Chapter 7 Clusters and Repeats 7 1 Introduction

















- Slides: 52

Chapter 7 Clusters and Repeats

7. 1 Introduction • gene family – A set of genes within a genome that encode related or identical proteins or RNAs. – The members were derived by duplication of an ancestral gene followed by accumulation of changes in sequence between the copies. – Most often the members are related but not identical.

7. 1 Introduction • pseudogenes – Inactive but stable components of the genome derived by mutation of an ancestral active gene. – Usually they are inactive because of mutations that block transcription or translation or both. • gene cluster – A group of adjacent genes that are identical or related.

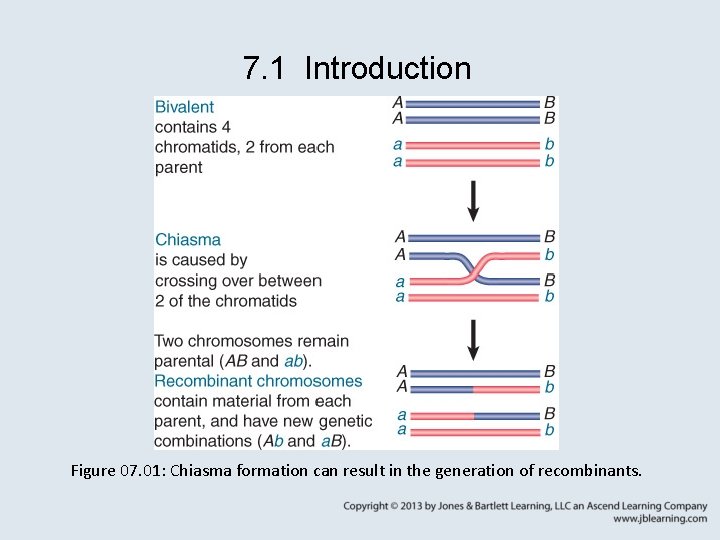
7. 1 Introduction Figure 07. 01: Chiasma formation can result in the generation of recombinants.

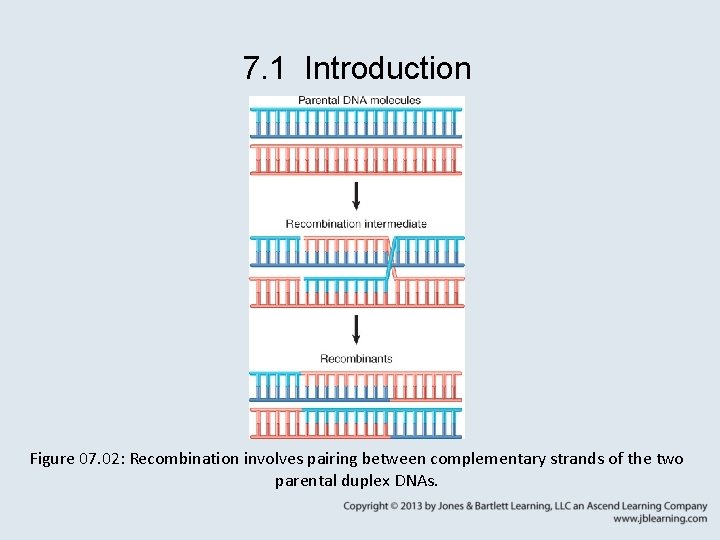
7. 1 Introduction Figure 07. 02: Recombination involves pairing between complementary strands of the two parental duplex DNAs.

7. 1 Introduction • unequal crossing over (nonreciprocal recombination) – Unequal crossing over results from an error in pairing and crossing over in which nonequivalent sites are involved in a recombination event. – It produces one recombinant with a deletion of material and one with a duplication. Figure 07. 03: Unequal crossing over results from pairing between nonequivalent repeats in regions of DNA consisting of repeating units.

7. 1 Introduction • satellite DNA – DNA that consists of many tandem repeats (identical or related) of a short basic repeating unit.

7. 1 Introduction • minisatellite – DNAs consisting of tandemly repeated copies of a short repeating sequence, with more repeat copies than a microsatellite but fewer than a satellite. – The length of the repeating unit is measured in tens of base pairs. – The number of repeats varies between individual genomes.

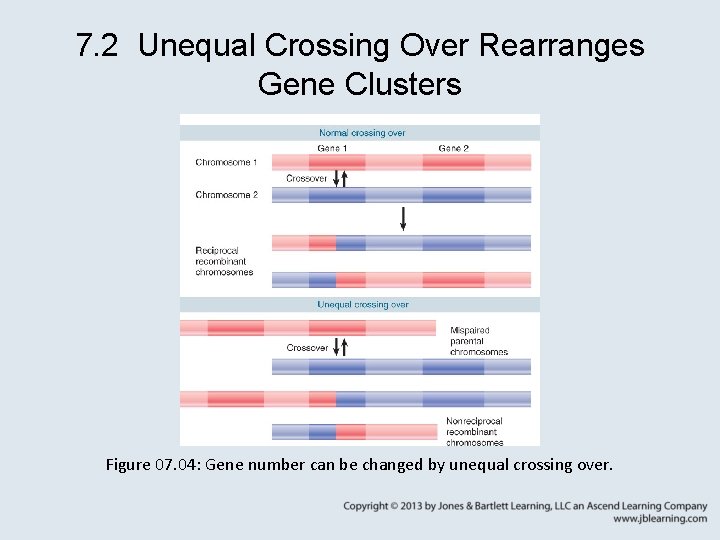
7. 2 Unequal Crossing Over Rearranges Gene Clusters • When a genome contains a cluster of genes with related sequences, mispairing between nonallelic loci can cause unequal crossing over. – This produces a deletion in one recombinant chromosome and a corresponding duplication in the other.

7. 2 Unequal Crossing Over Rearranges Gene Clusters Figure 07. 04: Gene number can be changed by unequal crossing over.

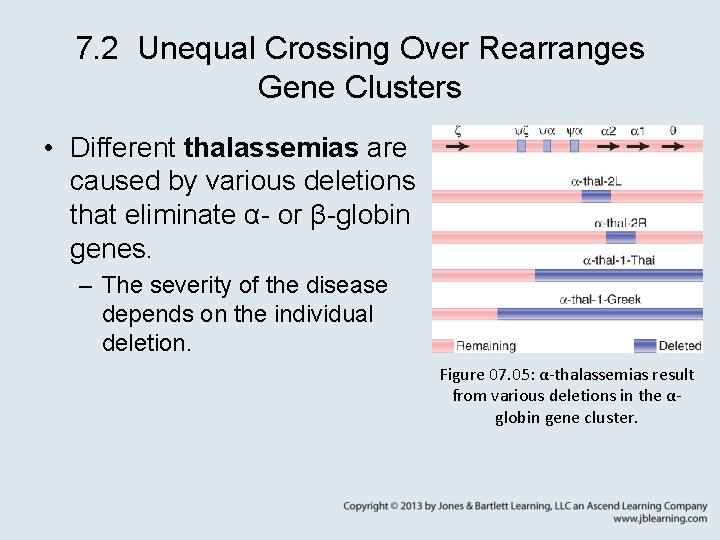
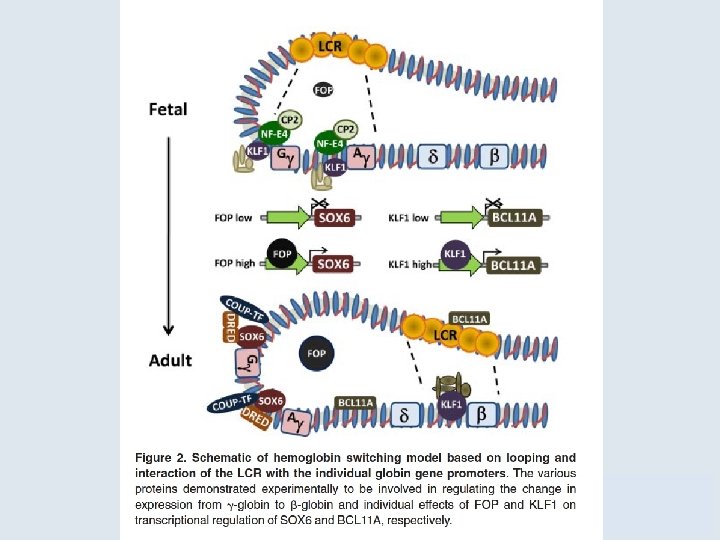
7. 2 Unequal Crossing Over Rearranges Gene Clusters • Different thalassemias are caused by various deletions that eliminate α- or β-globin genes. – The severity of the disease depends on the individual deletion. Figure 07. 05: α-thalassemias result from various deletions in the αglobin gene cluster.

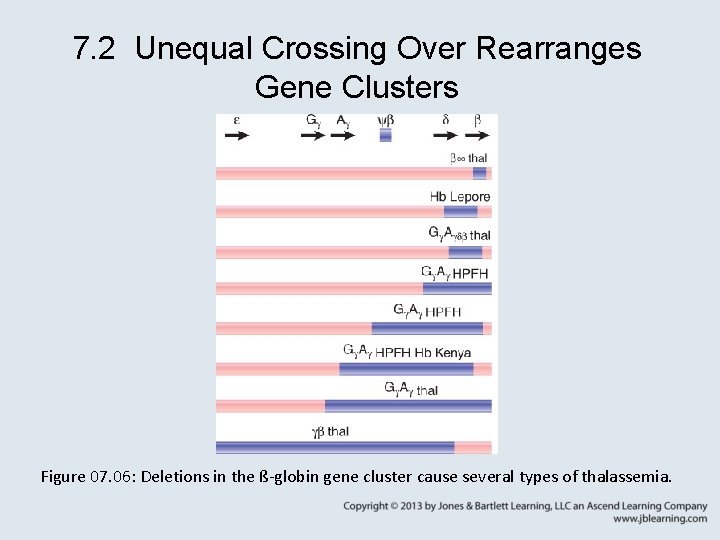
7. 2 Unequal Crossing Over Rearranges Gene Clusters Figure 07. 06: Deletions in the ß-globin gene cluster cause several types of thalassemia.

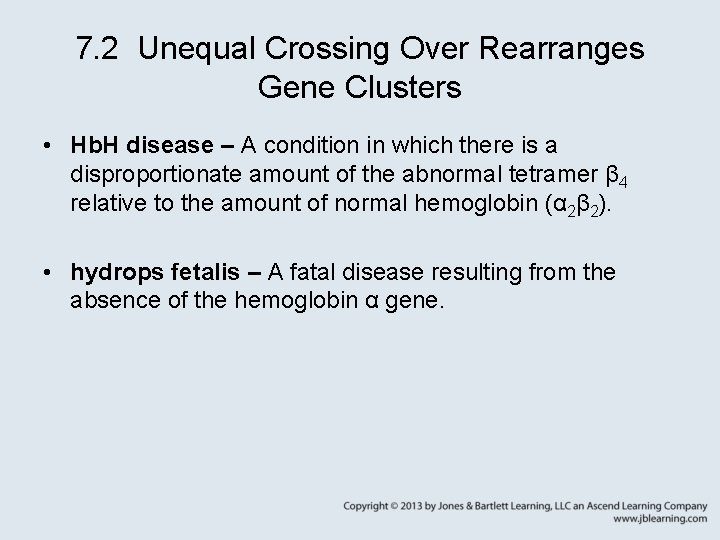
7. 2 Unequal Crossing Over Rearranges Gene Clusters • Hb. H disease – A condition in which there is a disproportionate amount of the abnormal tetramer β 4 relative to the amount of normal hemoglobin (α 2β 2). • hydrops fetalis – A fatal disease resulting from the absence of the hemoglobin α gene.

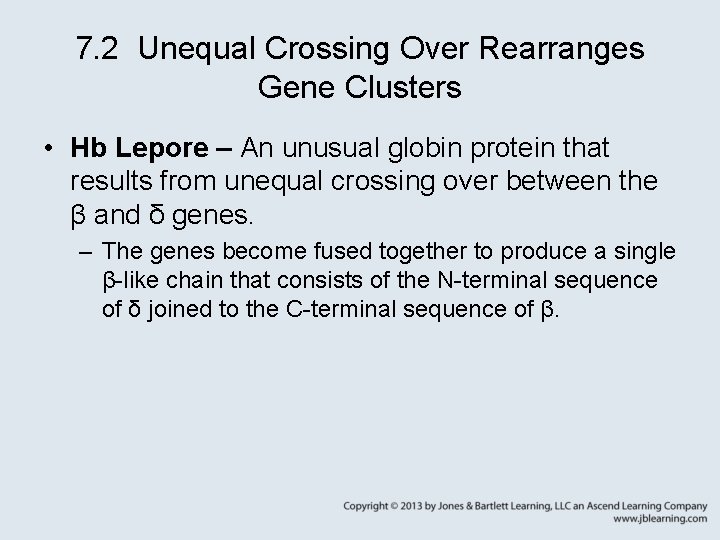
7. 2 Unequal Crossing Over Rearranges Gene Clusters • Hb Lepore – An unusual globin protein that results from unequal crossing over between the β and δ genes. – The genes become fused together to produce a single β-like chain that consists of the N-terminal sequence of δ joined to the C-terminal sequence of β.

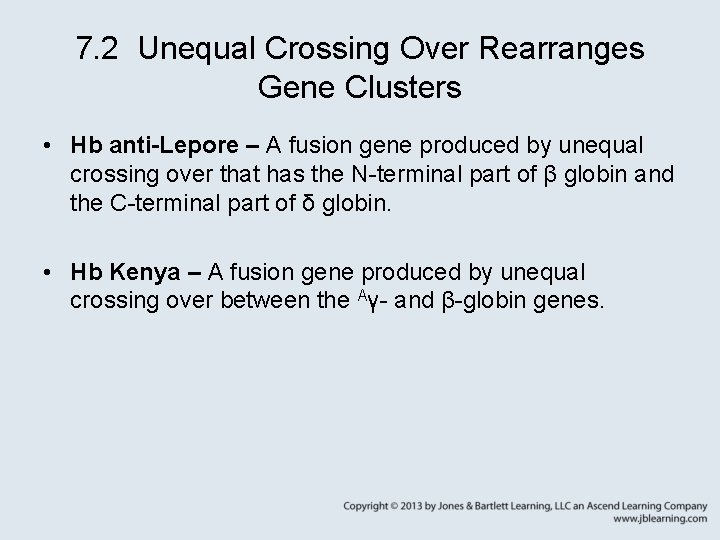
7. 2 Unequal Crossing Over Rearranges Gene Clusters • Hb anti-Lepore – A fusion gene produced by unequal crossing over that has the N-terminal part of β globin and the C-terminal part of δ globin. • Hb Kenya – A fusion gene produced by unequal crossing over between the Aγ- and β-globin genes.


















7. 3 Genes for r. RNA Form Tandem Repeats Including an Invariant Transcription Unit • Ribosomal RNA is encoded by a large number of identical genes that are tandemly repeated to form one or more clusters. • Each r. DNA cluster is organized so that transcription units giving a joint precursor to the major r. RNAs alternate with nontranscribed spacers. • The genes in an r. DNA cluster all have an identical sequence.

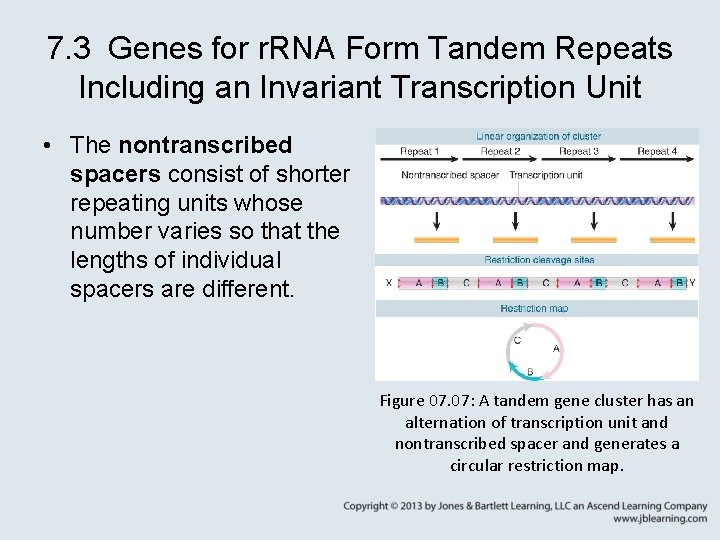
7. 3 Genes for r. RNA Form Tandem Repeats Including an Invariant Transcription Unit • The nontranscribed spacers consist of shorter repeating units whose number varies so that the lengths of individual spacers are different. Figure 07. 07: A tandem gene cluster has an alternation of transcription unit and nontranscribed spacer and generates a circular restriction map.

7. 3 Genes for r. RNA Form Tandem Repeats Including an Invariant Transcription Unit • nucleolus – A discrete region of the nucleus where ribosomes are produced. • nucleolar organizer – The region of a chromosome carrying genes encoding r. RNA.

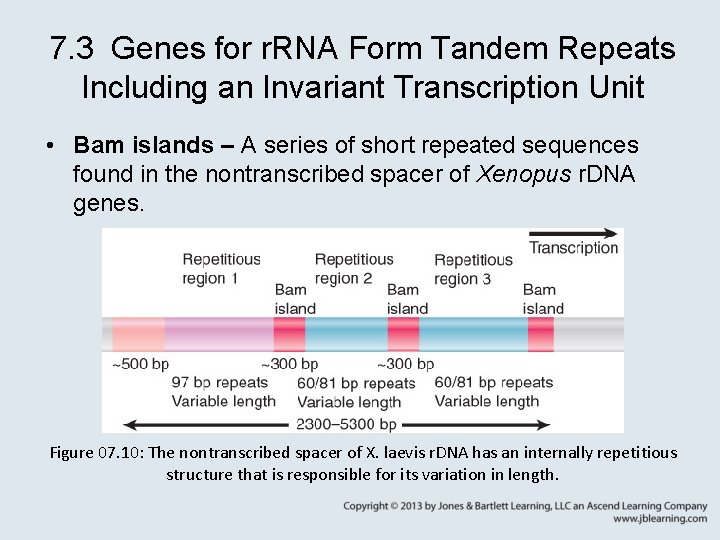
7. 3 Genes for r. RNA Form Tandem Repeats Including an Invariant Transcription Unit • Bam islands – A series of short repeated sequences found in the nontranscribed spacer of Xenopus r. DNA genes. Figure 07. 10: The nontranscribed spacer of X. laevis r. DNA has an internally repetitious structure that is responsible for its variation in length.

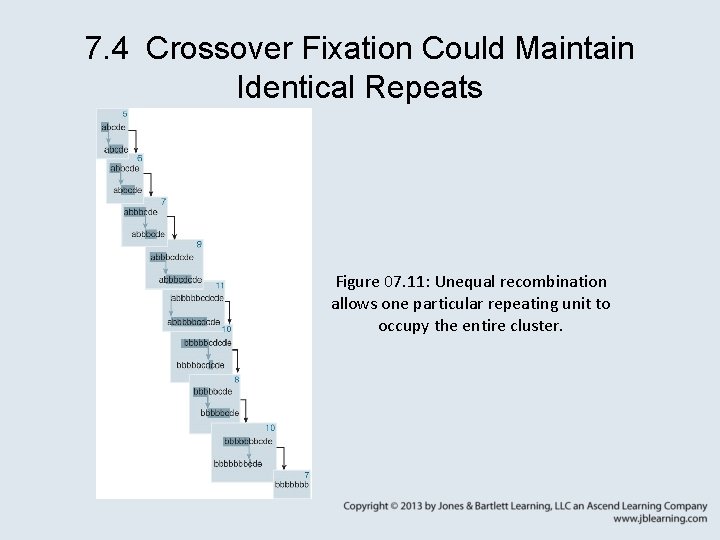
7. 4 Crossover Fixation Could Maintain Identical Repeats • Unequal crossing over changes the size of a cluster of tandem repeats. • Individual repeating units can be eliminated or can spread through the cluster. • concerted evolution (coincidental evolution) – The ability of two or more related genes to evolve together as though constituting a single locus.

7. 4 Crossover Fixation Could Maintain Identical Repeats • gene conversion – The alteration of one strand of a heteroduplex DNA to make it complementary with the other strand at any position(s) where there were mispaired bases. • crossover fixation – A possible consequence of unequal crossing over that allows a mutation in one member of a tandem cluster to spread through the whole cluster (or to be eliminated).

7. 4 Crossover Fixation Could Maintain Identical Repeats Figure 07. 11: Unequal recombination allows one particular repeating unit to occupy the entire cluster.

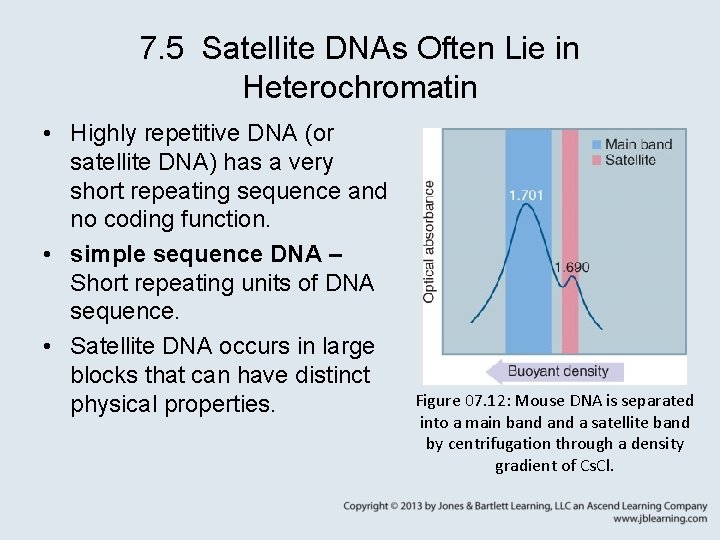
7. 5 Satellite DNAs Often Lie in Heterochromatin • Highly repetitive DNA (or satellite DNA) has a very short repeating sequence and no coding function. • simple sequence DNA – Short repeating units of DNA sequence. • Satellite DNA occurs in large blocks that can have distinct physical properties. Figure 07. 12: Mouse DNA is separated into a main band a satellite band by centrifugation through a density gradient of Cs. Cl.

7. 5 Satellite DNAs Often Lie in Heterochromatin • cryptic satellite – A satellite DNA sequence not identified as such by a separate peak on a density gradient. – It remains present in main-band DNA.

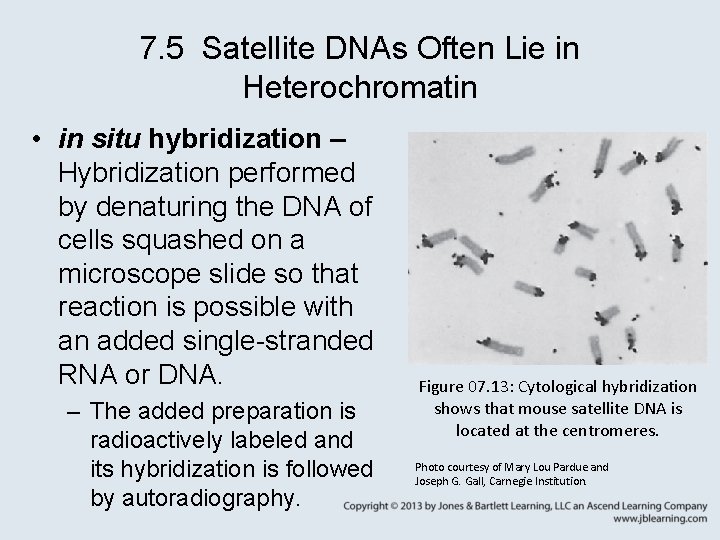
7. 5 Satellite DNAs Often Lie in Heterochromatin • in situ hybridization – Hybridization performed by denaturing the DNA of cells squashed on a microscope slide so that reaction is possible with an added single-stranded RNA or DNA. – The added preparation is radioactively labeled and its hybridization is followed by autoradiography. Figure 07. 13: Cytological hybridization shows that mouse satellite DNA is located at the centromeres. Photo courtesy of Mary Lou Pardue and Joseph G. Gall, Carnegie Institution.

7. 5 Satellite DNAs Often Lie in Heterochromatin • Satellite DNA is often the major constituent of centromeric heterochromatin. • euchromatin – Regions that comprise most of the genome in the interphase nucleus are less tightly coiled than heterochromatin, and contain most of the active or potentially active single-copy genes.

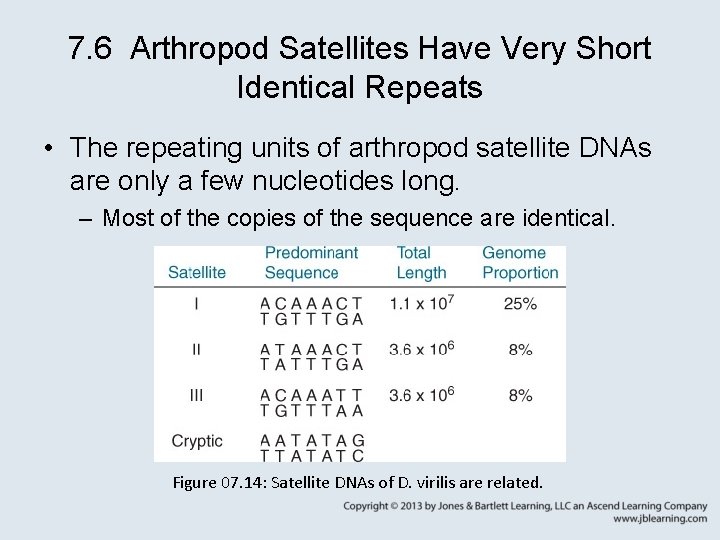
7. 6 Arthropod Satellites Have Very Short Identical Repeats • The repeating units of arthropod satellite DNAs are only a few nucleotides long. – Most of the copies of the sequence are identical. Figure 07. 14: Satellite DNAs of D. virilis are related.

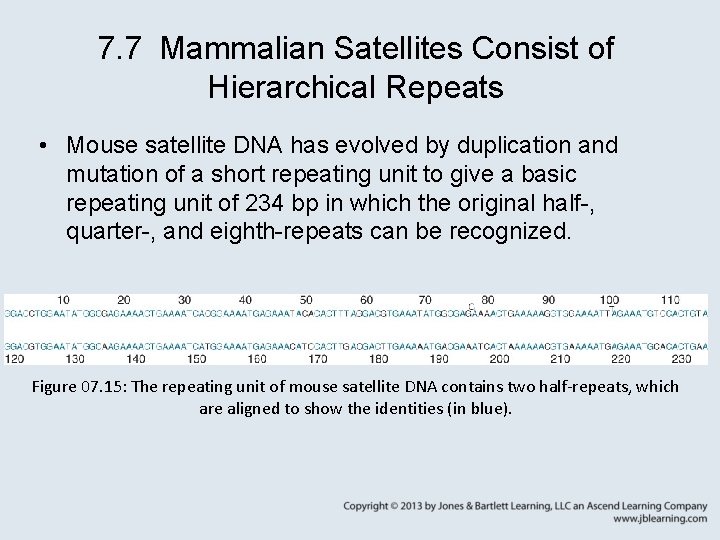
7. 7 Mammalian Satellites Consist of Hierarchical Repeats • Mouse satellite DNA has evolved by duplication and mutation of a short repeating unit to give a basic repeating unit of 234 bp in which the original half-, quarter-, and eighth-repeats can be recognized. Figure 07. 15: The repeating unit of mouse satellite DNA contains two half-repeats, which are aligned to show the identities (in blue).

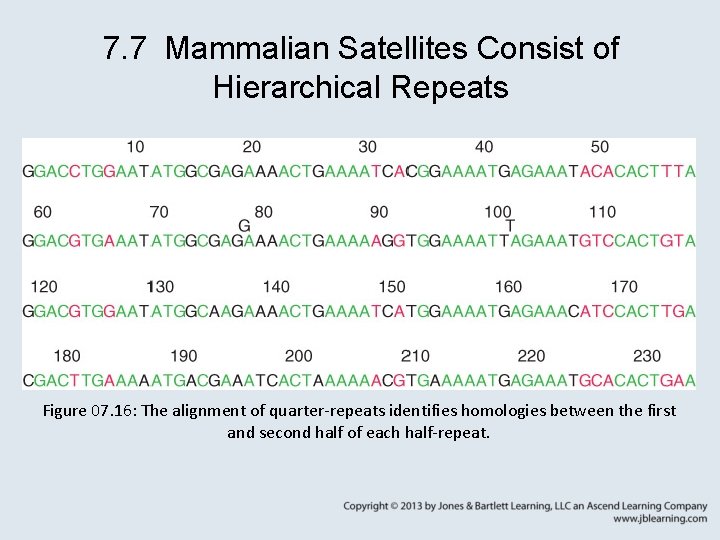
7. 7 Mammalian Satellites Consist of Hierarchical Repeats Figure 07. 16: The alignment of quarter-repeats identifies homologies between the first and second half of each half-repeat.

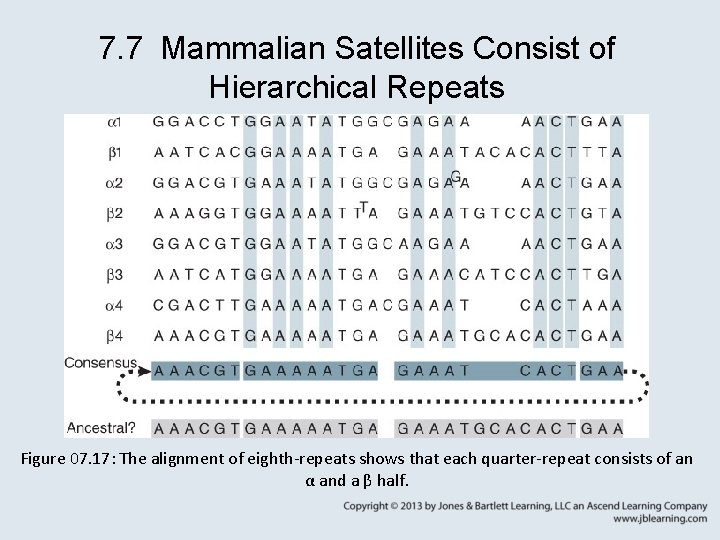
7. 7 Mammalian Satellites Consist of Hierarchical Repeats Figure 07. 17: The alignment of eighth-repeats shows that each quarter-repeat consists of an α and a β half.

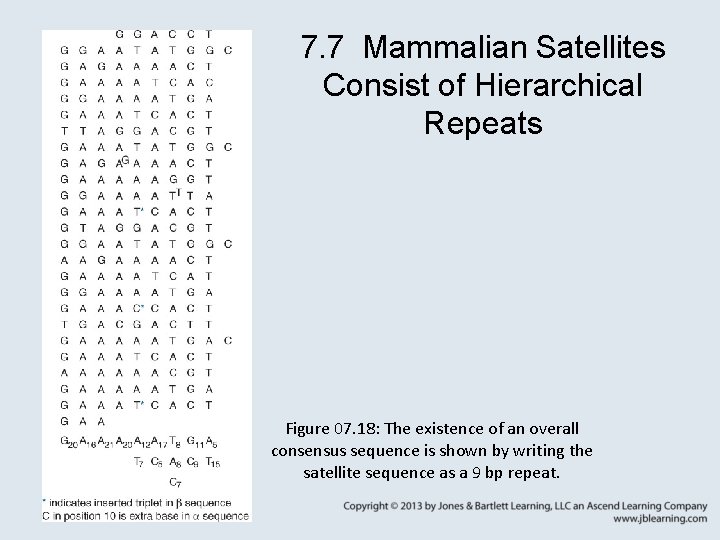
7. 7 Mammalian Satellites Consist of Hierarchical Repeats Figure 07. 18: The existence of an overall consensus sequence is shown by writing the satellite sequence as a 9 bp repeat.

7. 8 Minisatellites Are Useful for Genetic Mapping • The variation between microsatellites or minisatellites in individual genomes can be used to identify heredity unequivocally by showing that 50% of the bands in an individual are inherited from a particular parent. • variable number tandem repeat (VNTR) – Very short repeated sequences, including microsatellites and minisatellites.

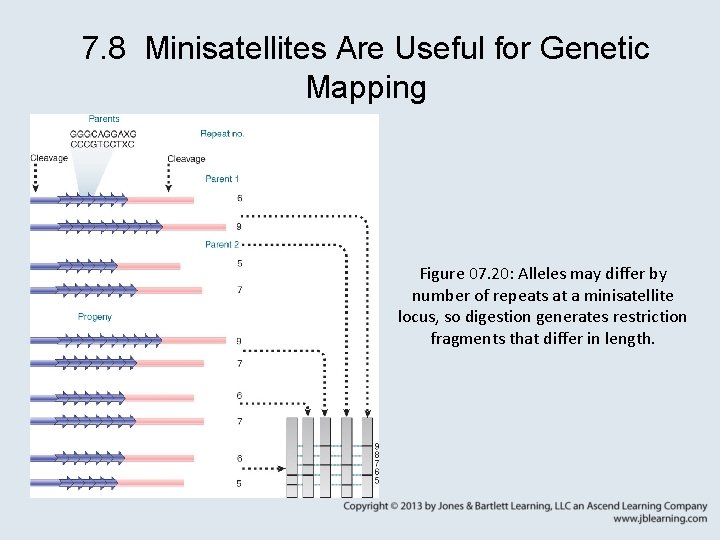
7. 8 Minisatellites Are Useful for Genetic Mapping Figure 07. 20: Alleles may differ by number of repeats at a minisatellite locus, so digestion generates restriction fragments that differ in length.

7. 8 Minisatellites Are Useful for Genetic Mapping • DNA fingerprinting – Analysis of the differences between individuals of restriction fragments that contain short repeated sequences, or by PCR. – The lengths of the repeated regions are unique to every individual, so the presence of a particular subset in any two individuals shows their common inheritance (e. g. , a parent–child relationship).

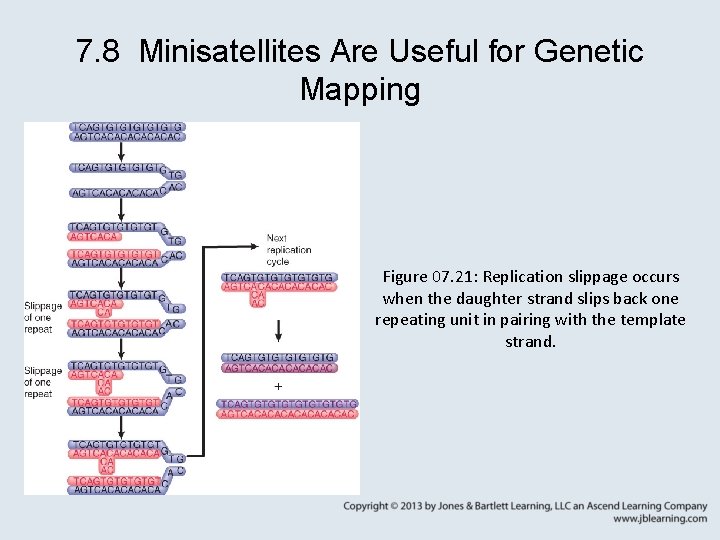
7. 8 Minisatellites Are Useful for Genetic Mapping Figure 07. 21: Replication slippage occurs when the daughter strand slips back one repeating unit in pairing with the template strand.