Chromatin Structure Gene Expression n The Histone Code

- Slides: 29

Chromatin Structure & Gene Expression n The Histone Code

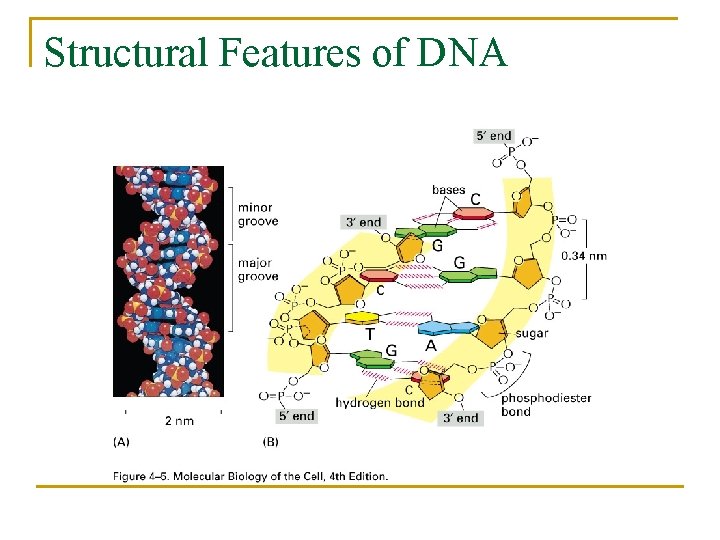
Structural Features of DNA

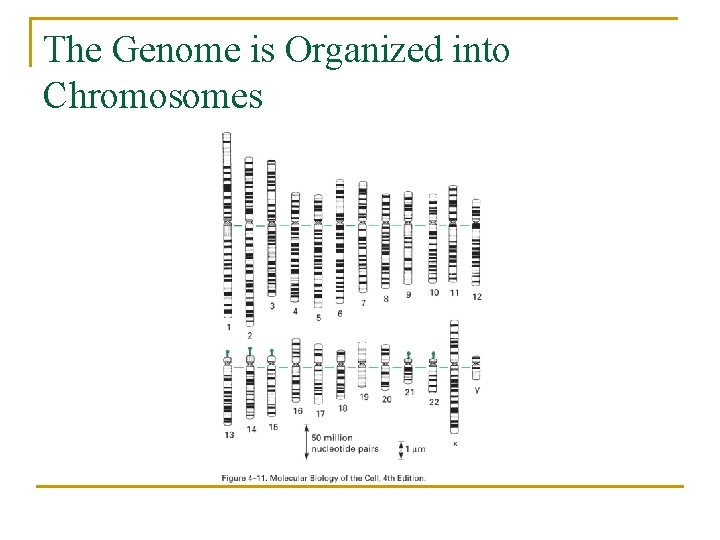
The Genome is Organized into Chromosomes

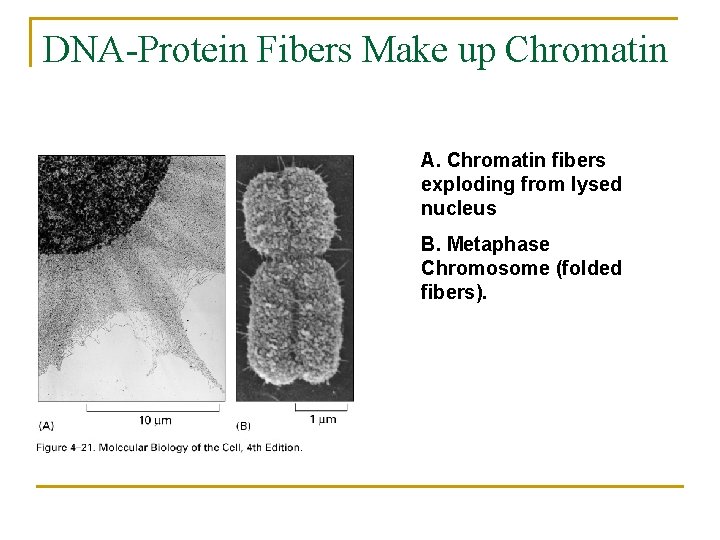
DNA-Protein Fibers Make up Chromatin A. Chromatin fibers exploding from lysed nucleus B. Metaphase Chromosome (folded fibers).

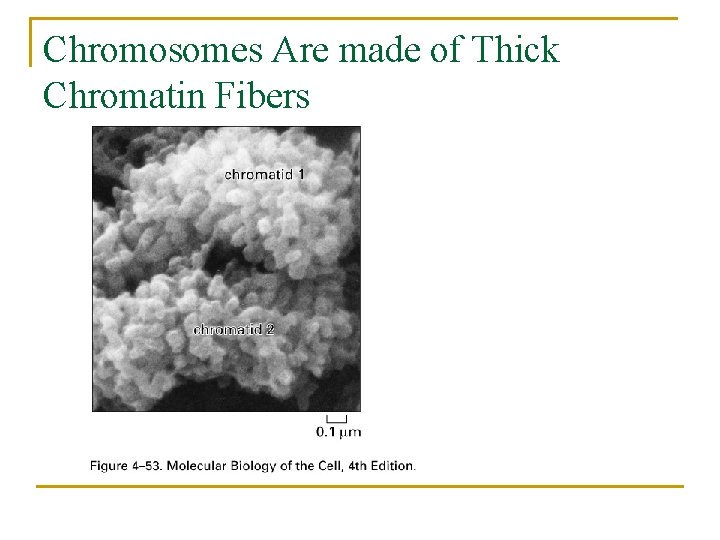
Chromosomes Are made of Thick Chromatin Fibers

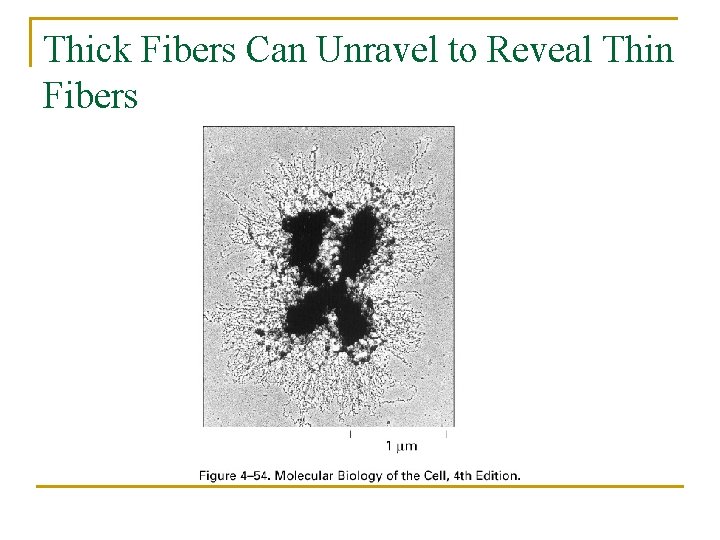
Thick Fibers Can Unravel to Reveal Thin Fibers

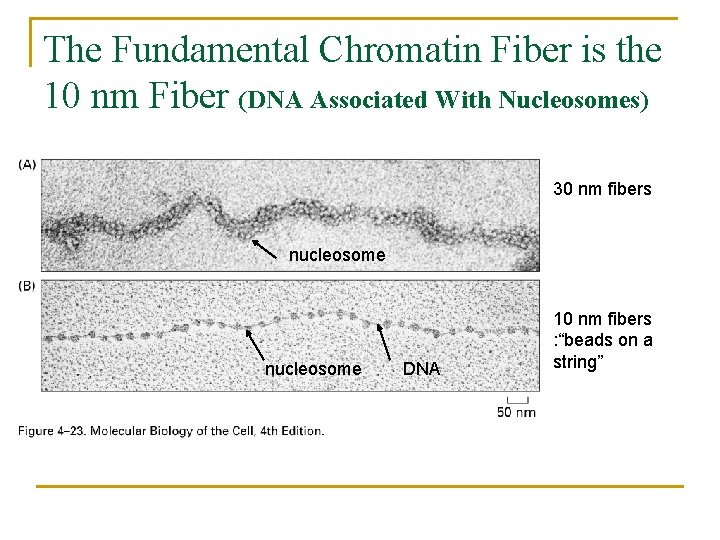
The Fundamental Chromatin Fiber is the 10 nm Fiber (DNA Associated With Nucleosomes) 30 nm fibers nucleosome DNA 10 nm fibers : “beads on a string”

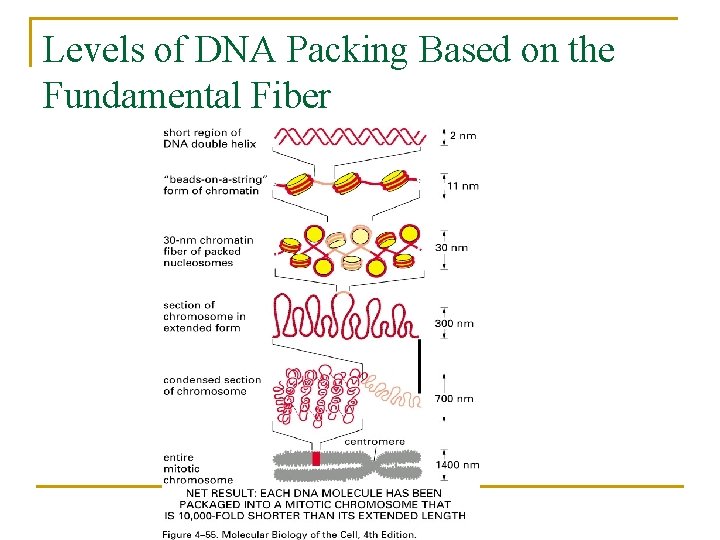
Levels of DNA Packing Based on the Fundamental Fiber

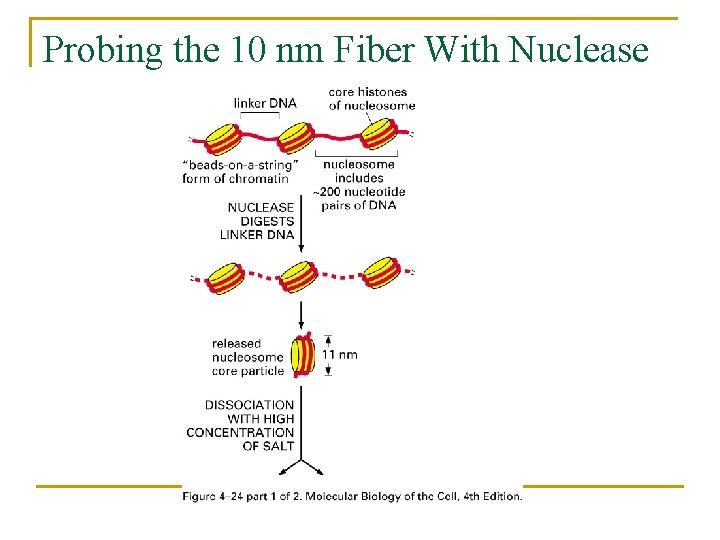
Probing the 10 nm Fiber With Nuclease

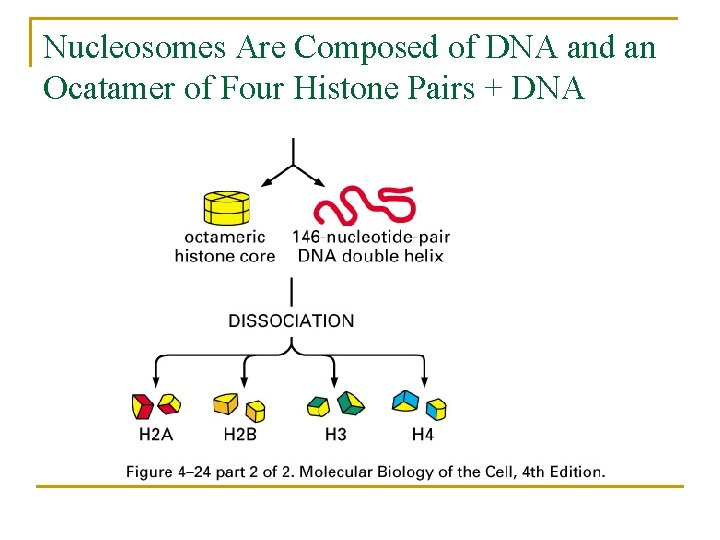
Nucleosomes Are Composed of DNA and an Ocatamer of Four Histone Pairs + DNA

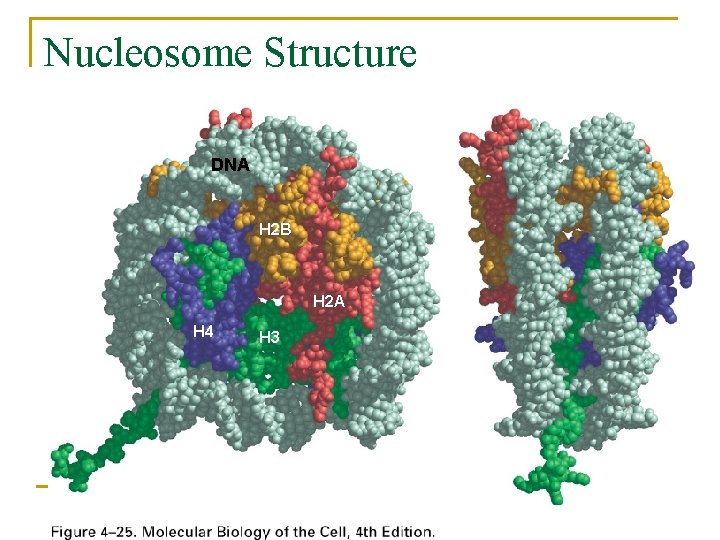
Nucleosome Structure DNA H 2 B H 2 A H 4 H 3

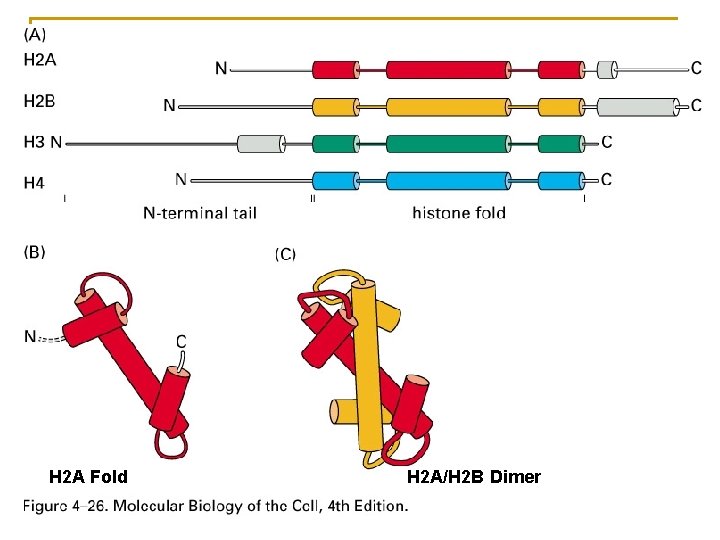
H 2 A Fold H 2 A/H 2 B Dimer

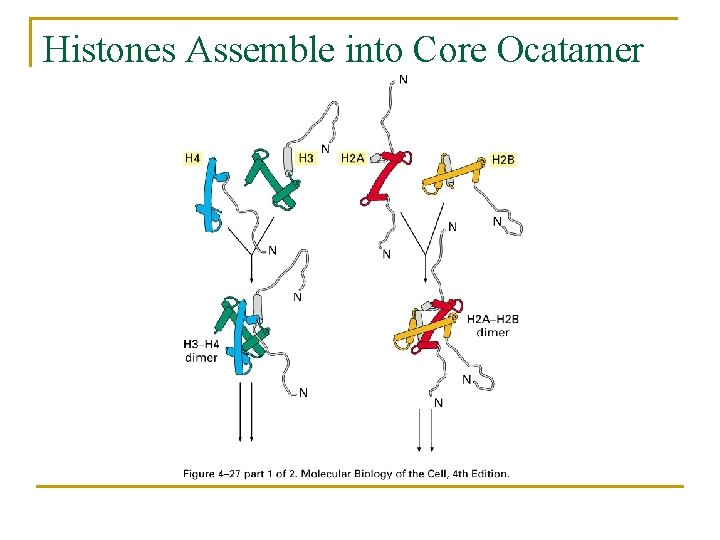
Histones Assemble into Core Ocatamer

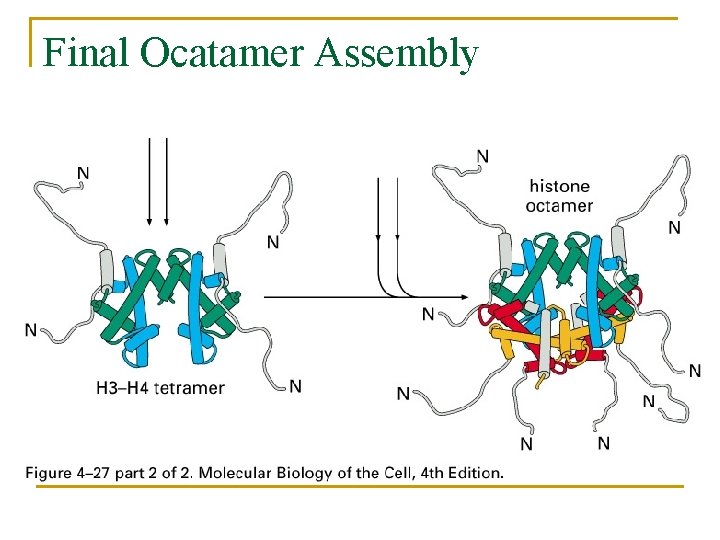
Final Ocatamer Assembly

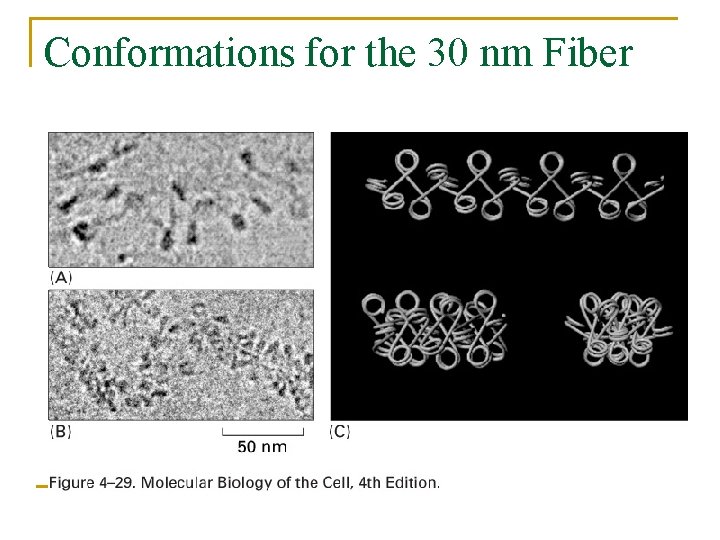
Conformations for the 30 nm Fiber

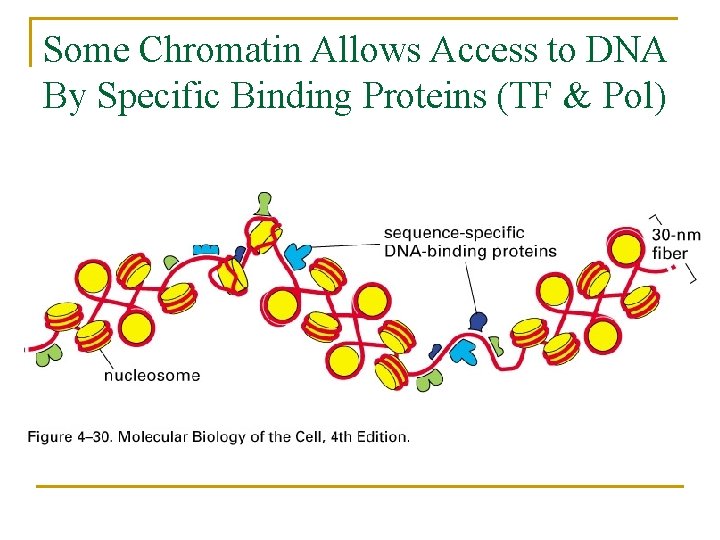
Some Chromatin Allows Access to DNA By Specific Binding Proteins (TF & Pol)

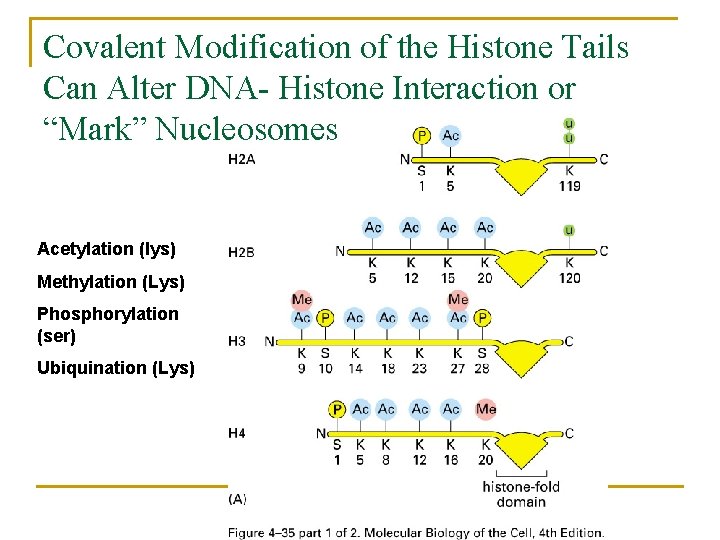
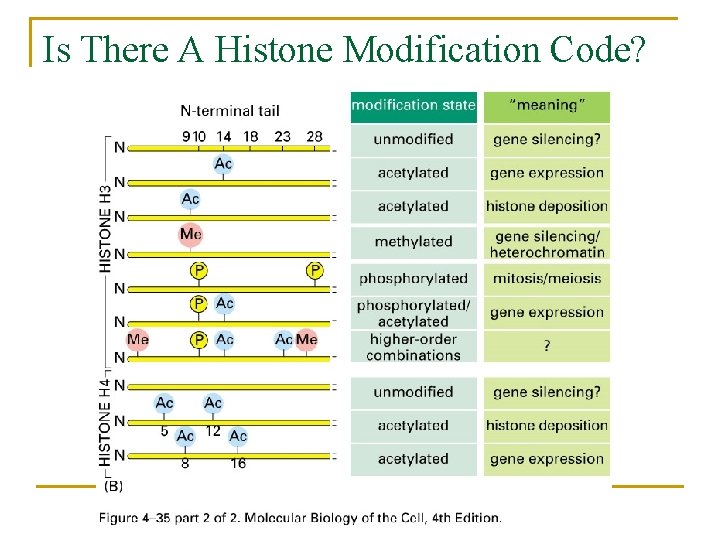
Covalent Modification of the Histone Tails Can Alter DNA- Histone Interaction or “Mark” Nucleosomes Acetylation (lys) Methylation (Lys) Phosphorylation (ser) Ubiquination (Lys)

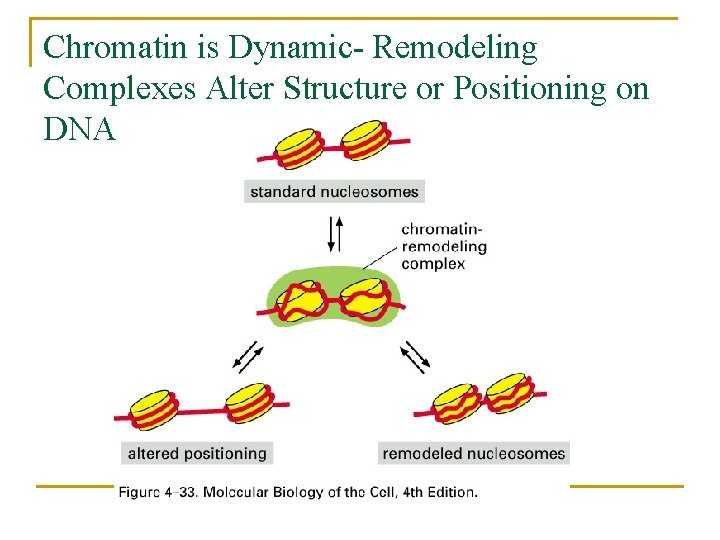
Chromatin is Dynamic- Remodeling Complexes Alter Structure or Positioning on DNA

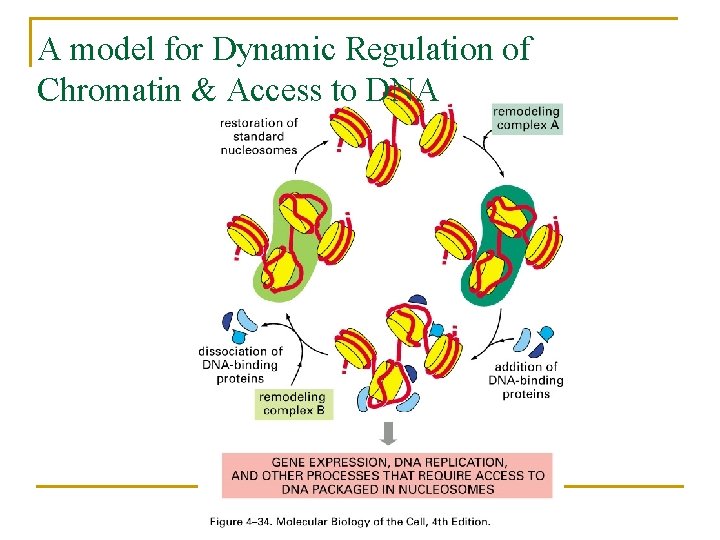
A model for Dynamic Regulation of Chromatin & Access to DNA

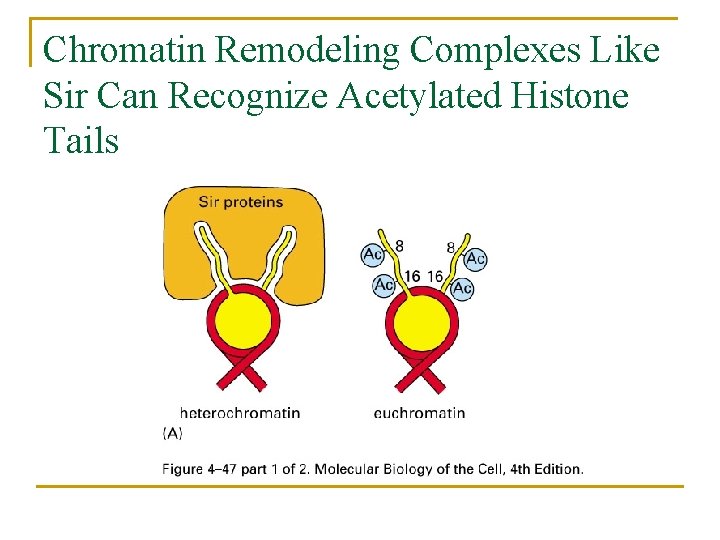
Chromatin Remodeling Complexes Like Sir Can Recognize Acetylated Histone Tails

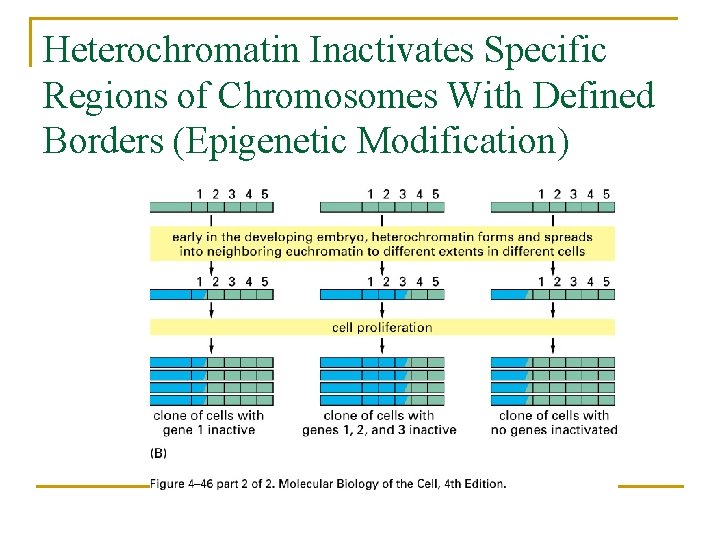
Heterochromatin Inactivates Specific Regions of Chromosomes With Defined Borders (Epigenetic Modification)

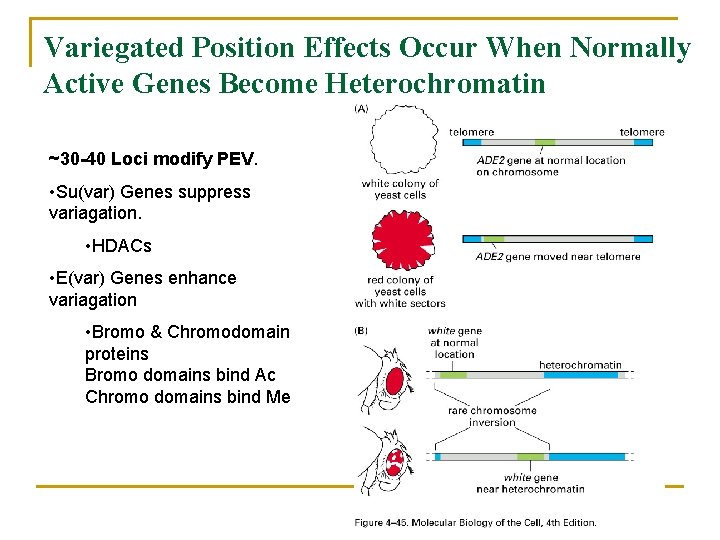
Variegated Position Effects Occur When Normally Active Genes Become Heterochromatin ~30 -40 Loci modify PEV. • Su(var) Genes suppress variagation. • HDACs • E(var) Genes enhance variagation • Bromo & Chromodomain proteins Bromo domains bind Ac Chromo domains bind Me

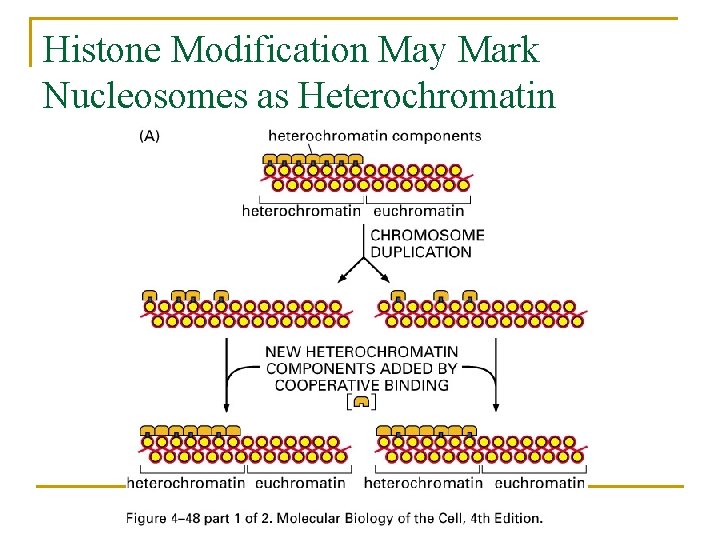
Histone Modification May Mark Nucleosomes as Heterochromatin

Is There A Histone Modification Code?

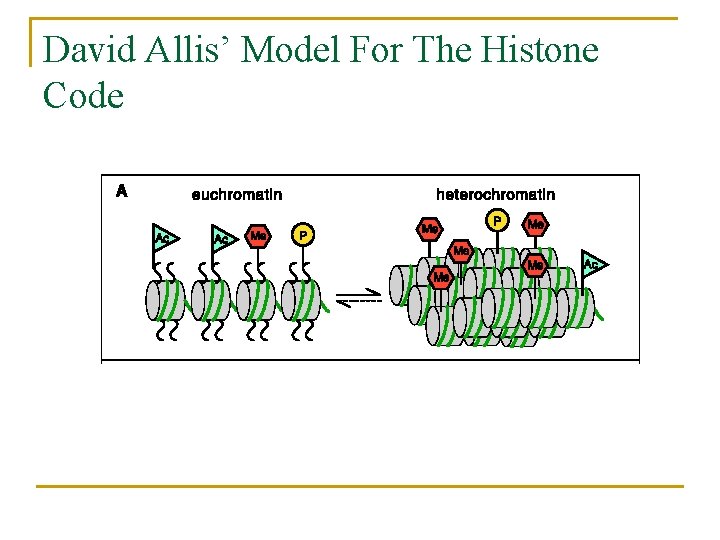
David Allis’ Model For The Histone Code

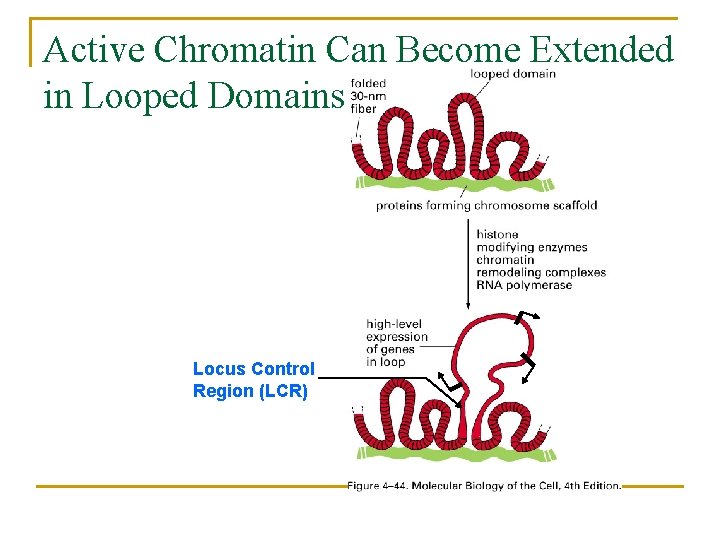
Active Chromatin Can Become Extended in Looped Domains Locus Control Region (LCR)

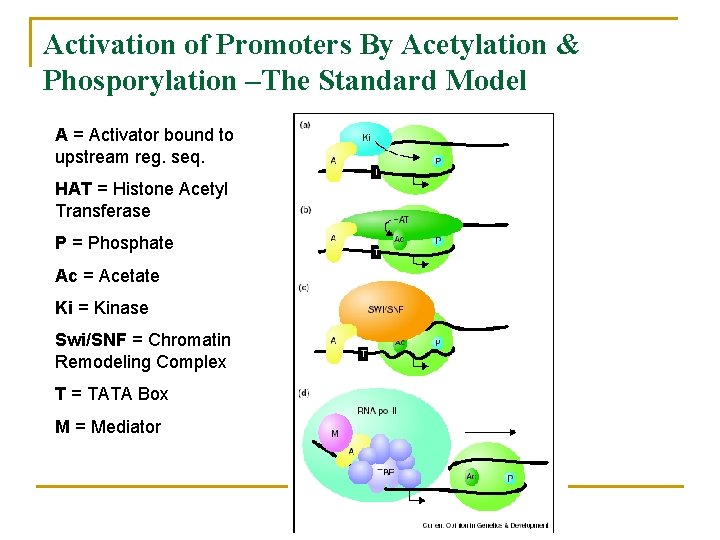
Activation of Promoters By Acetylation & Phosporylation –The Standard Model A = Activator bound to upstream reg. seq. HAT = Histone Acetyl Transferase P = Phosphate Ac = Acetate Ki = Kinase Swi/SNF = Chromatin Remodeling Complex T = TATA Box M = Mediator

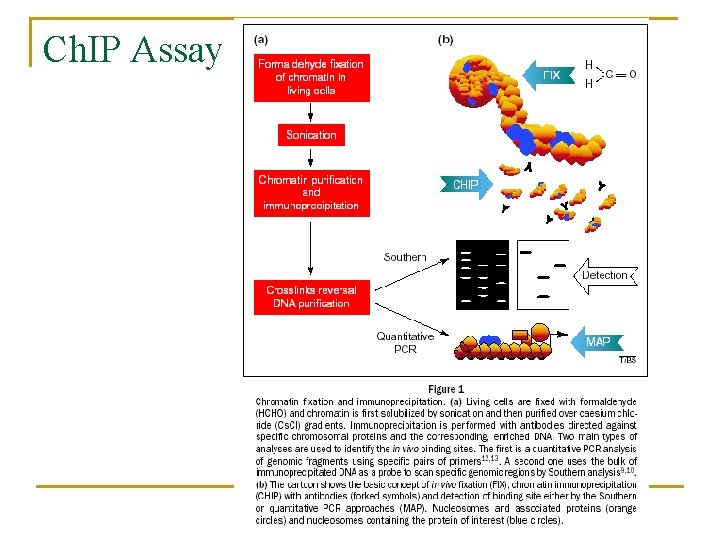
Ch. IP Assay

The Histone Code: What do we know for sure? n n n The information required for proper gene transcription is not just the promotors enhancers and transcription factors Chromatin structure regulates gene expression Chromatin structure is regulated at different levels q q n n n Heterochromatin vs. Euchromatin Nucleosome structure at promoters and other regulatory sequences Histone modification plays a role in the regulation of chromatin structure Modification of transcription factors also plays a role We can’t read the code yet!