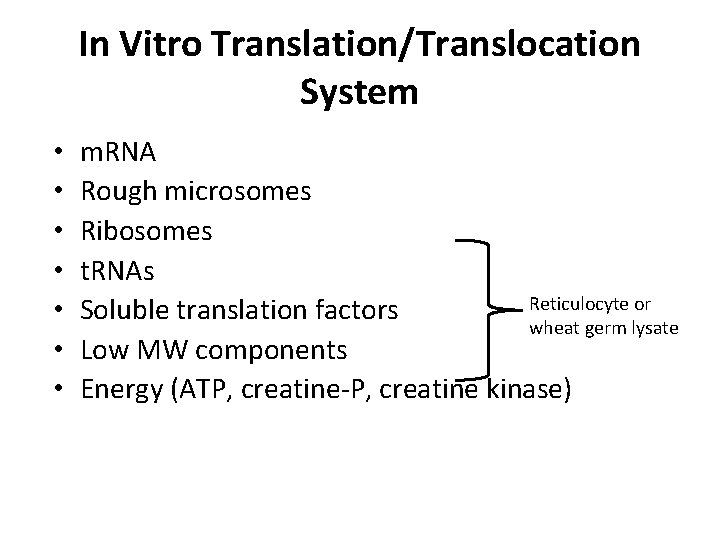
In Vitro Translation/Translocation System • • m. RNA Rough microsomes Ribosomes t. RNAs Reticulocyte or Soluble translation factors wheat germ lysate Low MW components Energy (ATP, creatine-P, creatine kinase)

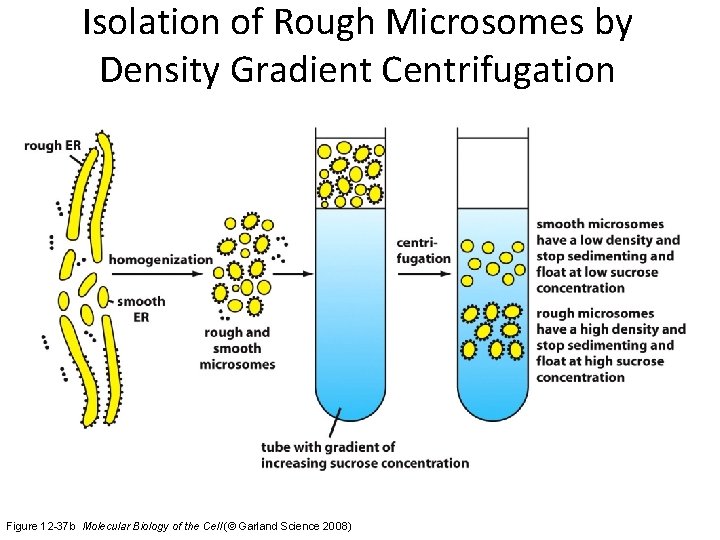
Isolation of Rough Microsomes by Density Gradient Centrifugation Figure 12 -37 b Molecular Biology of the Cell (© Garland Science 2008)

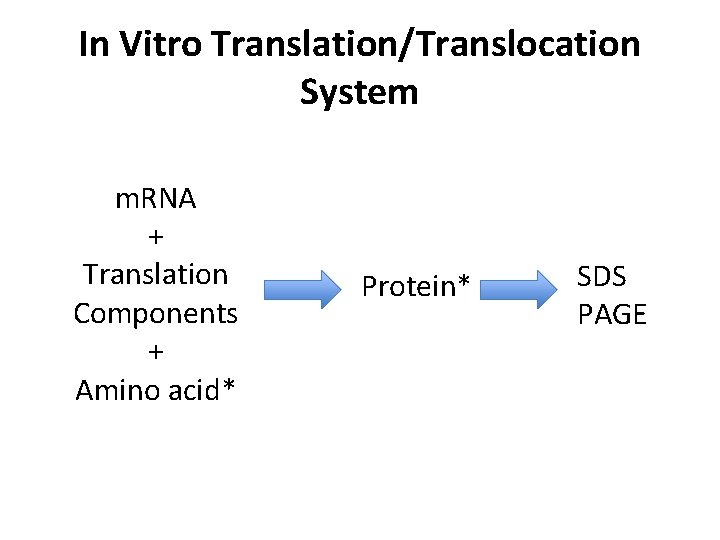
In Vitro Translation/Translocation System m. RNA + Translation Components + Amino acid* Protein* SDS PAGE

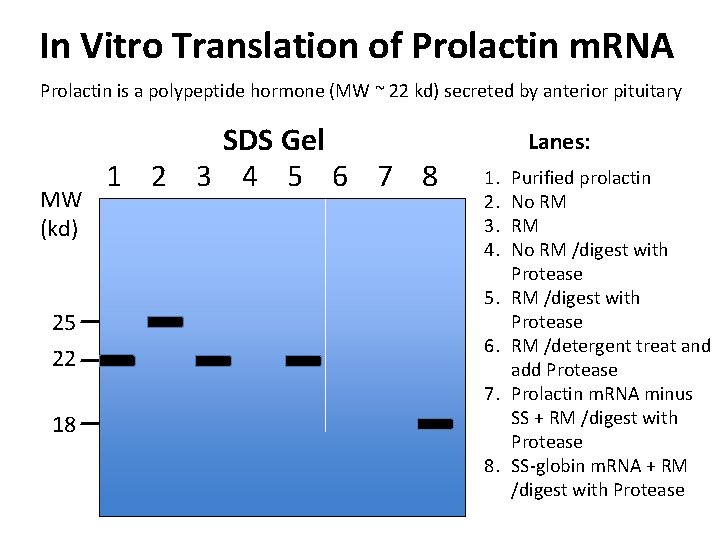
In Vitro Translation of Prolactin m. RNA Prolactin is a polypeptide hormone (MW ~ 22 kd) secreted by anterior pituitary MW (kd) 25 22 SDS Gel 1 2 3 4 5 6 7 8 Lanes: 1. 2. 3. 4. 5. 6. 7. 18 8. Purified prolactin No RM RM No RM /digest with Protease RM /detergent treat and add Protease Prolactin m. RNA minus SS + RM /digest with Protease SS-globin m. RNA + RM /digest with Protease

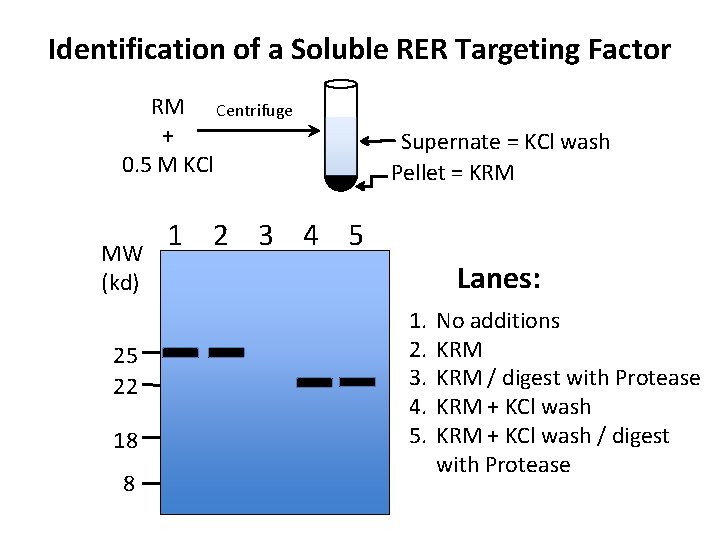
Identification of a Soluble RER Targeting Factor RM Centrifuge + 0. 5 M KCl MW (kd) 25 22 18 8 Supernate = KCl wash Pellet = KRM 1 2 3 4 5 Lanes: 1. 2. 3. 4. 5. No additions KRM / digest with Protease KRM + KCl wash / digest with Protease

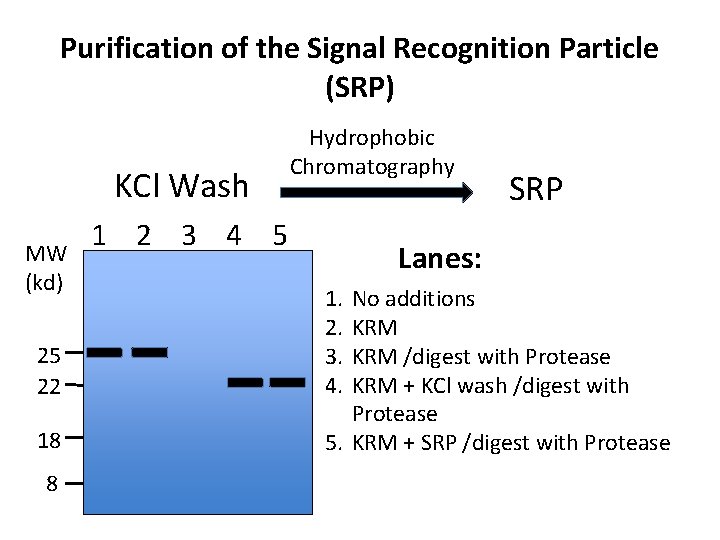
Purification of the Signal Recognition Particle (SRP) KCl Wash MW (kd) 25 22 18 8 Hydrophobic Chromatography 1 2 3 4 5 SRP Lanes: 1. 2. 3. 4. No additions KRM /digest with Protease KRM + KCl wash /digest with Protease 5. KRM + SRP /digest with Protease

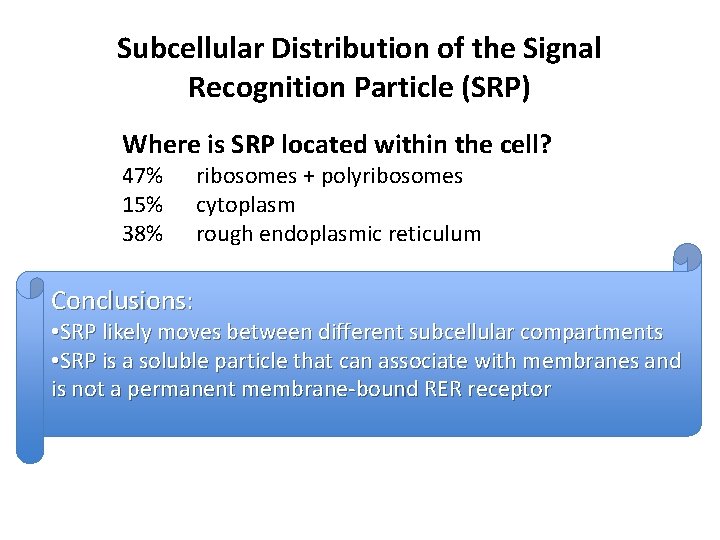
Subcellular Distribution of the Signal Recognition Particle (SRP) Where is SRP located within the cell? 47% 15% 38% Conclusions: ribosomes + polyribosomes cytoplasm rough endoplasmic reticulum • SRP likely moves between different subcellular compartments • SRP is a soluble particle that can associate with membranes and is not a permanent membrane-bound RER receptor

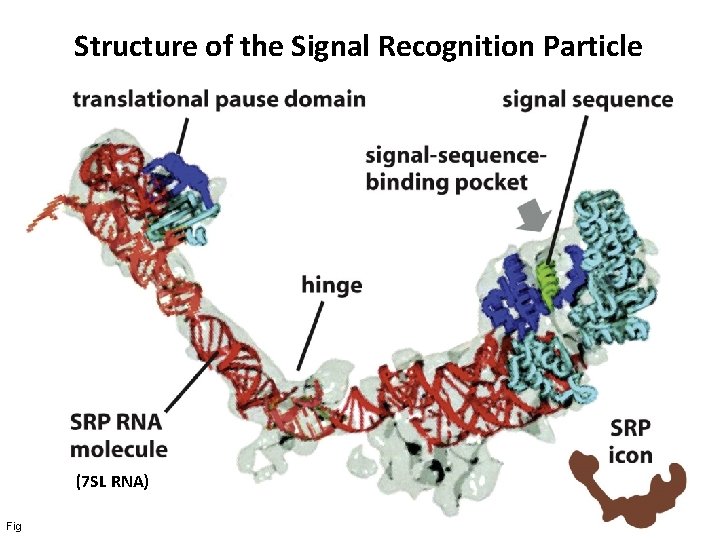
Structure of the Signal Recognition Particle (7 SL RNA) Figure 12 -39 a Molecular Biology of the Cell (© Garland Science 2008)

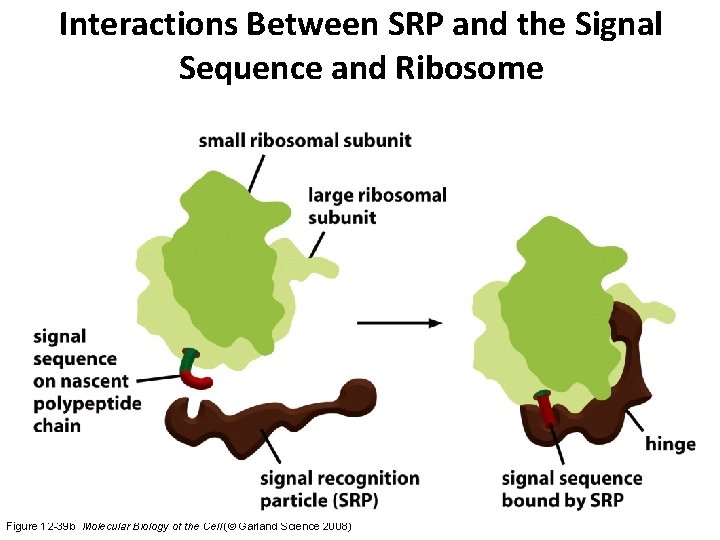
Interactions Between SRP and the Signal Sequence and Ribosome Figure 12 -39 b Molecular Biology of the Cell (© Garland Science 2008)

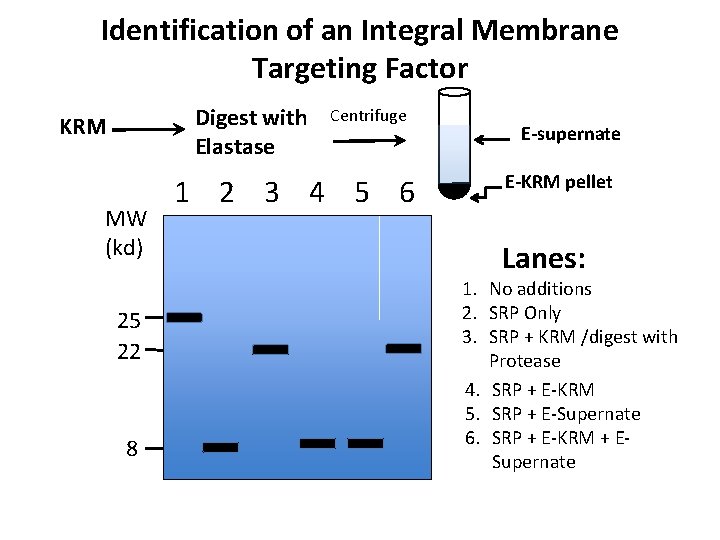
Identification of an Integral Membrane Targeting Factor Digest with Elastase KRM MW (kd) 25 22 8 Centrifuge 1 2 3 4 5 6 E-supernate E-KRM pellet Lanes: 1. No additions 2. SRP Only 3. SRP + KRM /digest with Protease 4. SRP + E-KRM 5. SRP + E-Supernate 6. SRP + E-KRM + ESupernate

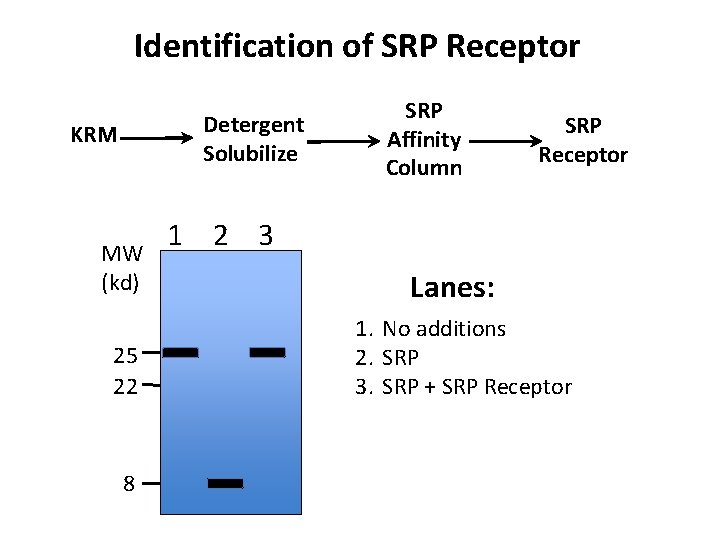
Identification of SRP Receptor Detergent Solubilize KRM MW (kd) 25 22 8 SRP Affinity Column SRP Receptor 1 2 3 Lanes: 1. No additions 2. SRP 3. SRP + SRP Receptor

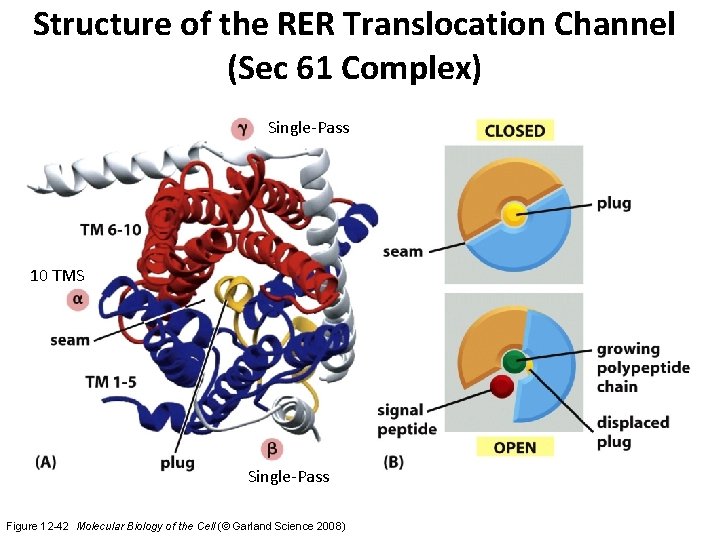
Structure of the RER Translocation Channel (Sec 61 Complex) Single-Pass 10 TMS Single-Pass Figure 12 -42 Molecular Biology of the Cell (© Garland Science 2008)

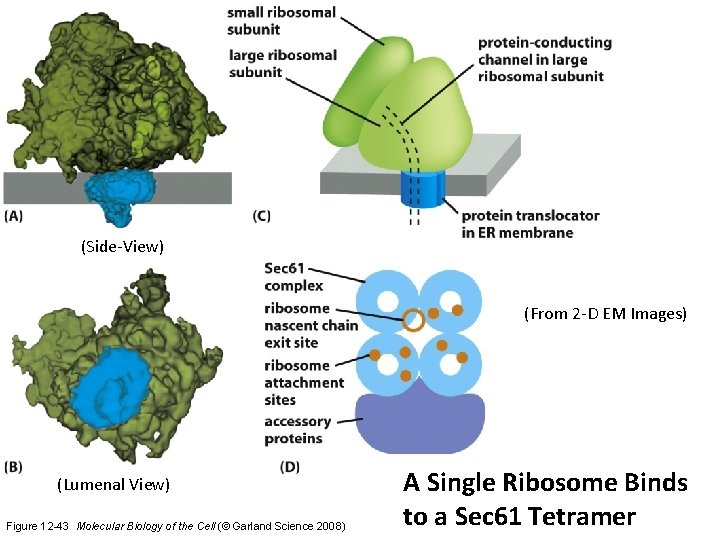
(Side-View) (From 2 -D EM Images) (Lumenal View) Figure 12 -43 Molecular Biology of the Cell (© Garland Science 2008) A Single Ribosome Binds to a Sec 61 Tetramer

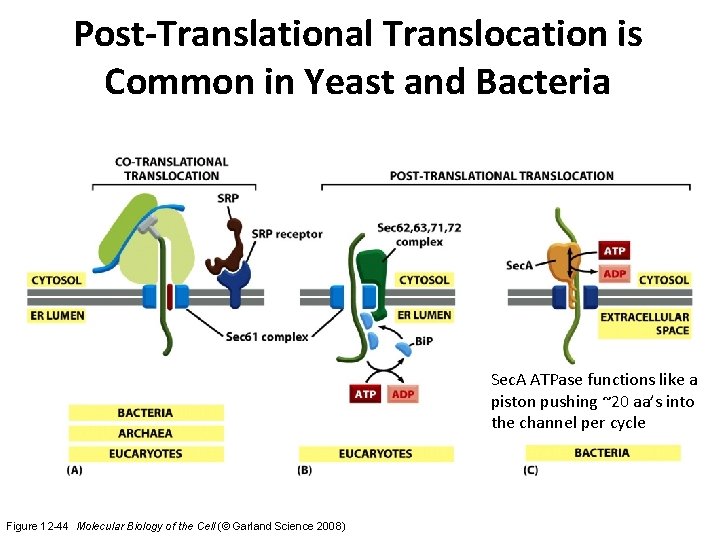
Post-Translational Translocation is Common in Yeast and Bacteria Sec. A ATPase functions like a piston pushing ~20 aa’s into the channel per cycle Figure 12 -44 Molecular Biology of the Cell (© Garland Science 2008)

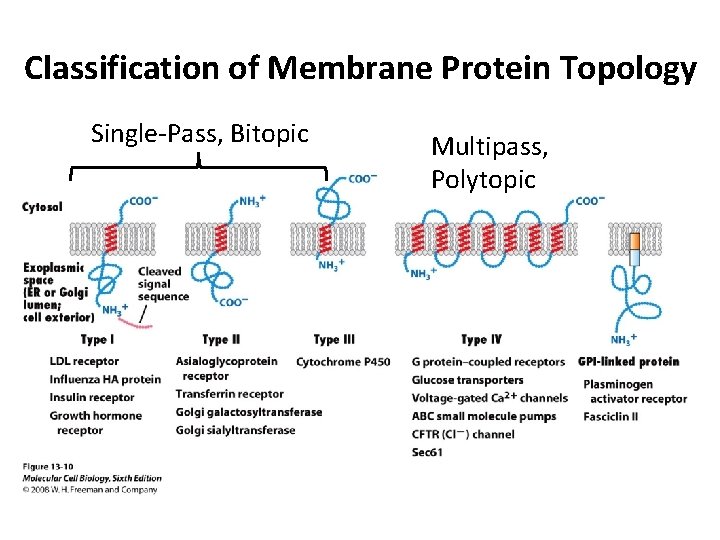
Classification of Membrane Protein Topology Single-Pass, Bitopic Multipass, Polytopic

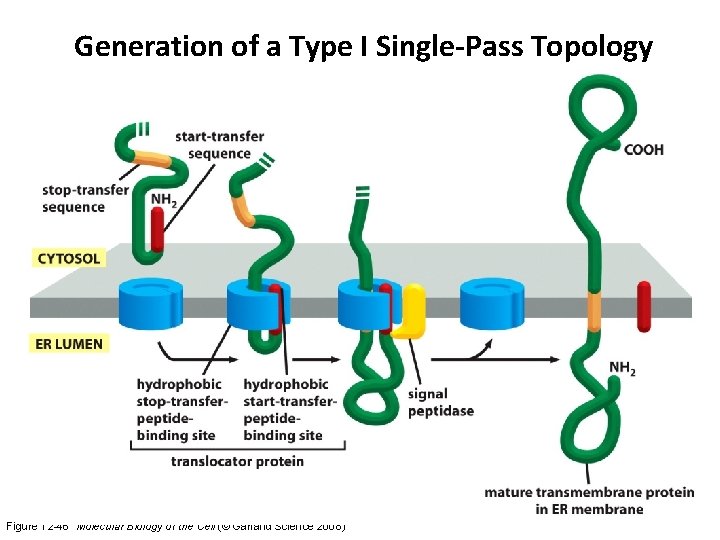
Generation of a Type I Single-Pass Topology Figure 12 -46 Molecular Biology of the Cell (© Garland Science 2008)

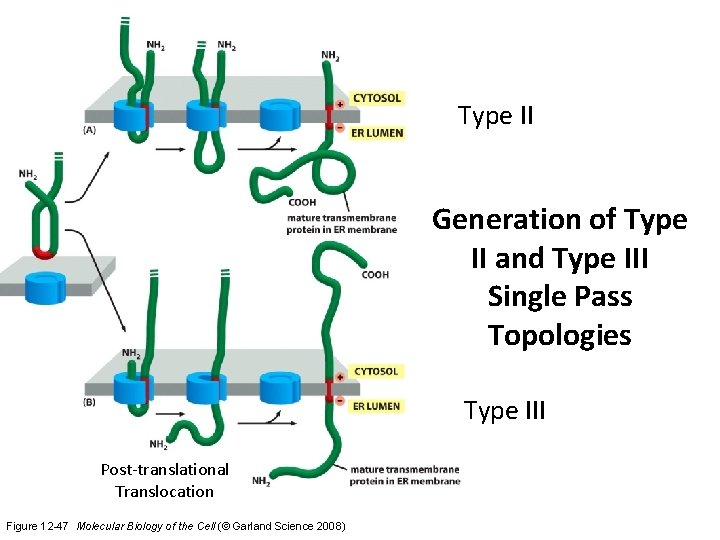
Type II Generation of Type II and Type III Single Pass Topologies Type III Post-translational Translocation Figure 12 -47 Molecular Biology of the Cell (© Garland Science 2008)

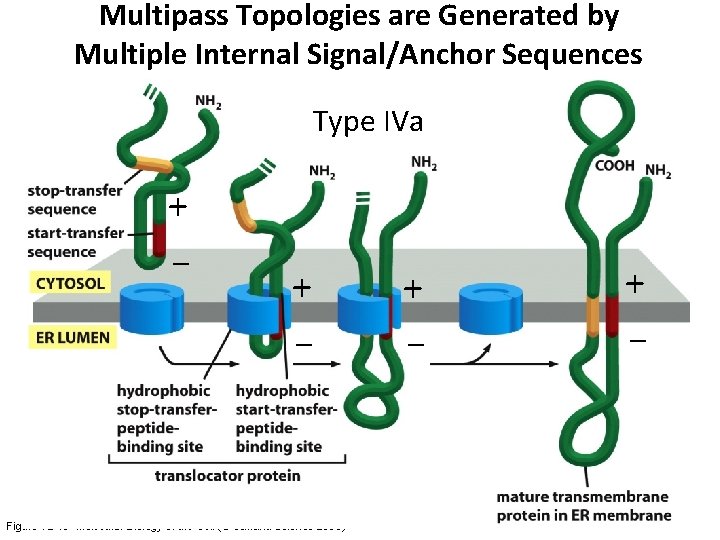
Multipass Topologies are Generated by Multiple Internal Signal/Anchor Sequences Type IVa + – + + + – – – Figure 12 -48 Molecular Biology of the Cell (© Garland Science 2008)

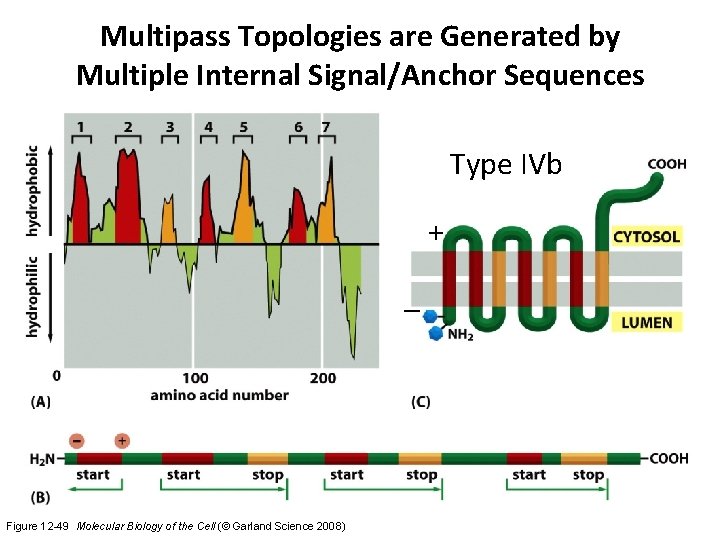
Multipass Topologies are Generated by Multiple Internal Signal/Anchor Sequences Type IVb + – Figure 12 -49 Molecular Biology of the Cell (© Garland Science 2008)

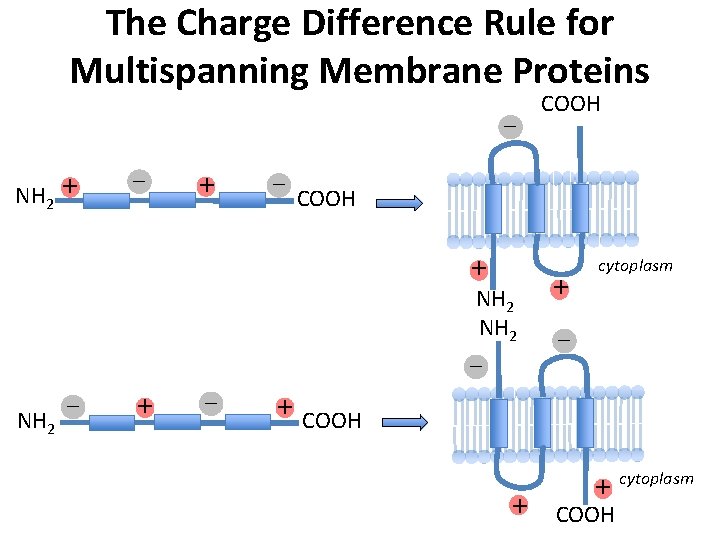
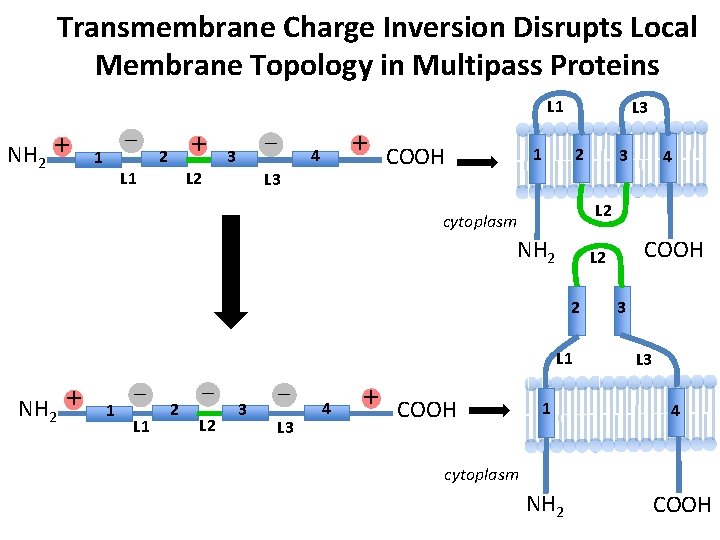
The Charge Difference Rule for Multispanning Membrane Proteins – NH 2 + – COOH + NH 2 – + cytoplasm – + COOH + NH 2 – COOH + cytoplasm COOH

Transmembrane Charge Inversion Disrupts Local Membrane Topology in Multipass Proteins L 1 NH 2 + – 1 + 2 – 3 L 2 L 1 + 4 L 3 COOH L 3 1 2 NH 2 L 1 1 – L 1 2 – L 2 3 – L 3 4 + COOH L 2 2 + 4 L 2 cytoplasm NH 2 3 1 3 L 3 4 cytoplasm NH 2 COOH

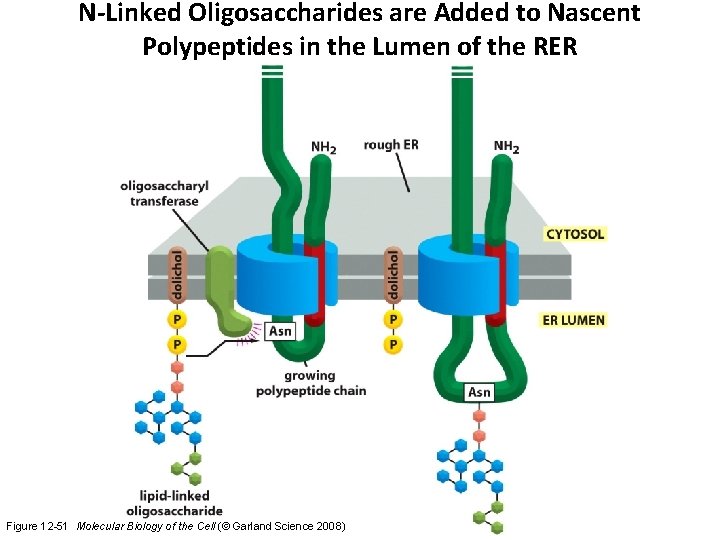
N-Linked Oligosaccharides are Added to Nascent Polypeptides in the Lumen of the RER Figure 12 -51 Molecular Biology of the Cell (© Garland Science 2008)

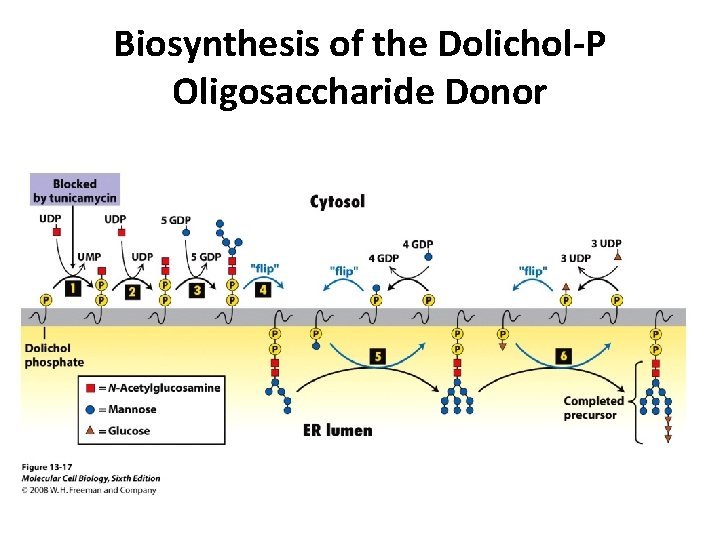
Biosynthesis of the Dolichol-P Oligosaccharide Donor

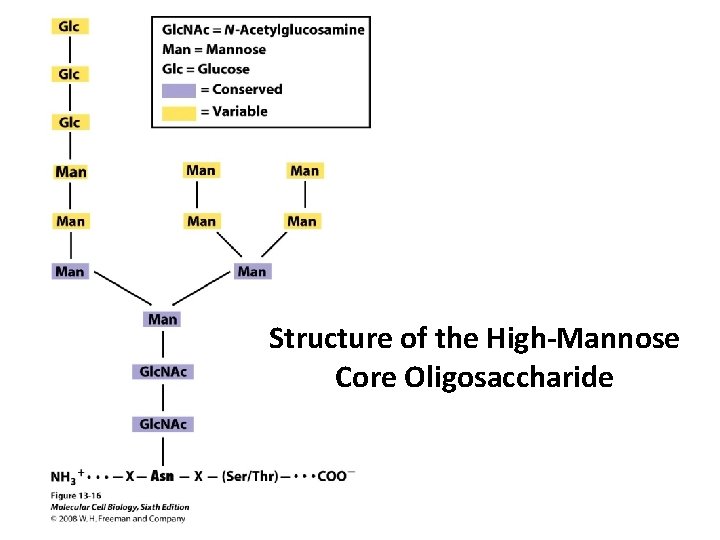
Structure of the High-Mannose Core Oligosaccharide

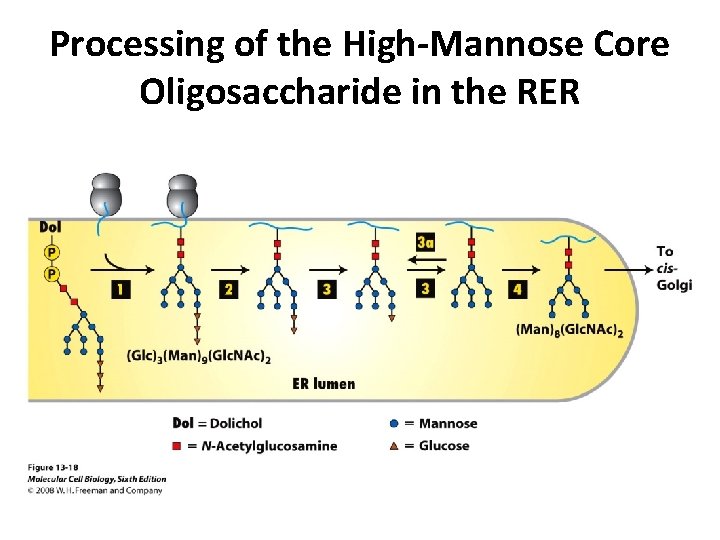
Processing of the High-Mannose Core Oligosaccharide in the RER

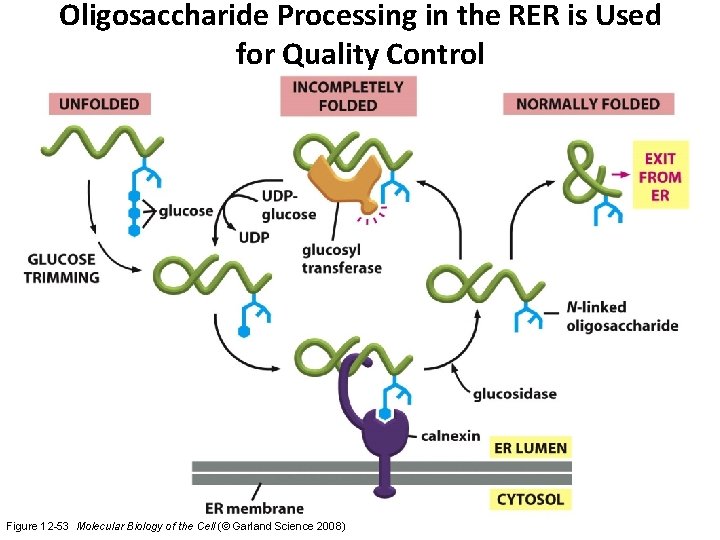
Oligosaccharide Processing in the RER is Used for Quality Control Figure 12 -53 Molecular Biology of the Cell (© Garland Science 2008)

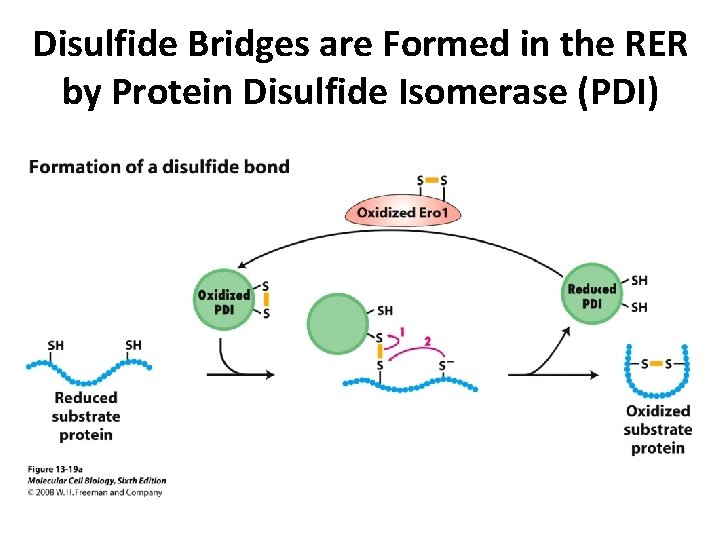
Disulfide Bridges are Formed in the RER by Protein Disulfide Isomerase (PDI)
 Baseband signal and bandpass signal
Baseband signal and bandpass signal Baseband signal and bandpass signal
Baseband signal and bandpass signal What is the product of an even signal and odd signal
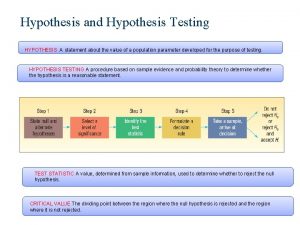
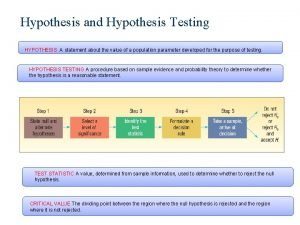
What is the product of an even signal and odd signal Hypothesis testing mean
Hypothesis testing mean Null and alternative hypothesis examples
Null and alternative hypothesis examples Protoplanet hypothesis weakness
Protoplanet hypothesis weakness Inorganic emulsifying agent
Inorganic emulsifying agent In-situ soap method of emulsion preparation
In-situ soap method of emulsion preparation Dry gum method of emulsion preparation
Dry gum method of emulsion preparation Wet gum method examples
Wet gum method examples Digital signal as a composite analog signal
Digital signal as a composite analog signal Signal hypothesis steps
Signal hypothesis steps Alternative hypothesis
Alternative hypothesis Market segmentation and targeting ppt
Market segmentation and targeting ppt Avon market segmentation
Avon market segmentation International market segmentation
International market segmentation Targeting and segmentation
Targeting and segmentation Sony market segmentation, targeting and positioning
Sony market segmentation, targeting and positioning Positioning involves meeting consumers lower performance
Positioning involves meeting consumers lower performance Sony market segmentation, targeting and positioning
Sony market segmentation, targeting and positioning Presciber segmentation
Presciber segmentation Nokia target market
Nokia target market Southwest airlines
Southwest airlines Global marketing segmentation
Global marketing segmentation Process of market segmentation
Process of market segmentation Marketing segmentation and targeting
Marketing segmentation and targeting Market segmentation, targeting and positioning
Market segmentation, targeting and positioning Linterland
Linterland Segmentation targeting positioning of amul
Segmentation targeting positioning of amul Benefits sought segmentation
Benefits sought segmentation Segmentation targeting differentiation and positioning
Segmentation targeting differentiation and positioning Concentrated marketing
Concentrated marketing Stp example
Stp example What is global market segmentation
What is global market segmentation International positioning strategies
International positioning strategies Marketing segmentation table
Marketing segmentation table